Significance
Transposable elements (TEs) are mobile genetic elements that colonize the nuclei of all organisms. Although TEs can be detrimental, they are considered important evolutionary forces. We discovered a domesticated TE in the mating-type locus of the yeast Kluyveromyces lactis. K. lactis hobo/Activator/Tam3 (hAT) transposase 1 (Kat1) mobilizes this TE from the genome by inducing DNA double-strand breaks followed by gene conversion, resulting in a switch of mating type. Hence, Kat1 triggers an adaptive genome rearrangement facilitating sexual differentiation. Surprisingly, the translation of Kat1 requires a programmed frameshift. The frameshift in the KAT1 gene dampens the activity of Kat1. In contrast, Kat1 is transcriptionally activated by nutrient limitation. Together our results reveal Kat1 as a highly regulated transposase that stimulates sexual reproduction.
Keywords: mating type, DNA double-strand break, transposable element, frameshift, DNA hairpin
Abstract
Transposable elements (TEs) have had a major influence on shaping both prokaryotic and eukaryotic genomes, largely through stochastic events following random or near-random insertions. In the mammalian immune system, the recombination activation genes1/2 (Rag1/2) recombinase has evolved from a transposase gene, demonstrating that TEs can be domesticated by the host. In this study, we uncovered a domesticated transposase, Kluyveromyces lactis hobo/Activator/Tam3 (hAT) transposase 1 (Kat1), operating at the fossil imprints of an ancient transposon, that catalyzes the differentiation of cell type. Kat1 induces mating-type switching from mating type a (MATa) to MATα in the yeast K. lactis. Kat1 activates switching by introducing two hairpin-capped DNA double-strand breaks (DSBs) in the MATa1–MATa2 intergenic region, as we demonstrate both in vivo and in vitro. The DSBs stimulate homologous recombination with the cryptic hidden MAT left alpha (HMLα) locus resulting in a switch of the cell type. The sites where Kat1 acts in the MATa locus most likely are ancient remnants of terminal inverted repeats from a long-lost TE. The KAT1 gene is annotated as a pseudogene because it contains two overlapping ORFs. We demonstrate that translation of full-length Kat1 requires a programmed −1 frameshift. The frameshift limited Kat1 activity, because restoring the zero frame causes switching to the MATα genotype. Kat1 also was transcriptionally activated by nutrient limitation via the transcription factor mating type switch 1 (Mts1). A phylogenetic analysis indicated that KAT1 was domesticated specifically in the Kluyveromyces clade of the budding yeasts. We conclude that Kat1 is a highly regulated transposase-derived endonuclease vital for sexual differentiation.
The mating types in budding yeasts are encoded by the mating type (MAT) loci MATa and MATα. Some yeasts have the unusual ability to change their mating type without going through mating or meiosis, a process called “mating-type switching.” In Saccharomyces cerevisiae, mating-type switching has been thoroughly explored (1). Switching is initiated when the homothallic switching (HO) endonuclease induces a DNA double-strand break (DSB) in the MAT locus (2). Next, the replacement is completed through a gene conversion, in which transcriptionally silent copies of MAT genes, known as “hidden MAT left, HMLα” and “hidden MAT right, HMRa,” are copied into the expressed MAT locus.
The related yeast Kluyveromyces lactis also uses gene conversion for mating-type switching. K. lactis has a nonfunctional copy of the HO gene (3), indicating that the common ancestor of K. lactis and S. cerevisiae used an HO-mediated switching mechanism but that K. lactis has acquired a new mechanism since then. Previously, we discovered a novel switching mechanism in which the MATα3 gene that resides in the MATα locus was the critical component (4). MATα3 shares homology with mutator-like transposable elements (MULEs), and regulated excision of this element from the genome initiates mating-type switching from MATα to MATa. Furthermore, a transcriptional regulator called “mating type switch 1” (Mts1; also known as “Rme1”) is another critical component for switching in K. lactis. Transcription of MTS1 is induced by nutrient limitation in a cAMP/Ras-dependent manner (5). Next, Mts1 binds to sites close to the MATα3 gene, stimulating the excision of the MULE and hence switching (4). Mts1 also induces switching from MATa to MATα, but in this case the molecular mechanism is unknown.
Transposable elements (TEs) and their remnants make up almost half of the human genome. Active TEs mobilize using either a cDNA/DNA copy of the element (copy and paste) or direct excision of the element from its original position and its insertion into a new position (cut and paste) (6). The former produces an additional copy of the TE, increasing the mutational load of the host genome. Nonautonomous TEs lack autonomous TE’s transposase gene but can still be mobilized from the genome using a transposase protein encoded by another TE.
A peculiar feature of many TEs is that they undergo translational frameshifts (7, 8). Programmed translational frameshifting is an alternate process of translation in which the ribosome slides back one nucleotide (−1 frameshifting) or skips one nucleotide (+1 frameshifting) (8, 9). The eukaryotic −1 frameshifting sites typically contain a “slippery” sequence (10) fitting the consensus motif X XXY YYZ, where XXX represents any three identical nucleotides; YYY represents AAA or UUU; Z represents A, C, or U; and spaces separate zero-frame codons. The tandem slippage model for frameshifting (11) posits that the peptidyl site (P-site) tRNA repairs from XXY to XXX, maintaining the codon–anticodon interaction except at the wobble position. Similarly, the aminoacyl site (A-site) tRNA repairs from YYZ to YYY. The relative frameshifting efficiency of such heptanucleotide slippery sequences is low (<1%), but additional stimulatory elements can increase frameshifting substantially. Such stimulatory elements, consisting of mRNA secondary structures such as pseudoknots (12), usually are on the 3′ side of the slippery sequence. It is believed that such mRNA structures induce a translational pause that allows time for codon–anticodon repairing.
We set out to elucidate the molecular mechanism underlying the switching from MATa to MATα in K. lactis. We discovered a protein related to hAT (hobo/Activator/Tam3) transposases that was essential for switching. This enzyme, K. lactis hAT transposase 1 (Kat1), cleaved the MATa locus in two different positions, resulting in DSBs that stimulated recombination. Kat1 activity was found to be highly regulated. Transcription of KAT1 was induced by Mts1, and translation of the KAT1 mRNA required a programmed −1 frameshift. The frameshift limited the Kat1 activity, thereby maintaining a balance between the MATa and MATα genotypes.
Results
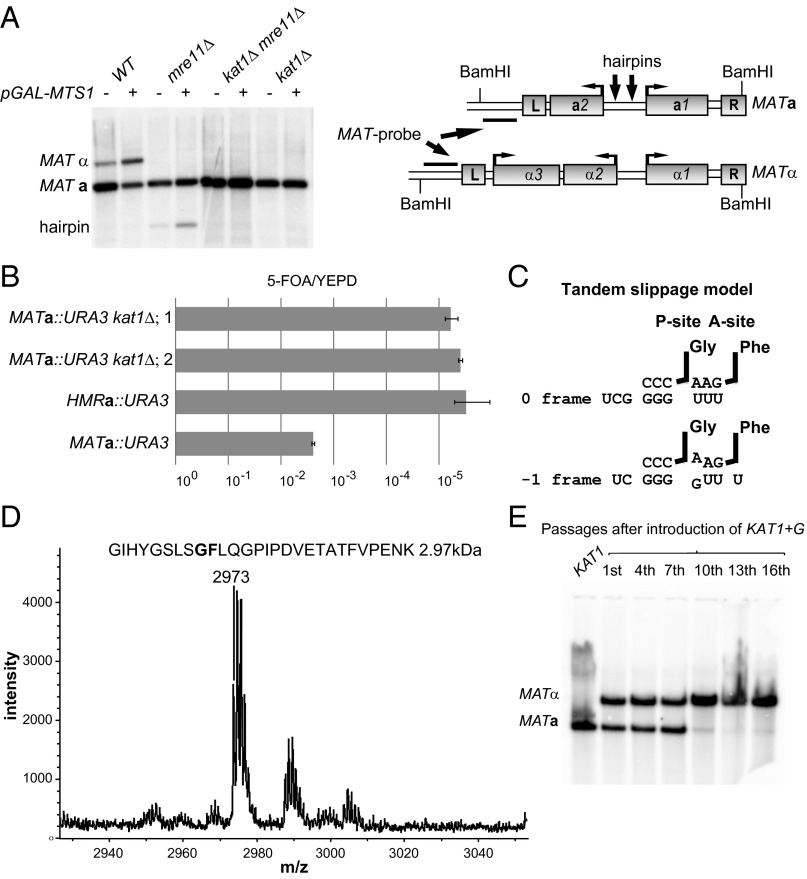
Hairpin-capped DSBs are observed in the MATa1–MATa2 intergenic region during mating-type switching in K. lactis (4). The Hermes transposase previously has been shown to generate hairpin-capped DSBs (13). Because Hermes belongs to the hAT superfamily of transposases (14), we hypothesized that a protein related to hAT transposases was cleaving the MATa locus. We searched the genome sequence translated in six frames using the hAT transposase from the fungus Fusarium oxysporum (Tfo1) as query (15). Two loci, one on chromosome A and one on chromosome D, shared homology with Tfo1 (Expect < 0.02) (Fig. S1). Both loci were deleted, and mating-type switching was assessed in the mutant strains by the overexpression of Mts1, followed by DNA blot analysis. The strain with the deletion on chromosome A switched mating type with normal efficiency, but the strain with the locus deletion on chromosome D displayed no detectable switching (Fig. 1A). The locus required for switching (KLLA0D05677g) was named “KAT1.” The kat1Δ strain displayed a normal growth rate.
Fig. 1.
Kat1 is essential for mating-type switching and is subjected to a programmed −1 frameshift. (A, Left) DNA blot analysis of BamHI-digested genomic DNA from WT, mre11Δ, kat1Δ, and mre11Δ kat1Δ containing plasmid alone (−) or pGAL–MTS1 (+), hybridized with a MAT-specific probe. The MATa, MATα, and hairpin bands are indicated. (Right) Schematic diagram of the MATa and MATα loci. Boxes denote genes and the Left (L) and Right (R) repetitive elements. The probe used, the BamHI sites, and approximate positions of hairpins (arrows) are indicated. (B) The ratios of 5-FOA/yeast extract peptone dextrose (YEPD) plating efficiencies were determined in MATa::URA3, HMRa::URA3, and two independently generated MATa::URA3 kat1Δ strains. Error bars represent the SEM from three independent measurements. (C) Tandem slippage model for frameshifting at the KAT1 slippery site. The ribosomal P- and A-sites harboring the tRNAGly and tRNAPhe are indicated. The zero-frame and −1 frame base pairings between the tRNAs and mRNA are shown. (D) MS analysis of a peptide obtained from a GST–Kat1 slippery site–MBP fusion protein. The predicted peptide (2.97 kDa) from the tandem-slippage hypothesis is shown above the graph. The x axis shows the molecular weight in daltons. (E, Lower) DNA blot analysis as described in A. DNA was prepared from a strain carrying the KAT1+G allele at the endogenous locus. (Upper) The number of overnight passages in synthetic complete medium after the generation of the KAT1+G strain.
Strains lacking the DNA repair protein meiotic recombination 11 (Mre11) accumulate a hairpin-capped DSB in MATa because Mre11 is required for opening hairpins (16, 17). An mre11Δ kat1Δ double mutant did not accumulate the hairpin (Fig. 1A), indicating that Kat1 was acting upstream of Mre11. To investigate if there was any residual switching in the kat1Δ strain, a more sensitive assay was used (5). This assay relies on a uracil requiring URA3 gene inserted into the MATa locus. Mating-type switching results in the loss of URA3 and resistance to 5-fluoroorotic acid (5-FOA). Because HMRa is used as a donor during switching, the HMRa::URA3 strain becomes 5-FOA resistant with a low frequency, thus serving as a control. This assay showed that the level of mating-type switching was ∼1,000-fold lower in the kat1Δ strains than in the WT strain (Fig. 1B) and was similar to the level in the HMRa::URA3 strain. Thus, mating-type switching was completely blocked in the kat1Δ strains.
The KAT1 locus is annotated as a pseudogene because it contains two long ORFs interrupted by a −1 frameshift. We hypothesized that the translation of Kat1 involved a programmed −1 frameshift. To identify a slippery sequence, the KAT1 loci from five closely related Kluyveromyces species (K. aestuarii, K. dobzhanskii, K. lactis, K. marxianus, and K. wickerhamii) were compared. The other yeast species contained a −1 frameshift in a position similar to KlKAT1. Multiple sequence alignments revealed an 8-bp sequence that was conserved in the overlapping sequence between the long ORFs (Fig. S2A). In K. lactis a predicted pseudoknot (Fig. S2B), known to stimulate frameshifting by inducing a translational pause (12), was located 5 bp downstream from the conserved octamer. The potential slippery sequence in KAT1 (CG GGG UUU) was not a typical slippery site, because the predicted A-site tRNAPhe would exhibit a first-position purine:purine clash after slipping one nucleotide in the 3′ direction (Fig. 1C). However, the ribosome can accommodate purine clashes in certain circumstances (18). To investigate this frameshifting further, we generated a frameshift reporter protein. A glutathione S-transferase (GST)–Kat1 slippery site–maltose binding protein (MBP) fusion protein, in which the MBP moiety was in the −1 frame compared with the GST moiety, was expressed in Escherichia coli because bacteria can use eukaryotic frameshifting signals (19). A GST–MBP fusion protein was readily obtained, indicating that the KAT1 slippery site was functional. Next, we analyzed trypsin-treated fusion protein by MS. The mass of the peptide covering the frameshift site (2.97 kDa) strongly suggested that the A-site tRNA during slippage indeed was tRNAPhe (Fig. 1D). (The split peaks close to the 2.97-kDa peak are caused by the isotopic contribution of C13.) In addition, an MS/MS analysis of the 2.97-kDa peptide confirmed that it covered the frameshift site (Fig. S3). A dual-luciferase assay was used to characterize the slippery site further, confirming that the sequence of the octamer was important for frameshifting (Fig. S2C). Hence, KAT1 translation was subjected to a programmed −1 frameshift involving an unusual purine:purine clash.
We reasoned that the frameshift in KAT1 could limit Kat1 activity, thereby restricting MATa-to-MATα switching. To test this idea, a single nucleotide was inserted into the slippery site, allowing KAT1 to be translated in a continuous frame. To address the cellular role of the frameshift, the frameshift-correcting mutation (KAT1+G) was introduced at the endogenous KAT1 locus in a MATa strain. Next, the resulting strain was grown for 16 overnight passages in nutrient-limited conditions (synthetic dextrose plus the required amino acids), and samples were collected for DNA blot analysis. After 10 passages the majority of the cells had switched to the MATα genotype (Fig. 1E). Introduction of a plasmid overexpressing Mts1 into the cells from the 16th passage resulted in a switch back to the MATa genotype, demonstrating that the phenotype is reversible (Fig. S2D). Overexpression of Kat1+G, but not Kat1, resulted in an almost complete conversion to MATα after only a single overnight passage (Fig. S4A). Hence, the frameshift in the KAT1 gene is critical to maintain the balance between the MATa and MATα cell-types.
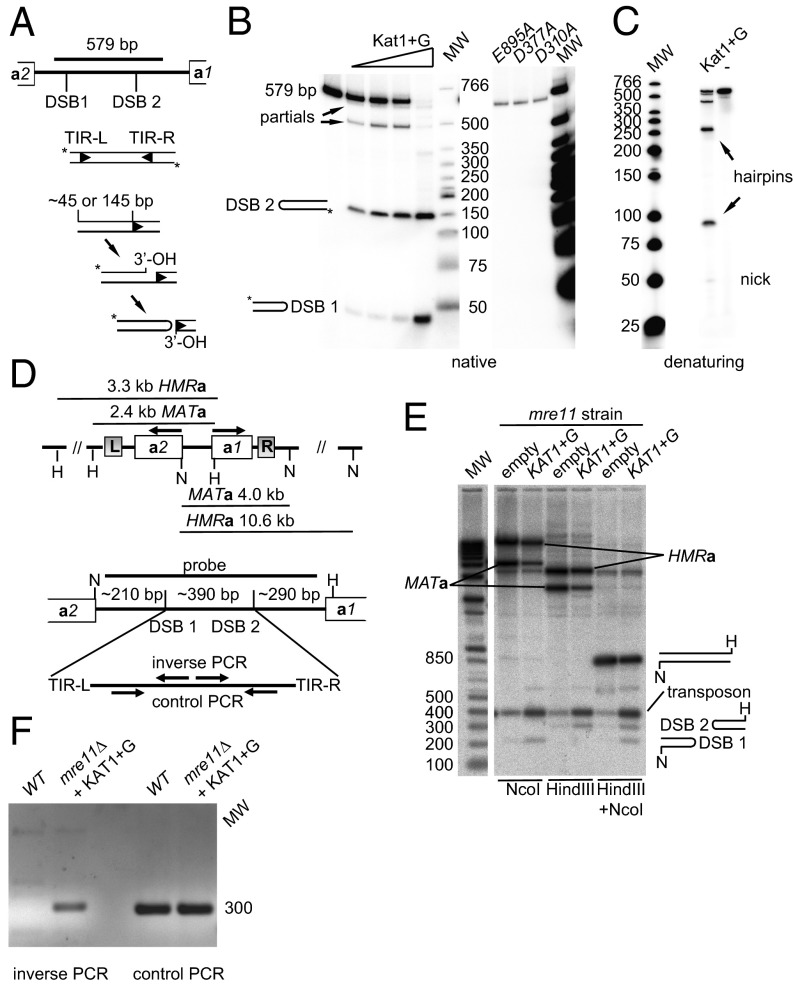
The proteins encoded by the recombination activating genes 1/2 (Rag1/2) promote V(D)J recombination in lymphoid cells and are known to generate hairpin-capped DSBs (20). The generation of hairpin-capped DSBs by hAT transposases and Rag1/2 involves a two-step reaction. First, the enzymes introduce a nick at (or close to) the boundary between the terminal inverted repeats (TIRs) and the host DNA. In the second step, the newly formed 3′ hydroxyl (OH) attacks a phosphodiester bond on the opposite strand, forming a covalently closed hairpin structure (Fig. 2A). To investigate if Kat1 shares this activity, GST–Kat1+G was purified (Fig. S4D) and incubated with a radiolabeled 579-bp DNA fragment covering most of the MATa1–MATa2 intergenic region (see Fig. S5 for details). The results showed a dose-dependent cleavage of the substrate. Kat1 generated two DSBs in the substrate, resulting in products of ∼45 and 145 bp (Fig. 2B). Because the substrate was end-labeled, the fragment between the sites was not observed as a product. In denaturing conditions, the labeled products migrated with an apparent size twice that on the native gel, showing that they contained hairpin-capped ends (Fig. 2C).
Fig. 2.
Kat1 is an endonuclease that generates two hairpin-capped DSBs in MATa. (A) Mechanism of DNA cleavage of hAT transposases. A schematic diagram of the 579-bp fragment used in the cleavage assays is shown with the DSB 1/2 sites indicated. First a nick is generated at or close to the TIR. Next, the exposed 3′ OH attacks the complementary strand to generate a hairpin, releasing the transposon end. The approximate lengths of the end-labeled products are shown. (B, Left) In vitro activity of Kat1. Increasing amounts of GST–Kat1+G were mixed with the 579-bp substrate, followed by separation on 6% PAGE using neutral conditions. (Right) The same assay using GST–Kat1+G with mutations in the DDE motif. The lengths of the products and DNA ladder are indicated. MW, molecular weight. (C) Same conditions as in B, but the products were separated in denaturing conditions. Nicked products and hairpins are indicated. (D, Upper) Schematic diagram of the MATa/HMRa loci showing the NcoI (N) and HindIII (H) sites and the lengths of restriction fragments. (Lower) The MATa1–MATa2 intergenic region, showing the probe used, restrictions enzyme sites, the approximate positions of DSBs, and lengths of generated fragments. (Lower) The primers for inverse/control PCR are depicted schematically. (E) DNA blot analysis using NcoI, HindIII, and double-digested DNA using the probe indicated in C. DNA was prepared from an mre11Δ strain containing plasmid alone or pKAT1+G. The MATa and HMRa bands are indicated. Bands induced by Kat1 overexpression were named “DSB 1,” “DSB 2,” and “transposon.” The molecular weight marker is shown on the left. (F) Genomic DNA from mre11Δ containing pKAT1+G or WT containing plasmid alone was subjected to inverse/control PCR using the primers depicted in C followed by agarose gel electrophoresis and ethidium bromide staining.
The active site of transposases contains a DDE motif (21), which is critical for the catalytic activity. A multiple sequence alignment between KAT1 and four hAT transposases (Fig. S1) identified residues predicted to be part of the DDE motif. Exchanging the residues in Kat1 that were predicted to define the DDE motif (D310, D377, and E895) for alanines abolished the Kat1 activity in vitro (Fig. 2B) and in vivo (Fig. S4B). The proteins were not unstable or poorly expressed, because the steady-state levels of the DDE mutants were similar to WT levels both in yeast (Fig. S4C) and as recombinant proteins (Fig. S4D).
To obtain evidence for Kat1 binding to the MATa1–MATa2 intergenic region in vivo, we performed ChIP of Myc–Kat1+G (Fig. S6). Because expression of WT Myc–Kat1+G results in a rapid switch of mating type to the MATα genotype, we used the DDE mutant kat1D310A+G for this experiment, assuming that Kat1D310A binds MATa with normal efficiency. The precipitated DNA was analyzed by quantitative PCR (qPCR) using two primer pairs in the MATa1–MATa2 intergenic region. The data were normalized to a primer pair from a locus not expected to be bound by Kat1. The Myc–Kat1 samples showed approximately sevenfold enrichment over the background signal, demonstrating that Myc–kat1D310A was binding to MATa in vivo.
To examine the activity of Kat1 in vivo, we overexpressed Kat1+G, followed by DNA blotting using a probe spanning the MATa1–MATa2 intergenic region. We used an mre11 strain because it is easier to detect hairpins in this background. In agreement with the in vitro results, we observed that Kat1+G overexpression induced two DSBs (Fig. 2 D and E). The endogenous KAT1 gene induced small amounts of DSBs in the absence of Kat1 overexpression. The DNA between the DSBs hybridized strongly with the probe, indicating that it was stabilized in vivo, possibly by circularization. To investigate this possibility, we performed inverse PCR (Fig. 2D and Fig. S5). Inverse PCR allows amplification of templates that are circular but cannot amplify templates that are hairpin-capped or linear. This analysis showed that the DNA between the DSBs was circular (Fig. 2F). We consider the circular DNA to be a nonautonomous transposon. Thus, Kat1 is an endonuclease that generates two DSBs in MATa and, furthermore, the DNA between the DSBs is circularized.
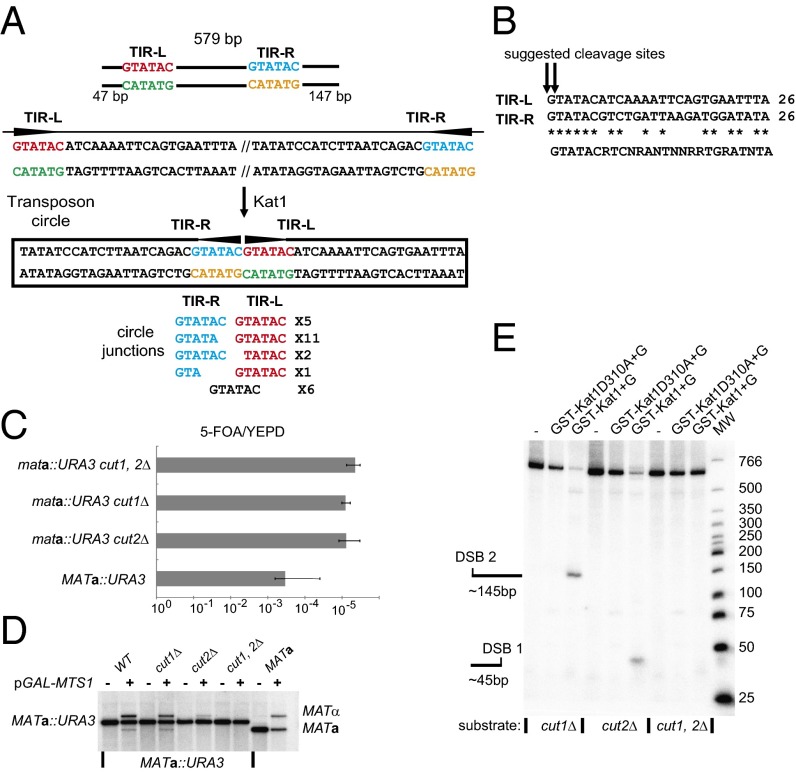
To pinpoint where Kat1 cuts the MATa locus, we sequenced the circular transposon. We performed inverse PCR using three different primers pairs and cloned and sequenced several independent clones from each. The sequencing analysis showed that the transposon was circularized at two sequences that were recognition sites for the AccI endonuclease (GTATAC) (Fig. 3A). Among the 25 clones sequenced, we obtained five different circle junctions. In 18 (72%) of the clones, the two AccI sites either were intact or lacked a single base pair, indicating that Kat1 cleaves close to the G residue in the AccI sites. In seven clones there were further deletions. We speculate that the shorter junctions may result from the processing of the transposon ends in vivo. Moreover, the G residues in the AccI sites were located 47 bp and 147 bp (Fig. 3A and Fig. S5), respectively, from the ends of the 579-bp fragment used in the cleavage assay (Fig. 2B). Therefore, the Kat1 cutting sites suggested by sequencing of the circle junctions were consistent with the sizes of the products in the in vitro assay.
Fig. 3.
Identification of TIRs in the MATa1–MATa2 intergenic region. (A, Top) Diagram of the 579-bp PCR fragment used in the cleavage assay showing the distance from the ends of the fragment to TIR-L and TIR-R flanked by a pair of AccI sites. The sites are colored for clarity. (Middle) DNA sequence of the TIRs and their relative orientations in linear and circular forms. (Bottom) The sequence of cloned inverse PCR fragments covering the circle junctions. The number of clones representing each sequence is shown. (B) Alignment of 26 bp of DNA from TIR-L with the reverse complement of TIR-R. Identical bases are indicated by asterisks, and a putative consensus Kat1 site is depicted below. The tentative Kat1 cleavage sites are indicated by the arrows. (C) The ratio of 5-FOA/YEPD plating efficiencies was determined in MATa::URA3, mata::URA3-cut1Δ, mata::URA3-cut2Δ, and mata::URA3-cut1,2Δ strains. The cut1Δ and cut2Δ mutations denote deletions of the two GTATAC sites. Error bars represent the SEM from three independent measurements. (D) DNA blot analysis of BamHI-digested genomic DNA from the strains in C and a MATa strain containing plasmid alone (−) or pGAL–MTS1 (+), hybridized with a MAT-specific probe. The MATa, MATa::URA3, and MATα bands are indicated. (E) In vitro nuclease assay using 579-bp substrates with deletions of the two AccI sites, either singly or in combination, labeled as in Fig. 2B.
DNA transposases normally act on TIRs that are longer than 6 bp. In addition, a recognition site of only 6 bp seems too short to mediate specificity to Kat1 cleavage. An alignment of the sequences flanking the two AccI sites (using one in the inverse orientation) revealed two imperfect 26-bp-long TIRs (designated “TIR-L” and “TIR-R”, suggesting that the Kat1 recognition site is extended (Fig. 3B).
To explore if the TIRs are important in vivo, we generated strains carrying exact deletions of the AccI sites in TIR-L and/or TIR-R, called “cut1Δ” and “cut2Δ,” respectively. Deletion of either of the sites reduced mating-type switching to background levels in the assay involving MATa::URA3 strains without overexpression of Mts1. (Fig. 3C). However, upon Mts1 overexpression, the strains in which only one of the sites was deleted still switched to MATα (Fig. 3D). Thus both sites are essential for switching under normal conditions, but either one of them suffices when switching is induced by Mts1 overexpression. In the in vitro cleavage assay, the substrates containing deletion of one of the sites were cut at the other site, and the substrate containing deletions of both sites was not cut at all (Fig. 3E), confirming that the TIRs are crucial for the endonuclease activity of Kat1.
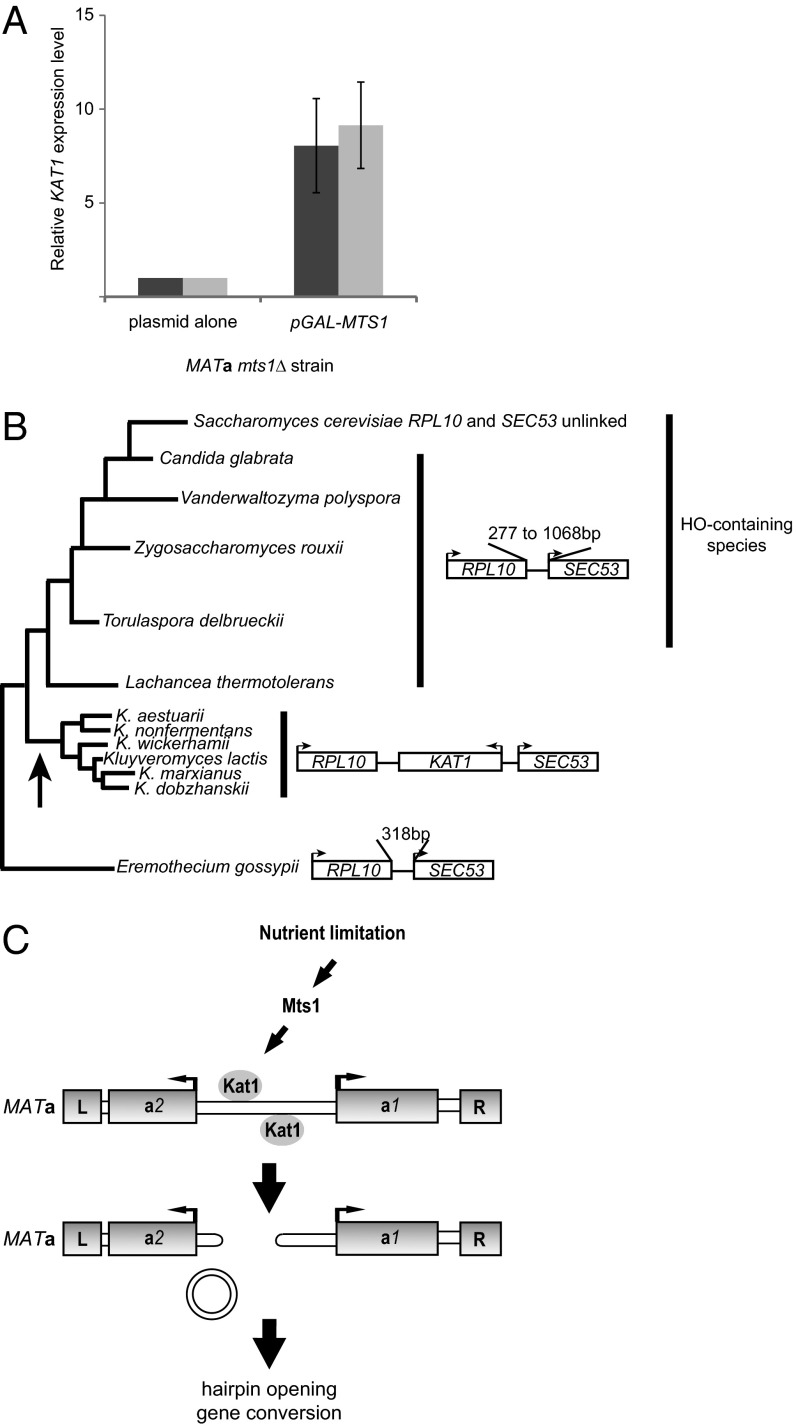
To examine the regulation of the cleavage event, we analyzed data from a genome-wide analysis of Mts1-binding sites (22) and observed a peak of Mts1 binding upstream from the KAT1 gene (Fig. S7). Closer scrutiny of this upstream region revealed a consensus Mts1-binding site, which was conserved in K. dobzhanskii. qRT-PCR showed that overexpression of Mts1 resulted in a KAT1 mRNA level approximately ninefold higher than seen in the same strain lacking Mts1 expression (Fig. 4A). Hence, Mts1 activates KAT1 transcription, presumably by binding to the KAT1 regulatory region.
Fig. 4.
Kat1 regulation, phylogeny of KAT1, and model for mating-type switching. (A) RT-qPCR using two primer pairs (light and dark gray bars) amplifying the KAT1 cDNA. Messenger RNA was prepared from an mts1Δ strain containing empty plasmid or pGAL–MTS1. The y axis shows the relative expression level of the KAT1 mRNA. The level in the strain containing empty plasmid was set to 1.0. The expression level was normalized to actin (ACT1) using the comparative threshold (Ct) method. Error bars indicate the SEM from three independent experiments. (B) Phylogenetic tree of the ascomyceteous lineage showing selected species. The length of the branches is not drawn to scale. The distance between SEC53 and RPL10 genes in the different species is shown at right. Also, the presence of long (>700-aa) HO-encoding genes in the species is shown. The arrow denotes the likely acquisition of the KAT1 gene in the common ancestor of the Kluyveromyces yeasts. (C) Diagram showing the MATa locus with the genes boxed. The L and R boxes depict repeats that flank all mating-type loci, presumably used as sequences to resolve recombination intermediates. Nutrient limitation activates switching via stimulation of MTS1/KAT1 transcription. Kat1 acts on two sites in the MATa1–MATa2 intergenic region, generating hairpin-capped ends, and the intervening DNA is joined into a circle. Next, a gene conversion takes place using the HMLα locus as donor.
Discussion
We hypothesized that KAT1 originated from an ancient transposition event and investigated the RPL10-SEC53 intergenic region from related yeasts whose genome sequences were known. Fig. 4B shows a phylogenetic tree (23) and the distance between the RPL10 and SEC53 genes in selected budding yeasts. In yeasts belonging to the same clade as K. lactis, close homologs to KAT1 were found in the RPL10–SEC53 intergenic region. RPL10 and SEC53 were closely linked (<1 kb) in the other species investigated, except in S. cerevisiae, where these genes reside on different chromosomes. The lack of synteny indicated that KAT1 originated from a transposition event that occurred in the common ancestor of the Kluyveromyces yeasts.
We also investigated the presence of an HO gene in these yeast species. BLAST searches revealed genes encoding long (>700-aa) proteins highly homologous to S. cerevisiae HO in four species (Fig. 4B). The KAT1 and HO genes did not coexist in the same genomes, as is consistent with the idea that a KAT1-mediated switching mechanism has replaced an ancestral HO-mediated mechanism.
We suggest a model of how K. lactis switches mating type from MATa to MATα (Fig. 4C). Nutrient limitation induces the transcription of Mts1 (4, 22), which in turn activates KAT1 transcription. Translation of Kat1 also is regulated by a −1 frameshift, probably acting as a general repressor of Kat1 synthesis. Next, Kat1 generates two hairpin-capped DSBs in the MATa locus, and the DSBs promote a gene conversion using HMLα as donor.
The KAT1 translational slippery site is unusual because it involves a purine:purine clash in the first position of the A-site after the −1 frameshift took place. Recent evidence shows that the ribosomal decoding center can accommodate purine:purine mismatches under certain circumstances (18) and therefore may be more flexible than previously thought. Another unusual feature of the KAT1 slippery site is that the conserved sequence (an octamer) is 1 bp longer than the common heptameric slippery sequences (9). We have found that the identity of the first base of the octamer is important for frameshifting efficiency, because a mutation of the first C to T reduces frameshifting by ∼20-fold in the dual-luciferase assay (Fig. S2C). The reduced frameshifting in the C to T mutant suggests a potential role for the exit-site tRNA in KAT1 frameshifting, but further studies are required to confirm this observation.
There are numerous examples of TEs becoming domesticated to perform functions that are beneficial for the host (24). A thoroughly studied example is the RAG1/2 proteins essential for V(D)J-recombination in lymphoid cells (20). RAG1/2 originated from a TE related to transib elements (25). Other examples are the transposase-related genes required for large-scale genome rearrangements in different ciliate species (26–28). Remarkably, Drosophila species use domesticated retrotransposons called “HeT-A” and “TART” for maintaining telomeres (29).
Nutrient limitation induces mating-type switching through transcriptional activation of the Mts1 gene. Previous studies have shown that TEs can be activated by different cellular stresses (30–32). An interesting example is provided by the fission yeast Tf2 retroelements that contain binding sites for Sre1, the yeast ortholog of sterol regulatory element binding protein, in their LTRs. Binding of Sre1 results in activation of Tf2 by oxygen deprivation. Sequences adjacent to the Tf2 LTRs also could become oxygen-regulated, showing that TEs can rewire transcription of host genes (33). Hence, the relationship between TEs and their host genomes is multifaceted.
Kat1 cleavage generates hairpin-capped ends similarly to RAG1/2 (34) and Hermes (13). In addition, during V(D)J recombination the signal joints are circularized, and circular forms of the hAT transposons restless, hobo, and Hermes have been observed (35, 36). However, an obvious difference between Kat1-mediated switching and RAG1/2-mediated recombination is the fate of the hairpin-capped DSBs (coding ends). During V(D)J recombination the hairpins are opened and then joined by nonhomologous end-joining (NHEJ) to generate mature V(D)J joints. In contrast, the hairpins in K. lactis are channeled into a gene-conversion pathway dependent on the RAD50 group of homologous recombination (HR) genes (37). The differential pathway usage might reflect the general activity of NHEJ and HR in yeasts and mammals, NHEJ being the main DSB-repair pathway in mammals, and HR being the main DSB-repair pathway in yeasts.
Mating-type switching in K. lactis, and presumably all species in the Kluyveromyces clade, involves two domesticated transposases. In MATα cells, excision of the MATα3 MULE induces switching, and in MATa cells Kat1 induces switching. It seems that the activity of α3 and Kat1 must be carefully balanced to ensure equal ratios of the MATa and MATα cell types. If the activity of one of the transposases were to dominate, then one cell type would take over in natural populations. Therefore, it was not surprising that the frameshift-correcting KAT1+G allele mediated a shift of cultures to the MATα cell type. Thus, a programmed frameshift regulates the ratios of mating types, which in turn control the likelihood of encountering a mating partner.
We speculate that in an ancestor of the Kluyveromyces yeasts an active KAT1-related TE was inserted between the MATa1 and MATa2 genes. This insertion may have rendered the ancestral HO gene redundant. Since then, the KAT1-related element has deteriorated, but the signals for excision have been maintained. In contrast, in another copy of the element (the KAT1 gene) the coding capacity has been maintained, but the signals for excision have deteriorated. The excision of the MATa-linked element has become regulated, stimulated by nutrient limitation through recruitment of an Mts1-mediated activation and repressed by a frameshift in the KAT1 gene. In this view, the MATa1–MATa2 intergenic region contains a nonautonomous TE that is mobilized by Kat1. Thus we have an intriguing example of a domesticated transposon promoting sexual differentiation of the host.
Materials and Methods
Yeast Strains.
The yeast strains used in this study are listed in Table S1. Gene deletions/insertions relied on HR using a one-step disruption/insertion procedure modified for K. lactis (38). See SI Materials and Methods for details.
Plasmids.
Cloning was performed using standard methods (39). The pGAL-MTS1 vector (pPMB35) and the K. lactis plasmids pCXJ18/20 were described previously (4, 40). Detailed procedures for plasmid construction are presented in SI Materials and Methods.
Methods.
DNA blots, DNA/RNA/protein preparations, and transformation of yeast and E. coli followed standard protocols (39, 41). Sequences of oligonucleotides used for generating probes for DNA blots, ChIP, and qPCR are available in Table S2. E. coli strain DE3 harboring GST–KAT1+G or DDE mutants was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (18 h at 18 °C). Proteins were purified from 1 L of culture using glutathione Sepharose 4B (GE Healthcare). The Kat1 cleavage reaction was performed by incubating the end-labeled 579-bp DNA substrate with increasing concentrations of Kat1+G in a buffer containing 50 mM Tris⋅HCl (pH 8.0), 0.5 mM EDTA, 1 mM DTT, 100 μg/mL BSA, and 5 mM MgCl2 at 30 °C for 3 h. The reactions were terminated by the addition of EDTA (10 mM) and were glutathione Sepharose 4B deproteinized by phenol:chloroform extraction followed by ethanol precipitation in presence of glycogen. The products were resolved on 6% native/denaturing PAGE.
Supplementary Material
Acknowledgments
We thank E. Barsoum and members of the S.U.Å. laboratory for helpful suggestions; Å. Engström for the MS analysis of the GST–Kat1–MBP peptides; J. F. Atkins and J. D. Dinman for kindly providing plasmids; and C. Andreasson, S. Bauer, M. Mannervik, J. Rine, and C. Samakovlis for critical reading of the manuscript. This work was supported by grants from the Swedish Cancer Society and the Carl Trygger Foundation (to S.U.Å.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406027111/-/DCSupplemental.
References
- 1.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strathern JN, et al. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 3.Fabre E, et al. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22(4):856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 4.Barsoum E, Martinez P, Aström SU. Alpha3, a transposable element that promotes host sexual reproduction. Genes Dev. 2010;24(1):33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barsoum E, Rajaei N, Aström SU. RAS/cyclic AMP and transcription factor Msn2 regulate mating and mating-type switching in the yeast Kluyveromyces lactis. Eukaryot Cell. 2011;10(11):1545–1552. doi: 10.1128/EC.05158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curcio MJ, Derbyshire KM. The outs and ins of transposition: From mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4(11):865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 7.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7(4):497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 8.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60(1):103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73(1):178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432(7020):995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 14.Arensburger P, et al. Phylogenetic and functional characterization of the hAT transposon superfamily. Genetics. 2011;188(1):45–57. doi: 10.1534/genetics.111.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda M, Ikeda K, Namiki F, Nishi K, Tsuge T. Tfo1: An Ac-like transposon from the plant pathogenic fungus Fusarium oxysporum. Mol Gen Genet. 1998;258(6):599–607. doi: 10.1007/s004380050773. [DOI] [PubMed] [Google Scholar]
- 16.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108(2):183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 17.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276(38):35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 18.Fernández IS, et al. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500(7460):107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yelverton E, Lindsley D, Yamauchi P, Gallant JA. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol Microbiol. 1994;11(2):303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: Transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 22.Booth LN, Tuch BB, Johnson AD. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature. 2010;468(7326):959–963. doi: 10.1038/nature09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachance MA. Current status of Kluyveromyces systematics. FEMS Yeast Res. 2007;7(5):642–645. doi: 10.1111/j.1567-1364.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 24.Biémont C. A brief history of the status of transposable elements: From junk DNA to major players in evolution. Genetics. 2010;186(4):1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowacki M, et al. A functional role for transposases in a large eukaryotic genome. Science. 2009;324(5929):935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baudry C, et al. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23(21):2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng CY, Vogt A, Mochizuki K, Yao MC. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell. 2010;21(10):1753–1762. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 30.Todeschini AL, Morillon A, Springer M, Lesage P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(17):7459–7472. doi: 10.1128/MCB.25.17.7459-7472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strand DJ, McDonald JF. Copia is transcriptionally responsive to environmental stress. Nucleic Acids Res. 1985;13(12):4401–4410. doi: 10.1093/nar/13.12.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandbastien MA, et al. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res. 2005;110(1-4):229–241. doi: 10.1159/000084957. [DOI] [PubMed] [Google Scholar]
- 33.Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3(8):e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152(3):417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brochta DA, et al. Transpositionally active episomal hAT elements. BMC Mol Biol. 2009;10:108. doi: 10.1186/1471-2199-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kempken F, Kück U. Evidence for circular transposition derivatives from the fungal hAT-transposon Restless. Curr Genet. 1998;34(3):200–203. doi: 10.1007/s002940050386. [DOI] [PubMed] [Google Scholar]
- 37.Malone RE, Esposito RE. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA. 1980;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kegel A, Martinez P, Carter SD, Aström SU. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 2006;34(5):1633–1645. doi: 10.1093/nar/gkl064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausubel FM. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. 4th Ed Wiley; New York: 1999. [Google Scholar]
- 40.Chen XJ. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene. 1996;172(1):131–136. doi: 10.1016/0378-1119(96)00125-4. [DOI] [PubMed] [Google Scholar]
- 41.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.