Significance
Because obese people are at an increased risk of many age-related diseases, it is a plausible hypothesis that obesity increases the biological age of some tissues and cell types. However, it has been difficult to detect such an accelerated aging effect because it is unclear how to measure tissue age. Here we use a recently developed biomarker of aging (known as “epigenetic clock”) to study the relationship between epigenetic age and obesity in several human tissues. We report an unexpectedly strong correlation between high body mass index and the epigenetic age of liver tissue. This finding may explain why obese people suffer from the early onset of many age-related pathologies, including liver cancer.
Keywords: obesity, epigenetics, aging, biological age, DNA methylation
Abstract
Because of the dearth of biomarkers of aging, it has been difficult to test the hypothesis that obesity increases tissue age. Here we use a novel epigenetic biomarker of aging (referred to as an “epigenetic clock”) to study the relationship between high body mass index (BMI) and the DNA methylation ages of human blood, liver, muscle, and adipose tissue. A significant correlation between BMI and epigenetic age acceleration could only be observed for liver (r = 0.42, P = 6.8 × 10−4 in dataset 1 and r = 0.42, P = 1.2 × 10−4 in dataset 2). On average, epigenetic age increased by 3.3 y for each 10 BMI units. The detected age acceleration in liver is not associated with the Nonalcoholic Fatty Liver Disease Activity Score or any of its component traits after adjustment for BMI. The 279 genes that are underexpressed in older liver samples are highly enriched (1.2 × 10−9) with nuclear mitochondrial genes that play a role in oxidative phosphorylation and electron transport. The epigenetic age acceleration, which is not reversible in the short term after rapid weight loss induced by bariatric surgery, may play a role in liver-related comorbidities of obesity, such as insulin resistance and liver cancer.
Dietary restriction in humans causes changes that protect against many age-related pathologies (1). In contrast, obesity increases the risk of chronic age-related diseases, such as type 2 diabetes, heart disease, osteoarthritis, and certain types of cancer, and thus constitutes a major and rising global health problem (2). It is a plausible hypothesis that obesity increases the risk of at least some of these diseases through accelerated tissue aging.
Leukocyte telomere length, which is a widely used biomarker of aging, has been found to be negatively correlated with body mass index (BMI) (3–7). Although the observed correlation between BMI and telomere length is relatively weak (r = 0.12) (4), it is remarkable that these studies demonstrated that BMI is associated with an age acceleration effect in blood.
Assessing tissue age poses a significant methodological challenge because it is not clear which biomarkers of aging are appropriate. There is a considerable debate in the literature as to what extent markers/causes of cellular senescence, such as telomere length, capture all aspects of tissue aging (8–10). Biomarkers of tissue age should ideally be validated in a wide range of tissues and cell types, provide a quantitative estimate of tissue age, and should be uniformly applicable to tissue samples from different tissue banks. S.H. recently developed a biomarker of aging (“epigenetic clock”) based on DNA methylation (DNAm) levels (11). This epigenetic clock is defined as a prediction method of chronological age based on the DNAm levels of 353 CpGs. The predicted (estimated) age resulting from the epigenetic clock is referred to as “DNAm age.” Here we use DNAm age as a proxy of epigenetic tissue age. DNAm age is highly (r = 0.96) correlated to chronological age across sorted cell types (CD4 T cells, monocytes, B cells, glial cells, neurons), complex tissues (e.g., blood), and organs (brain, breast, kidney, liver, lung) (11). The epigenetic clock is robust with respect to batch effects and can be applied to two commercially standardized platforms: the Illumina 450K array and the 27K array.
Results
DNAm age was calculated as described in ref. 11 from a total of 1,215 human samples profiled with the Illumina Infinium 450K and 27K arrays (n = 141 liver, n = 274 blood, n = 726 adipose tissue, n = 74 muscle). Methylation datasets were either obtained from public data repositories or—in the case of the replication dataset from liver, muscle, and adipose tissue—generated for the purposes of this study. To study the relationship between epigenetic age acceleration and transcriptional changes, we also generated a novel gene-expression dataset on the Human Gene 1.1 ST Array platform from Affymetrix. An overview of the datasets used is provided in Table 1 and in Methods.
Table 1.
Overview of the DNA methylation datasets
| Tissue source | Platform | n | Data ID | Source | Figure | cor with BMI |
| Discovery dataset | ||||||
| 1. Liver | Illum450K | 62 | GSE48325 | (12) | 1, 3, 4 | r = 0.42, P = 6.8×10−4 |
| 2. Adipose | Illum450K | 648 | E-MTAB1866 | (13) | 1 | r = −0.02, P = 0.68 |
| 3. Muscle | Illum450K | 48 | GSE50498 | (14) | 1 | r = −0.087, P = 0.56 |
| 4. Blood | Illum27K | 71 | GSE49909 | (15) | 1 | r = −0.066, P = 0.58 |
| 5. Blood | Illum27K | 92 | GSE37008 | (16) | S1 | r = 0.26, P = 0.012 |
| 6. Blood | Illum450K | 111 | GSE53840 | This study | S1 | r = −0.18, P = 0.12 |
| 7. Adipose | Illum450K | 46 | Reader comment | (20) | 4 | r = NA, P = NA |
| Replication dataset | ||||||
| 8. Liver | Illum450K | 79 | GSE61258 | This study | 2, 3 | r = 0.42, P = 1.2E-4 |
| 9. Adipose | Illum450K | 32 | GSE61257 | This study | 2 | r = 0.18, P = 0.32 |
| 10. Muscle | Illum450K | 26 | GSE61259 | This study | 2 | r = 0.085, P = 0.68 |
| Transcriptional data (messenger RNA) | ||||||
| 11. Liver | HuGen1.1ST | 134 | GSE61260 | This study | Table 3 | NA |
The rows correspond to the datasets used in this article. Columns report the tissue source, DNAm platform, number of subjects (n), access information and citation, and a reference to the use in this report. The last column reports the Pearson correlation coefficient between BMI and age acceleration, denoted as “cor,” and the corresponding P value. NA, not applicable.
Cross-Sectional Analysis of BMI and DNAm Age.
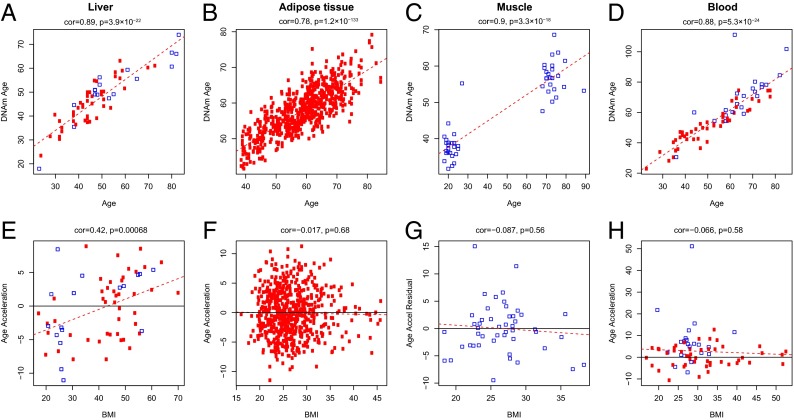
At the discovery stage, all publicly available Illumina DNAm datasets with phenotypic information on chronological age, BMI, and sex were analyzed. In these analyses of liver (12), adipose tissue (13), muscle (14), and blood (15, 16), a strong positive (and expected) correlation between chronological age and DNAm age was observed with correlation coefficients ranging from 0.78 to 0.90 (Fig. 1 A–D).
Fig. 1.
Discovery analysis. Analysis of the correlation of DNAm age to chronological age across the publicly available datasets: (A–D) This row shows the correlation of chronological age to the DNAm age in liver, adipose tissue, muscle, and blood. The red dashed line in these panels indicates the regression line. In all panels (A–H), each point corresponds to a human subject. Red circles indicate women and blue squares are used to denote male individuals. The age acceleration effect for each subject (point) corresponds to the vertical distance to the red regression line. (E–H) This row plots the relation of BMI and age acceleration in those tissues. The black horizontal line (y = 0) corresponds to an age acceleration of zero. It is evident that only liver tissue shows a significant correlation (r = 0.42, P = 6.8 × 10−4) to BMI.
Because DNAm age has a strong linear relationship with chronological age throughout tissues (11), we were able to define age acceleration as the residual resulting from a linear model that regressed DNAm age on chronological age. Thus, a liver that exhibits positive (negative) age acceleration appears to be older (younger) than expected. Interestingly, a significant age acceleration for BMI was only observed for liver (r = 0.42, P = 0.00068) (Fig. 1E), yielding ∼2.2 y of additional DNAm age for each 10 BMI units (kg/m2) in the linear model (Table 2). The correlation between BMI and epigenetic age acceleration in blood is negative and insignificant in two of three datasets [r = −0.066, P = 0.58 (Fig. 1H); r = 0.26, P = 0.012 (Fig. S1C), and r = −0.18, P = 0.12 (Fig. S1D)]. These negative results in blood echo those by Hannum et al. (17), who applied an alternative epigenetic biomarker of aging to a large (n = 656) blood methylation dataset. Our study demonstrates that it is critical to look at the appropriate tissue when it comes to detecting an increase of epigenetic age as a result of high BMI.
Table 2.
Estimation of the influence of BMI on liver DNAm age
| Variable | Discovery set | Replication set | Combined dataset | |||
| Estimate (SE) | P | Estimate (SE) | P | Estimate (SE) | P | |
| Chronological age | 0.77503 (0.04593) | <2 ×10−16 | 0.53518 (0.06043) | 2.5 ×10−13 | 0.62987 (0.04472) | <2 ×10−16 |
| BMI | 0.17352 (0.04616) | 3.9 ×10−4 | 0.23578 (0.06827) | 9.1 ×10−4 | 0.16789 (0.04603) | 3.8 ×10−4 |
| R2 | 0.83 | 0.51 | 0.61 | |||
| Age acceleration for 10-point increase in BMI | 2.2 y | 4.4 y | 2.7 y | |||
In multivariate regression models of DNAm age, chronological age, and BMI remain the only significant predictors (see Table S1 for full models). BMI is a significant covariate of DNAm age in both the discovery and replication dataset. The table reports estimates of the regression coefficients and corresponding SEs, Wald test P values. The last row reports the age acceleration associated with a 10-point increase in BMI. For example, a 10-point increase in BMI is associated with an increase of 2.2 y (= 0.17352 × 10/0.77503) in DNAm age in the discovery set.
Given the lack of significant correlations in the nonliver tissues, it is important to analyze the effect sizes (correlations) that could have been detected in these datasets. A sample size of n = 648 (adipose dataset 2) provides 87% power to reject the null hypothesis (correlation = 0) at a significance level of 0.05 if the absolute value of the true correlation coefficient |r| is larger than 0.12. A sample size of 71 (blood dataset 4) provides 81% power to detect a significant correlation if |r| is larger than 0.33. We briefly mention that BMI correlates with individual CpGs in blood and adipose tissue (18).
Replication and Validation of BMI-Related Epigenetic Age Acceleration in Liver.
To replicate the BMI-related epigenetic age acceleration in liver, we analyzed a second dataset (n = 79) of samples. The correlation of BMI with age acceleration was confirmed, yielding a similar effect size of r = 0.42 (P = 1.2 × 10−4) (Fig. 2 E and F) compared with the discovery dataset. The correlation between age acceleration and BMI (r = 0.42 in the discovery and r = 0.42 in the replication datasets) is unexpectedly high in comparison with the previously reported correlations (r = 0.12) between BMI and telomere length in blood (4). We think it unlikely that changes in telomere length could explain our results in liver because telomere length only has a weak negative correlation with DNAm age after correcting for chronological age (e.g., r = −0.28, P = 0.22 in adipose dataset 2). For a subset of the individuals from the liver datasets (discovery and replication combined), corresponding muscle (n = 26) and adipose tissue (n = 32) samples were available for analysis. In both nonliver tissues, no significant evidence of BMI-related DNAm age acceleration was observed (P = 0.32 and 0.68 for adipose and muscle tissue, respectively), thereby further supporting the liver-specificity of the age acceleration finding (Fig. 2 G and H).
Fig. 2.
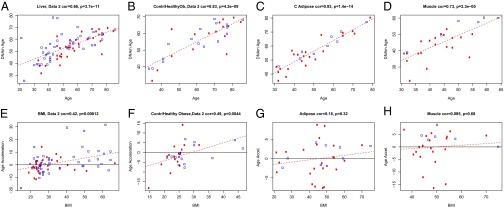
Replication analysis. Independent liver (A, B, E, F), adipose tissue (C and G), and muscle (D and H) datasets were analyzed for a correlation of chronological age and DNAm age and age acceleration to BMI. The data confirm the correlation of DNAm age acceleration and BMI in liver tissue (E and F), even if the analysis is restricted to individuals without histological evidence of NAFLD (i.e., controls and healthy obese subjects) (F) and the lack of this correlation in adipose and muscle tissue (G and H).
Post hoc analyses were performed in the combined liver datasets: First, the relation between BMI and age acceleration in the combined dataset was estimated as an increase of 2.7 y for each 10 BMI units in the linear model (Table 2). Second, BMI-related age acceleration was consistently observed both in women and in men and when restricting the analysis to one liver dataset at a time (Fig. S2 G, H, O, and P).
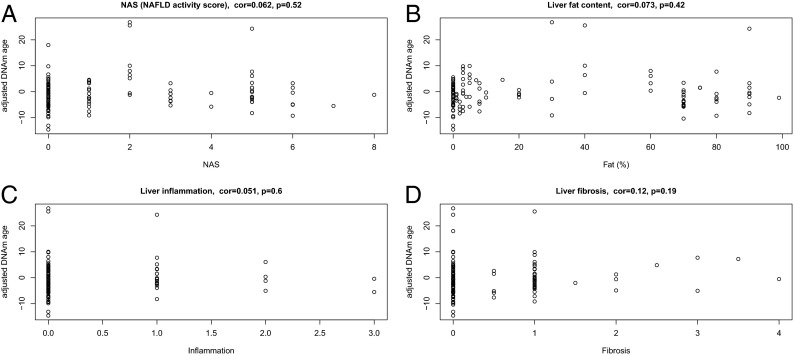
Increased BMI is the primary risk factor for nonalcoholic fatty liver disease (NAFLD), which ranges from simple liver steatosis to nonalcoholic steatohepatitis (NASH). Thus, further post hoc analyses using the combined liver datasets investigated whether the observed epigenetic age acceleration appears to be genuinely related to BMI or is primarily a marker of NAFLD. These analyses were enabled by the availability of a standardized histological scoring of all liver samples by a single pathologist. The following lines of evidence demonstrate that the observed age acceleration in liver is not mediated by NAFLD or NASH: First, when patients with histological evidence of NAFLD were excluded from the analysis (i.e., when the analysis was restricted to n = 32 controls and healthy obese subjects), BMI had a similar correlation with age acceleration (r = 0.49, P = 0.0044) (Fig. 2H) as in the analysis that included NAFLD subjects (r = 0.42, P = 1.2 × 10−4) (Fig. 2G). Second, we find that the detected age acceleration in liver is not associated with the NAFLD Activity Score (NAS) (19) or any of its component traits (steatosis, hepatocyte ballooning, inflammation) or fibrosis (Fig. 3 and Figs. S3 and S4). Third, a multivariate linear regression model of DNAm age on various covariates reveals that BMI remains a highly significant covariate even after adjusting for chronological, NAS component traits (steatosis, ballooning, inflammation), fat percentage, and fibrosis (Table S1).
Fig. 3.
Post hoc analyses of subgroups and histological characteristics of NASH. An adjusted measure of DNAm age is related to various measures of liver pathology. The adjusted measure of DNAm age acceleration was defined as residual from a regression model that regressed DNAm age on chronological age+BMI+sex. Note that this adjusted measure of age acceleration does not relate to (A) NAS, (B) fat percentage (steatosis), (C) inflammation, and (D) fibrosis. Each scatterplot reports the Pearson correlation coefficient and P value. Analogous results can also be found in the individual liver datasets (Figs. S3 and S4).
Other well-known risk factors of liver disease are unlikely to confound the relationship between BMI and age acceleration because subjects with evidence of viral hepatitis, hemochromatosis, or high alcohol consumption (greater than 20 g/d for women and 30 g/d for men) were excluded from this study. We do not find a significant association between type II diabetes status and age acceleration (P = 0.16) (Fig. S5 A and C), but these results should be interpreted with caution because a two-group comparison based on 23 diabetes cases and 103 controls only achieves sufficient statistical power (80% at a significance level of 0.05) for detecting moderate to large effects (mean differences of 0.7 SDs).
Furthermore, it is unlikely that smoking confounds the relationship between BMI and age acceleration because (i) we did not observe a significant relationship between BMI and smoking status in our liver data sets, and (ii) smoking status was not significantly related with age acceleration in liver, muscle, or blood tissue (Fig. S6).
Weight Loss Does Not Reverse Epigenetic Age Acceleration in the Short Term.
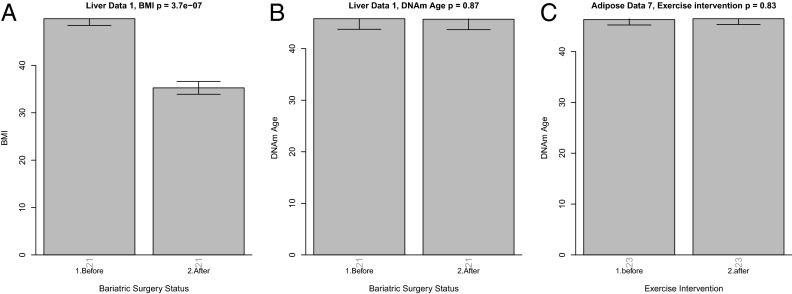
Bariatric surgery induces a profound weight loss and reverses a number of metabolic abnormalities associated with increased BMI. For 21 subjects from the combined liver datasets, liver tissue samples before and after bariatric surgery (collected within a 9-mo period after surgery) were available. As expected, bariatric surgery led to a significant decrease in BMI (ΔBMI = 14.6, P = 3.7 × 10−7) (Fig. 4A). However, bariatric surgery and rapid weight loss does not reverse the DNAm age in liver tissue within a 9-mo period (Fig. 4B).
Fig. 4.
Effect of weight loss and exercise intervention on DNAm age. For 21 subjects, liver methylation data were available before and after bariatric surgery. As expected, BMI drops significantly within 6–9 mo following bariatric surgery (A). However, the DNAm age of the liver tissue is unaffected (B). (C) DNAm age of adipose tissue is unaffected by a 6-mo exercise intervention (20).
Rönn et al. (20) studied whether a 6-mo physical exercise intervention affected DNA methylation levels in human adipose tissue. Although this exercise intervention did not lower the BMI of the subjects, the authors reported that 17,975 individual CpG sites on the Illumina 450K array showed altered DNAm levels. Using these DNAm data, we did not observe an effect of the exercise intervention on the DNAm age of adipose tissue (Fig. 4C).
Transcriptional Studies of Age Acceleration in Liver Tissue.
Using our gene-expression data from liver tissue, we identified 279 genes whose mRNA levels had a significant negative correlation with age acceleration at an uncorrected correlation test P-value threshold of 0.005 (corresponding to a false-discovery rate of 0.10). These 279 negatively related genes tend to be overexpressed in liver samples that do not exhibit age acceleration effects. Similarly, we identified 378 genes with a positive correlation with age acceleration in liver (i.e., these genes tend to be overexpressed in liver samples with a significant age acceleration effect). The results of a functional enrichment analysis with the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) (21) can be found in Table 3. The 279 negatively related genes are highly enriched with nuclear mitochondrial genes, which play a role in oxidative phosphorylation and electron transport. This finding recapitulates a previously observed conserved molecular signature of aging in which genes associated with these functions tend to be strongly underexpressed (22). The 378 positively related genes are known to play a role in nucleoside-triphosphatase regulator activity, cell adhesion, GTPase regulator activity, and response to wounding. Future mechanistic studies will be needed to dissect cause-and-effect relationships.
Table 3.
Functional enrichment of gene transcripts associated with age acceleration
| Category | Term | n | FE | P Bonferroni |
| 279 negatively correlated genes | ||||
| Cell Comp | Mitochondrion | 41 | 2.9 | 3.0 ×10−7 |
| KEGG | Oxidative phosphorylation | 11 | 6.5 | 3.7 ×10−4 |
| SP_PIR | Electron transport | 10 | 7.8 | 1.6 ×10−3 |
| 378 positively correlated genes | ||||
| Mol. Fnc. | Nucleoside-triphosphatase regulator activity | 27 | 3.2 | 2.1 ×10−4 |
| Biol. Proc. | Cell adhesion | 36 | 2.6 | 7.7 ×10−4 |
| Mol. Fnc. | GTPase regulator activity | 26 | 3.1 | 4.8 ×10−4 |
| Biol. Proc. | Response to wounding | 29 | 2.8 | 4.2 ×10−3 |
| SP_PIR | Guanine-nucleotide releasing factor | 13 | 5.7 | 1.0 ×10−3 |
| SP_PIR | Autophosphorylation | 9 | 9.8 | 1.2 ×10−3 |
| Biol. Proc. | Wound healing | 16 | 4.2 | 1.1 ×10−2 |
| Mol. Fnc. | GTPase binding | 12 | 5.3 | 7.5 ×10−3 |
| Biol. Pro. | Cell activation | 19 | 3.3 | 3.1 ×10−2 |
The upper part of the table reports the results from applying DAVID EASE to 279 genes whose expression levels had a significant negative correlation with age acceleration in liver. Similarly, the lower part reports the result for the 378 positively related genes. The table reports the gene ontology (GO) enrichment categories, number of genes (n), fold-enrichment (FE), and Bonferroni-corrected P value. Biol. Proc., biological process; Mol. Fnc., molecular function; SP_PIR, spliceosome protein–protein interaction resource.
Structural Equation Modeling Analysis of BMI, Age Acceleration, and Liver Traits.
Using a measure of age acceleration similar to that used in Fig. 1 (i.e., age acceleration is defined as residual from a regression model of DNAm age against chronological age in the combined dataset), we find that many liver traits correlate with age acceleration (Fig. S7). However, these significant relationships probably reflect confounding caused by BMI, as can be seen from a structural equation model analysis (SI Text and Table S2): an independence model (model 3: trait←BMI→age acceleration), which postulates that high BMI leads to liver pathology and age acceleration independently, achieves the best fit for the NAS score, hepatocyte ballooning, and liver inflammation. However, for steatosis, the reactive model (BMI→steatosis→age acceleration) provides a better fit. The results for fibrosis are ambiguous.
We caution the reader that structural equation models make various assumptions (SI Text) that may not be satisfied by our observational data. As the study design is primarily observational, we cannot establish causality but provide this as a speculative model.
Discussion
Aging of tissues is a complex, dynamic, and multifaceted process that is currently still poorly understood (23) because its investigation depends critically on the use of model organisms, and integration of findings with human physiology remains a challenge. Tissue aging has up until now predominantly been defined in single-tissue and organ studies, thus inhibiting global analyses of epidemiological factors and medical interventions. Genome-wide DNAm-based epigenetic analysis may provide a possible solution to this problem, as (i) epigenetic restructuring of the genome has been shown to play a key role in aging as demonstrated by altered global methylation levels, histone modifications and CpG methylation, and (ii) specific, age-related alterations in DNAm have been reported previously.
Many groups have reported sets of CpGs that correlate with age in multiple tissues (15, 24–28). Although these reports firmly establish the strong effect of age on epigenetic modifications, individual CpG sites are unsuitable for global comparisons between tissues in the context of epidemiological or interventional analysis, because the underlying methylation sites are highly tissue-specific. Thus, in this report, we use an aggregate measure of DNAm age that has been validated across sorted cell types (CD4 T cells, monocytes, B cells, glial cells, neurons), complex tissues, and organs (e.g., brain, breast, kidney, liver, lung) (11). This biomarker enabled (as far as we are aware) the first analysis of the relation between obesity and the epigenetic ages of different human tissues. Although the epigenetic clock stands out because of its high correlation with chronological age across most tissues (11), this biomarker of aging has a major limitation: it is not yet understood what it measures. Whereas many articles suggest that age-related changes in DNAm levels represent noise or epigenetic drift (29), DNAm age might measure the cumulative work of an epigenomic maintenance system (11). According to the gene-enrichment signature of epigenetic age acceleration in liver (Table 3), age acceleration relates to processes, such as oxidative stress and energy metabolism. Although this epigenomic maintenance system is as yet poorly defined, the continued nutritionally driven oxidative stress and metabolic pressure might point to the mechanisms leading to the liver-specific increase of DNAm age. Our data show that high BMI relates to increased DNAm age in a tissue-specific manner. Although BMI is widely used, it is a somewhat suboptimal measure of adiposity because it is correlated both with obesity and muscle mass. Thus, a possible association of DNAm age and muscle mass should be addressed in future studies. Examining the association between epigenetic age acceleration and tissue pathology may provide an important biological link between obesity and clinical manifestations of accelerated aging. The increased age of liver tissue in obese individuals may provide insights to liver-related comorbidities of obesity, such as insulin resistance and hepatocellular carcinoma (HCC).
Liver protein synthesis function, as measured on an organismal level, is quite plastic and not impaired in obesity in the absence of histopathological cirrhosis. However, even in the absence of structural liver disease, and in mild forms (such as early steatosis), the clinically relevant phenomena in these patients are insulin resistance (with its hepatic component very poorly understood) and an increased risk of HCC as the most tangible manifestations of obesity. Aging and cancer are not only intricately linked to each other (30, 31) but also to obesity and metabolic syndrome (32). Obesity is an established risk factor for many types of cancers, particularly for HCC, because of its carcinogenic potential and the association with NAFLD (33). In cohort studies in men, metabolic syndrome was most strongly associated with liver cancer followed by colorectal cancer (34). Future studies will be needed to test whether tissues with increased DNAm age have a lower threshold to carcinogenesis. Although epigenetic aging may help understand carcinogenesis, it may also have practical consequences. NASH is a risk factor for liver cancer even in the absence of cirrhosis (32) and is thus recommended as a high-risk group for screening in the current American Association for the Study of Liver Diseases guidelines (35). Future HCC screening recommendations may be influenced by our findings because BMI-induced increased tissue age (i) is independent of the presence of overt NASH and (ii) is irreversible, at least in the short term.
The increased epigenetic age of liver tissue in obese individuals should provide insights into common liver-related comorbidities of obesity, such as insulin resistance and liver cancer. These findings support the hypothesis that obesity is associated with accelerated aging effects (3) and stresses once more the importance of maintaining a healthy weight.
Methods
A detailed description of the datasets and statistical methods can be found in SI Text. All data presented in this article are available in public data repositories. Accession numbers are presented in Table 1.
Public DNAm Datasets.
All genome-wide human methylation datasets with BMI, age, and sex information publicly available at the time of analysis were used (datasets 1–6). Such datasets were available from liver, adipose tissue, muscle, and blood (Table 1). Postbariatric patients were excluded from the liver discovery set analysis.
Novel Replication Datasets: Samples.
Liver samples were collected from German patients (all Caucasians) under the same protocol and ethics approval as reported previously (12). In brief, liver samples were obtained percutaneously for patients undergoing liver biopsy for suspected NAFLD or intraoperatively for assessment of liver histology. Normal control samples were recruited from samples obtained for exclusion of liver malignancy during major oncological surgery. A percutaneous follow-up biopsy was obtained in consenting bariatric patients 5–9 mo after surgery. All patients provided written, informed consent. The study protocol was approved by the institutional review board (“Ethics commission of the Medical Faculty, University of Kiel” D425/07, A111/99) before the commencement of the study (SI Text). During surgery, in addition to liver tissue, subcutaneous fat and muscle tissue were obtained (Table 1, datasets 7–9) in a subset of patients. A standardized histological scoring of all liver samples by the same single pathologist (C.R.) according to the NAS score (19), who performed the histological scoring of the discovery dataset, thereby allowing pooling during post hoc analyses.
Ideally, one would want to carry out longitudinal studies of liver function to understand how liver function and DNAm changes with age and predicts functional change. The patients biopsied in our study are in a long-term follow up program and may help to provide such answers, which we regrettably cannot provide at this point.
Preprocessing of DNAm Data.
Samples were processed as reported previously (12). In brief, bisulfite conversion using the Zymo EZ DNA Methylation Kit (ZymoResearch), as well as subsequent hybridization of the HumanMethylation450K Bead Chip (Illumina) and scanning (iScan, Illumina), were performed according to the manufacturers protocols by applying standard settings. DNA methylation levels (β-values) were determined by calculating the ratio of intensities between methylated (signal A) and unmethylated (signal B) alleles. Specifically, the β-value was calculated from the intensity of the methylated (M corresponding to signal A) and unmethylated (U corresponding to signal B) alleles, as the ratio of fluorescent signals β = Max(M,0)/[Max(M,0) + Max(U,0) + 100]. Thus, β-values range from 0 (completely unmethylated) to 1 (completely methylated) (36).
Transcriptomic Data.
Novel liver messenger RNA expression datasets from the German patients were generated on the HuGene 1.1 ST gene array (Affymetrix) (Dataset S1).
Statistical Analysis.
Epigenetic age was calculated as reported previously. The epigenetic clock is defined as a prediction method of age based on the DNAm levels of 353 CpGs. Predicted age, referred to as DNAm age, correlates with chronological age in sorted cell types (CD4 T cells, monocytes, B cells, glial cells, neurons) and tissues and organs, including whole blood, brain, breast, kidney, liver, lung, saliva (11). Mathematical details and software tutorials for the epigenetic clock can be found in the supplemental files of ref. 11. Many authors have described methods for dealing with the two types of probes found on the Illumina 450K array (37–39). This is not a concern for the epigenetic clock because it only involves type II probes. However, our software implements a data normalization step that repurposes the BMIQ normalization method from Teschendorff et al. (38) so that it automatically references each sample to a gold standard based on type II probes (details can be found in supplemental file 2 in ref. 11).
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (NIA/NIH 5R01AG042511-02), the German ministry of education and research (BmBF) through the Virtual Liver Network (VLN, #0315763), and through institutional funds from the Christian-Albrechts-University Kiel and the University Hospitals Schleswig-Holstein and Dresden in Germany.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61256).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412759111/-/DCSupplemental.
References
- 1.Fontana L, Partridge L, Longo VD. Extending healthy life span—From yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haslam DW, James WPT. Obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 3.Valdes AM, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366(9486):662–664. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 4.Nordfjäll K, et al. Telomere length is associated with obesity parameters but with a gender difference. Obesity (Silver Spring) 2008;16(12):2682–2689. doi: 10.1038/oby.2008.413. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, et al. Obesity and weight gain in adulthood and telomere length. Cancer Epidemiol Biomarkers Prev. 2009;18(3):816–820. doi: 10.1158/1055-9965.EPI-08-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M, Martin H, Firpo MA, Demerath EW. Inverse association between adiposity and telomere length: The Fels Longitudinal Study. Am J Hum Biol. 2011;23(1):100–106. doi: 10.1002/ajhb.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Calzón S, et al. Longitudinal association of telomere length and obesity indices in an intervention study with a Mediterranean diet: The PREDIMED-NAVARRA trial. Int J Obes (Lond) 2014;38(2):177–182. doi: 10.1038/ijo.2013.68. [DOI] [PubMed] [Google Scholar]
- 8.Campisi J. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Chen J-H, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007;35(22):7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10) R115:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahrens M, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Grundberg E, et al. Multiple Tissue Human Expression Resource Consortium Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93(5):876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zykovich A, et al. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014;13(2):360–366. doi: 10.1111/acel.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day K, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013;14(9):R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam LL, et al. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci USA. 2012;109(Suppl 2):17253–17260. doi: 10.1073/pnas.1121249109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick KJ, et al. DNA methylation and body-mass index: A genome-wide analysis. Lancet. 2014;383(9933):1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 19.Kleiner DE, et al. Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 20.Rönn T, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25(7):875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berdasco M, Esteller M. Hot topics in epigenetic mechanisms of aging: 2011. Aging Cell. 2012;11(2):181–186. doi: 10.1111/j.1474-9726.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez DG, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20(6):1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grönniger E, et al. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6(5):e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: Implications, and a case of epigenetic thrift? Hum Mol Genet. 2013;22(R1):R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448(7155):767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 31.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75(1):685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J Hepatol. 2012;56(6):1384–1391. doi: 10.1016/j.jhep.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 33.Vanni E, Bugianesi E. Obesity and liver cancer. Clin Liver Dis. 2014;18(1):191–203. doi: 10.1016/j.cld.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Esposito K, Chiodini P, Colao A, Lenzi A, Giugliano D. Metabolic syndrome and risk of cancer: A systematic review and meta-analysis. Diabetes Care. 2012;35(11):2402–2411. doi: 10.2337/dc12-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruix J, Sherman M. American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: An update. Hepatology. 2011;53(3):1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavaré S, Ritchie ME. Statistical issues in the analysis of Illumina data. BMC Bioinformatics. 2008;9(1):85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yousefi P, et al. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics. 2013;8(11):1141–1152. doi: 10.4161/epi.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.