Significance
Epigenetic inactivation of X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is frequently observed in multiple human malignancies. However, the mechanisms underlying its tumor-suppression function remain largely undefined. Here, we identify XAF1 as a positive feedback regulator of p53, which directs the apoptotic switch of p53 signaling. As a unique transcription target of p53 in signaling apoptosis, XAF1 acts as a competitor of E3 ubiquitin ligase MDM2 in binding to p53 and thus disrupts the p53–MDM2 regulatory loop. Moreover, XAF1 appears to promote homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation by interrupting HIPK2-targeting function of E3 ubiquitin ligase Siah2 and promotes zinc finger protein 313 (ZNF313)-induced p21WAF1 ubiquitination. XAF1 thus represents one critical regulator of p53’s cell-fate decision function, suggesting that restoring the p53–XAF1 feedback loop could be an attractive avenue for the therapeutic intervention of malignant tumor progression.
Keywords: XAF1, p53, HIPK2, Siah2, ZNF313
Abstract
X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is a tumor suppressor that is frequently inactivated in many human cancers. However, the molecular mechanism underlying its growth-inhibitory function remains largely unknown. Here, we report that XAF1 forms a positive feedback loop with p53 and acts as a molecular switch in p53-mediated cell-fate decisions favoring apoptosis over cell-cycle arrest. XAF1 binds directly to the N-terminal proline-rich domain of p53 and thus interferes with E3 ubiquitin ligase MDM2 binding and ubiquitination of p53. XAF1 stimulates homeodomain-interacting protein kinase 2 (HIPK2)-mediated Ser-46 phosphorylation of p53 by blocking E3 ubiquitin ligase Siah2 interaction with and ubiquitination of HIPK2. XAF1 also steps up the termination of p53-mediated cell-cycle arrest by activating zinc finger protein 313 (ZNF313), a p21WAF1-targeting ubiquitin E3 ligase. XAF1 interacts with p53, Siah2, and ZNF313 through the zinc finger domains 5, 6, and 7, respectively, and truncated XAF1 isoforms preferentially expressed in cancer cells fail to form a feedback loop with p53. Together, this study uncovers a novel role for XAF1 in p53 stress response, adding a new layer of complexity to the mechanisms by which p53 determines cell-fate decisions.
The tumor suppressor p53 elicits a wide array of cytostatic or cytotoxic responses to intrinsic and exogenous stresses, resulting in cell-cycle arrest, DNA repair, apoptosis, or senescence (1). The choice of p53 between life and death is dictated by its ability to switch on particular subsets of genes, and multiple molecular mechanisms contribute to its target gene selectivity (2–4). This process entails stress-dependent and site-specific phosphorylation by several DNA-damage–activated protein kinases, including ataxia telangiectasia mutated (ATM) and ATM and Rad3-related, which ultimately determine the DNA-binding specificity of p53 (5). In particular, phosphorylation at Ser-46 by homeodomain-interacting protein kinase 2 (HIPK2) is a specific modification that can induce changes in p53 affinity for preferential induction of proapoptotic target promoters (6–8). The p53 pathway is highly connected to several signaling systems and tightly controlled by a large variety of negative- and positive-feedback loops (9).
X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is a tumor suppressor gene, which encodes 33.1-kDa protein with seven zinc fingers (10, 11). Loss or reduction of XAF1 expression due to aberrant promoter hypermethylation is associated with the advanced stage and high grade of many cancers (12–14). A recent study demonstrated that PTEN-null mouse prostate tumors showing resistance to androgen-deprivation therapy have reduced levels of XAF1, and its reduction is associated with recurrence and metastasis in human samples (15). Among at least five distinct XAF1 transcripts (XAF1A–E) expressed in normal tissues, the full-length transcript (XAF1A) is preferentially inactivated in human tumors, whereas truncated short isoforms are rather increased, suggesting that these variants may function differentially or elicit a dominant-negative action (16, 17). XAF1 was originally identified as a nuclear protein that could bind and interfere with anticaspase function of XIAP by sequestering XIAP protein to the nucleus (11). It was thus proposed that loss of XAF1 may increase the functional pool of cytoplasmic XIAP, which in turn deregulates the apoptotic process and contributes to tumor progression (18, 19). However, XAF1 has been shown to evoke an apoptotic effect in XIAP−/− cells to the extent comparable in XIAP+/+ cells, indicating that its function is not solely dependent on the XIAP-interfering activity (14, 20). XAF1 was also identified as an IFN-stimulated gene that contributes to IFN-dependent sensitization of cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis (21–23). Despite previous reports showing the XAF1 implication in p53-mediated apoptosis, the molecular basis for the interplay between p53 and XAF1 and its significance in tumorigenesis has not been understood (13, 24).
In this study, we present evidence that XAF1 is a feedback activator of p53 that plays a crucial role in p53-mediated cell-fate decisions through the modulation of the E3 ubiquitin ligases MDM2, Siah2, and ZNF313.
Results
A p53–XAF1 Loop Drives Apoptotic Switch of p53 Signaling.
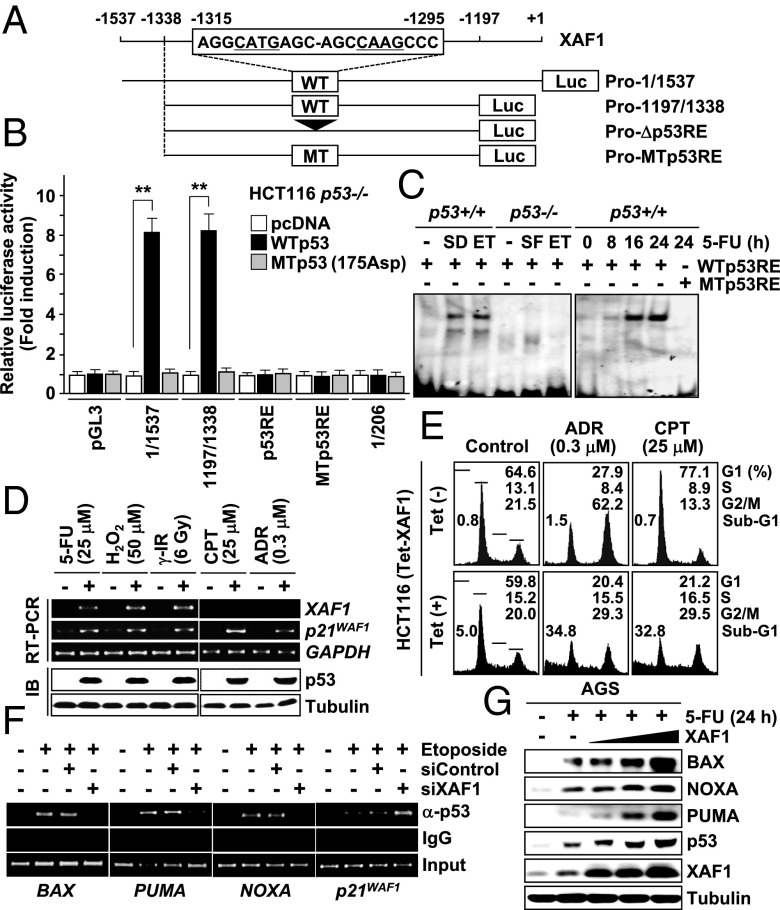
To understand the p53–XAF1 interplay in stress response, we examined whether XAF1 is regulated by p53. XAF1A (hereafter referred to as XAF1) expression was activated by restoration of wild-type (WT) p53 in p53-null Calu-1 cells (Fig. S1A). Both mRNA and protein levels of XAF1 were greatly elevated in WTp53 but not p53-deficient cells exposed to etoposide, 5-fluorouracil (5-FU), γ-irradiation (IR), H2O2, and hypoxic stress while this induction was blocked by pretreatment with the p53 inhibitor pifithrin-α or siRNA-mediated p53 depletion (Fig. S1 B–D). A putative p53 response element (p53RE) was identified in the 5′ upstream region (nucleotides −1,315/−1,295 relative to ATG) (Fig. 1A). The reporters containing this p53RE showed an approximately eightfold increase in luciferase activity in response to p53 transfection, whereas reporters omitting the p53RE (ProΔp53RE) or containing a mutated p53RE (Pro–MTp53RE) failed to respond to p53 (Fig. 1B and Fig. S1E). Electrophoretic mobility-shift assay (EMSA) and chromatin immunoprecipitation (ChIP) assay revealed that the p53RE is occupied by endogenous p53 within living cells (Fig. 1C and Fig. S1 F and G). Recently, p53 was reported to repress XAF1 transcription through the interaction with the +5 to +157 region of the gene (24). However, we could not detect this interaction and repression (Fig. 1B and Fig. S1H). In AGS cells exposed to etoposide, induction of XAF1 mRNA was recognized at ∼12 h posttreatment, later than p53 proarrest targets (p21WAF1 and p53R2) but slightly ahead of proapoptotic targets, such as Bcl2-associated X protein (BAX), phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1/NOXA), and p53 upregulated modulator of apoptosis (PUMA) (Fig. S1I). We next sought whether XAF1 affects outcomes of p53 activation using HCT116 cells that manifest predominantly cytostatic response to adriamycin (ADR) and cisplatin (CPT). XAF1 was induced by cytotoxic damage, such as 5-FU, H2O2, and γ-IR, but not affected by ADR and CPT, whereas p53 and p21WAF1 were activated by both types of stresses (Fig. 1D and Fig. S1 J and K). However, the cellular response to ADR and CPT was switched to apoptosis when XAF1 is induced using a tetracyclin-inducible system (Tet–XAF1) (Fig. 1E and Fig. S1 L and M). Although XAF1 depletion led to a significant reduction of p53-induced apoptosis, its expression accelerated p53 activation of BAX, PUMA, and NOXA promoters but debilitated p53 activation of the p21WAF1 promoter, indicating that XAF1 provokes opposite effects on p53 transactivation of proapoptotic and proarrest targets (Fig. 1F, Table S1, and Fig. S2 A–D). It was also found that XAF1 expression stimulates p53 accumulation in stressed cells (Fig. 1G and Fig. S2E). Together, these results indicate that XAF1 is a bona fide transcription target of p53, which directs apoptotic switch of p53 signaling through the feedback loop formation with p53.
Fig. 1.
Identification of a p53–XAF1 feedback loop. (A) A p53RE in the XAF1 promoter and reporter construction for luciferase assay. (B) p53 activation of the XAF1 promoter. Data represent the mean ± SD. **P < 0.01 (Student t test). (C) EMSA for p53 interaction with the p53RE. SD, serum deprivation (48 h); ET, etoposide (50 μM, 48 h). (D) Cytotoxic damage-specific XAF1 induction in HCT116 cells. IB, immunoblot. (E) Effect of XAF1 on HCT116 cell response to cytostatic damages. (F) Effect of XAF1 on p53 binding to its target promoters in HCT116 treated with etoposide (50 μM, 24 h). (G) XAF1 activation of p53 and its proapoptotic targets.
XAF1 Stabilizes p53 Through Direct Binding to the N-Terminal Proline-Rich Domain.
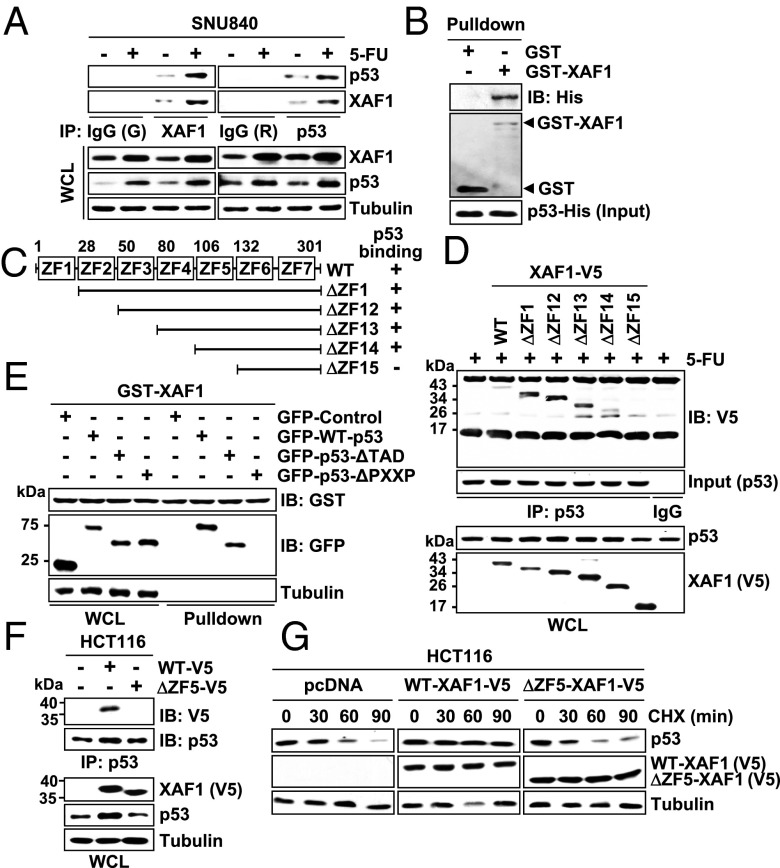
To understand the mechanistic basis for the XAF1 activation of p53, we tested whether XAF1 binds to p53. Immunofluorescence assays showed that XAF1 proteins are distributed both in the cytoplasm and the nucleus and colocalized with nuclear p53 (Fig. S3 A and B). Immunoprecipitation and pull-down assays also revealed that XAF1 interacts directly with p53 (Fig. 2 A and B and Fig. S3 C and D). Using a series of deletion mutants, we identified that the zinc finger domain 5 (ZF5; amino acids 106–132) of XAF1 binds to the N-terminal PXXP repeats-containing proline-rich domain (amino acids 62–92) of p53 (Fig. 2 C–E and Fig. S3 E–G). As predicted, a mutant XAF1 lacking the ZF5 domain (ΔZF5–XAF1) failed to interact with and stabilize p53 (Fig. 2 F and G). Compared with WT–XAF1, ΔZF5–XAF1 evoked markedly low apoptosis-promoting effect in stressed cells (Fig. S3 H and I).
Fig. 2.
XAF1 binds directly to p53. (A) Immunoprecipitation assay of the XAF1–p53 interaction in SNU480 cells. G, goat; R, rabbit; WCL, whole-cell lysate. (B) GST pull-down assay for the XAF1–p53 interaction. (C) XAF1 constructs and its p53-binding status. (D) Identification of the ZF5 domain as a p53-binding region of XAF1 in HCT116 cells. (E) A crucial role for p53 PXXP domain in binding to XAF1. (F) Loss of p53-binding activity of XAF1 by deletion of the ZF5 domain. (G) CHX chase experiment showing the loss of p53-stabilizing activity of ΔZF5–XAF1.
XAF1 Interferes with MDM2 Binding and Ubiquitination of p53.
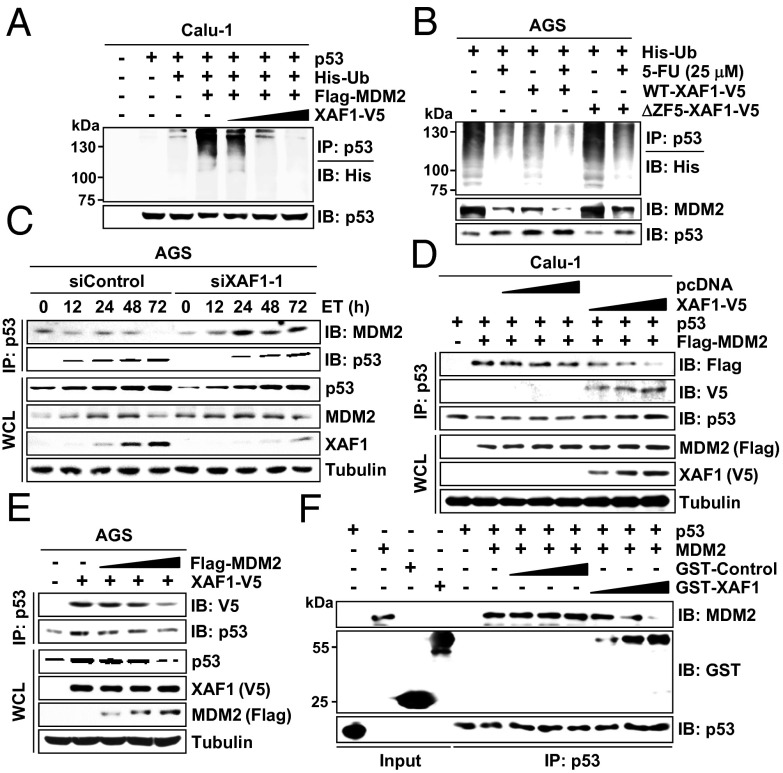
Given that MDM2, a major p53-targeting ubiquitin E3 ligase, binds to the N terminus of p53, we asked whether XAF1 competes with MDM2 in binding to p53 and consequently antagonizes p53-targeting activity of MDM2. p53 ubiquitination was markedly increased in XAF1-depleted cells, and MDM2-induced p53 ubiquitination was suppressed by XAF1 expression in a dose-dependent manner (Fig. 3A and Fig. S4 A–C). XAF1 did not interact with MDM2 but impaired the MDM2–p53 interaction (Fig. S4D). Unlike WT–XAF1, ΔZF5–XAF1 failed to inhibit MDM2 interaction with and ubiquitination of p53, indicating that XAF1 interferes with the MDM2–p53 interaction through its p53-binding property (Fig. 3B and Fig. S4E). Consistently, p53–MDM2 interaction was up- and down-regulated by XAF1 depletion and expression, respectively, whereas p53–XAF1 interaction was suppressed by MDM2 expression in a dose-associated manner (Fig. 3 C–E and Fig. S4F). Furthermore, an in vitro binding assay using recombinant p53, MDM2, and GST–XAF1 proteins revealed that XAF1–p53 interaction blocks the p53–MDM2 interaction (Fig. 3F). These data indicate that XAF1 antagonizes the p53-targeting activity of MDM2 by hindering its interaction with p53.
Fig. 3.
XAF1 protects p53 from MDM2-induced ubiquitination. (A) XAF1 inhibition of MDM2-induced p53 ubiquitination. (B) Loss of the p53 ubiquitination-protecting function of XAF1 by deletion of the ZF5 domain. (C) Increased p53–MDM2 interaction in XAF1-depleted cells. (D) XAF1 inhibition of the MDM2–p53 interaction. (E) MDM2 inhibition of the XAF1–p53 interaction. (F) In vitro binding assay showing the competition of XAF1 and MDM2 in the interaction with p53.
XAF1 Stimulates Ser-46 Phosphorylation of p53 via HIPK2 Activation.
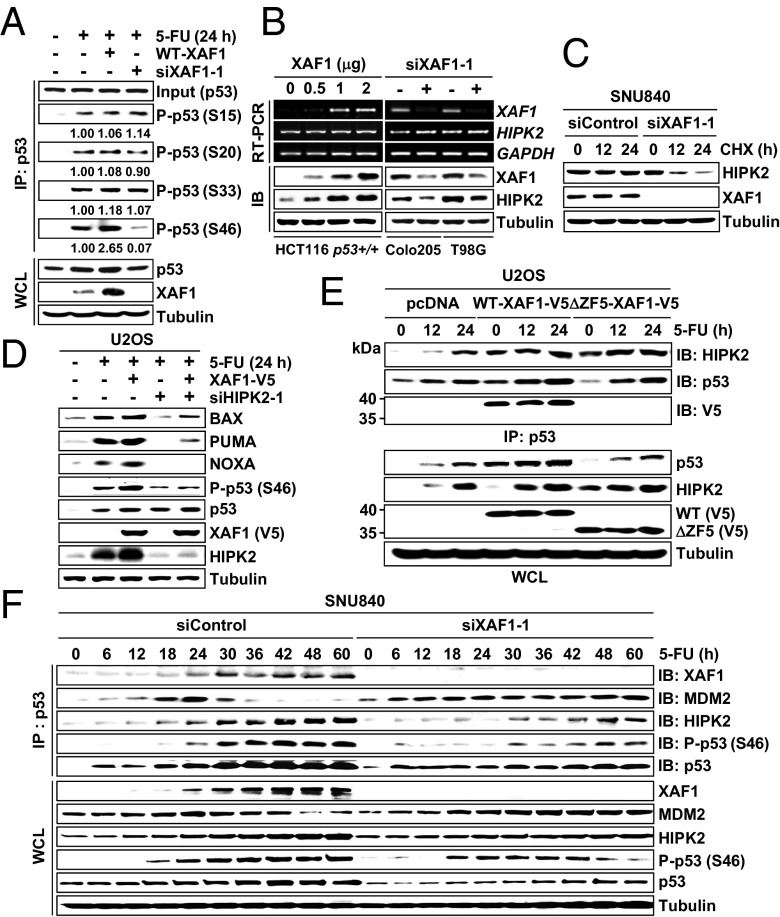
Next, we investigated whether XAF1 influences posttranslational modification of p53 to promote apoptosis. XAF1 induction in HCT116 (Tet–XAF1) cells led to an overall increment of phosphorylated or acetylated p53 levels (Fig. S5A). To exclude the effect of total p53 increase, we performed immunoprecipitation using equal input of p53 proteins and found that phosphorylation on Ser-46 residue is specifically increased by XAF1 (Fig. 4A). Moreover, XAF1 failed to activate the BAX–Luc promoter in Calu-1 cells expressing Δ46–p53 (a mutant p53 lacking the codon 46), indicating that XAF1 activation of p53 signaling is linked to its property to stimulate Ser-46 phosphorylation (Fig. S5B). The serine/threonine kinase HIPK2 has been known to play a key role for Ser-46 phosphorylation of p53 (6–8). Intriguingly, we found that XAF1 increases HIPK2 by enhancing its protein stability and that XAF1 activation of p53 Ser-46 phosphorylation, target promoters, and apoptosis is considerably debilitated in HIPK2-depleted cells (Fig. 4 B–D and Fig. S5 C and D). In a microscopic assay of HCT116 cells, some of the nuclear XAF1 proteins appeared to colocalize with HIPK2 (Fig. S5E). However, XAF1 was coprecipitated with HIPK2 in p53+/+ but not in p53−/− cells, and ΔZF5–XAF1 failed to interact with HIPK2 in WTp53 cells, indicating that XAF1 and HIPK2 are brought to protein complex through association with a common partner p53 (Fig. 4E and Fig. S5 F and G). Furthermore, ΔZF5–XAF1 was shown to retain the HIPK2-stabilizing activity and evoke HIPK2-dependent apoptotic effect, indicating that XAF1 activates HIPK2 in a p53-independent fashion (Fig. 4E and Fig. S5H). We finally elicited the role of stress-induced XAF1 in HIPK2 regulation of p53. In 5-FU–exposed SNU840 cells, the XAF1–p53 interaction was apparently increased at ∼18 h after treatment, which was accompanied by elevated HIPK2–p53 interaction and p53 Ser-46 phosphorylation (Fig. 4F and Fig. S5I). However, when XAF1 induction was blocked, the HIPK2–p53 interaction and p53 Ser-46 phosphorylation were markedly attenuated, whereas the MDM2–p53 interaction was drastically increased.
Fig. 4.
XAF1 stimulates HIPK2-mediated p53 phosphorylation. (A) XAF1 induction of p53 Ser-46 phosphorylation in HCT116 cells. P-p53, phosphorylated p53; S, serine. Numbers indicate relative levels of P-p53. (B) XAF1 induction of HIPK2 protein level. (C) Reduction of HIPK2 stability by XAF1 depletion. (D) HIPK2-dependent activation of p53 by XAF1. (E) Comparison of WT– and ΔZF5–XAF1 effect on the HIPK2–p53 interaction. (F) Effect of XAF1 depletion on the HIPK2–p53 interaction and p53 Ser-46 phosphorylation in 5-FU–exposed cells.
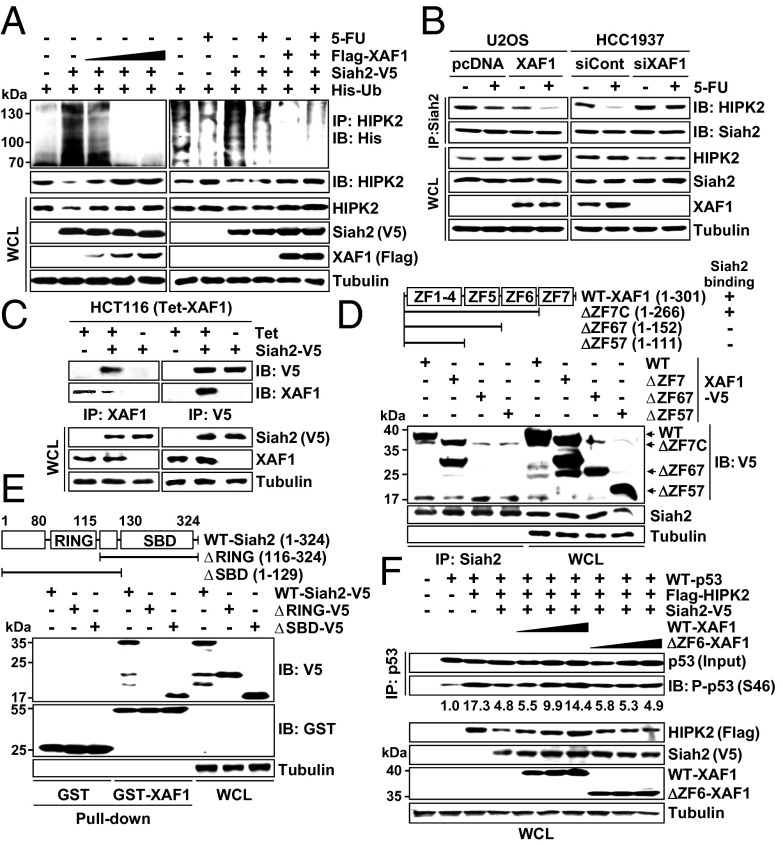
XAF1 Protects HIPK2 Ubiquitination by Antagonizing Siah2 E3 Ligase Activity.
To elicit the mechanism underlying HIPK2 induction by XAF1, we asked whether XAF1 regulates HIPK2-destabilizing factors. Among six known HIPK2 antagonists (Siah1, Siah2, SENP1, HMGA1, WSB1, and MDM2) we tested, Siah1 and Siah2 showed detectable HIPK2-reducing effect, and XAF1 suppressed Siah2-mediated HIPK2 reduction, although it did not affect expression of all six antagonists (Fig. S6 A–C). XAF1 blocked Siah2-mediated HIPK2 ubiquitination and also disrupted its inhibitory effect on HIPK2-mediated p53 phosphorylation (Fig. 5A and Fig. S6D). Consistently, Siah2-bound HIPK2 level was down- and up-regulated by XAF1 expression and depletion, respectively (Fig. 5B). Furthermore, coimmunoprecipitation and pull-down assays revealed that the ZF6 domain of XAF1 binds directly to the really interesting new gene (RING) domain of Siah2 (Fig. 5 C–E). The XAF1–Siah2 interaction was supported by our finding of Siah2 as one of XAF1-interacting proteins in a yeast two-hybrid assay (Fig. S6E). Unlike WT–XAF1, ΔZF6–XAF1 failed to suppress Siah2–HIKP2 interaction, protect HIPK2 from Siah2-induced degradation, and stimulate p53 Ser-46 phosphorylation (Fig. 5F and Fig. S6 F and G). As predicted, ΔZF6–XAF1 failed to rescue the p53-activating function of HIPK2 suppressed by Siah2 (Fig. 5F and Fig. S6 H and I). These results indicate that XAF1 activates the HIPK2–p53 axis by blocking the Siah2–HIPK2 interaction.
Fig. 5.
XAF1 suppresses HIPK2-targeting function of Siah2. (A) XAF1 inhibition of Siah2-mediated HIPK2 ubiquitination in HCT116 cells treated with 5-FU (25 μM, 24 h). (B) XAF1 inhibition of Siah2 interaction with HIPK2. (C) Immunoprecipitation assay showing XAF1 interaction with Siah2. (D and E) Identification of the ZF6 domain as a Siah2-binding region of XAF1 and the RING domain as a XAF1-binding region of Siah2. SBD, substrate-binding domain. (F) Comparison of WT– and ΔZF6–XAF1 effect on Siah2 inhibition of HIPK2-mediated p53 phosphorylation in Calu-1 cells.
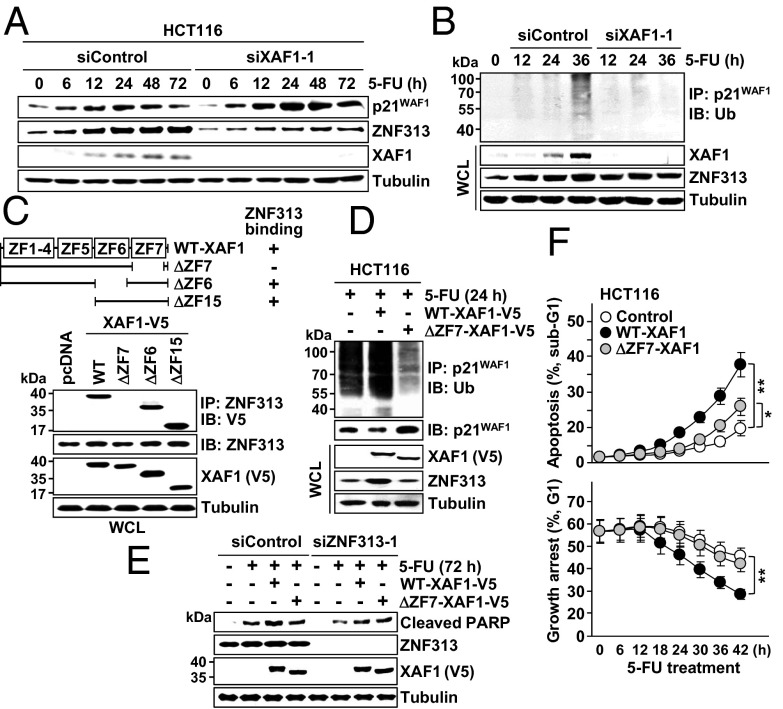
XAF1 Stimulates p21WAF1 Degradation via ZNF313 Activation.
Based on our recent finding of ZNF313 as a novel XAF1-binding, p21WAF1-targeting ubiquitin E3 ligase, we explored the possible implication of ZNF313 in XAF1 regulation of p53 signaling (25). ZNF313 level was increased in cells exposed to 5-FU or etoposide, and this induction was abolished if XAF1 expression was depleted (Fig. 6A and Fig. S7A). XAF1 was found to increase ZNF313 protein stability and attenuated stress-mediated p21WAF1 induction in a ZNF313-dependent manner, indicating that XAF1 promotes the turnover of p21WAF1 via ZNF313 induction (Fig. S7 B–D). In cells treated with 5-FU, an increment of p21WAF1 ubiquitination was apparent at ∼36 h after treatment, but this increase was not detected when XAF1 induction was blocked (Fig. 6B). We found that XAF1 interacts with ZNF313 through the ZF7 domain, and a ZF7-deleted mutant (ΔZF7–XAF1) has no activity to increase ZNF313 stability and p21WAF1 ubiquitination (Fig. 6 C and D and Fig. S7 E–H). Compared with WT–XAF1, ΔZF7–XAF1 was less potent in apoptosis promotion, but this difference was not recognized in ZNF313-depleted cells (Fig. 6E and Fig. S7I). Furthermore, XAF1-driven switch of cellular response to 5-FU from G1 cell-cycle arrest to apoptosis was considerably attenuated when ZNF313 was depleted (Fig. S7J). Consistently, ΔZF7–XAF1 was profoundly less potent than WT–XAF1 in the stimulation of the arrest to apoptosis switch (Fig. 6F and Fig. S7K). These data indicate that the ZNF313–p21WAF1 axis plays a key role in the XAF1-driven apoptotic switch of p53 signaling.
Fig. 6.
XAF1 stimulates p21WAF1 ubiquitination via ZNF313 induction. (A and B) XAF1-dependent ZNF313 induction and p21WAF1 ubiquitination in cells treated with 5-FU (25 μM). (C) Identification of the ZF7 domain as a ZNF313-binding region of XAF1. (D) Loss of p21WAF1-degrading function of XAF1 by deletion of the ZF7 domain. (E) Comparison of the WT– and ΔZF7–XAF1 effect on 5-FU–induced apoptosis in AGS cells. (F) Comparison of WT– and ΔZF7–XAF1 effect on cellular response to 5-FU (25 μM). Data represent the mean ± SD. *P < 0.05; **P < 0.01.
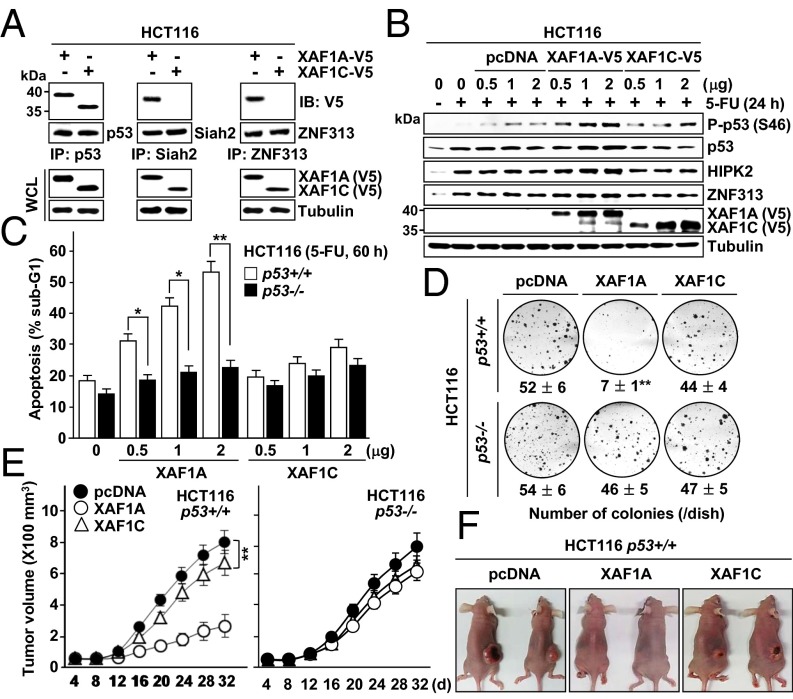
Isoform Switch of XAF1 Causes Loss of the p53–XAF1 Feedback Loop in Cancer Cells.
We next evaluated the relative contribution of the ZF5-7 domains to the XAF1-driven switch of p53 function. ΔZF5–XAF1 failed to increase p53 but retained the ability to interact with Siah2 and promote p53 phosphorylation, whereas ΔZF6–XAF1 failed to bind to Siah2 but retained p53-stabilizing activity (Fig. S8 A and B). Albeit profoundly less potent than WT–XAF1, ΔZF5–XAF1, but not ΔZF6–XAF1, evoked an apoptosis-inducing effect in HCT116 cells exposed to a cytostatic dose of ADR or CPT (Fig. S8 C and D). Compared with WT–XAF1, ΔZF5–, ΔZF6–, and ΔZF7–XAF1 showed ∼61%, 86%, and 49% reduction of apoptosis-promoting activity, respectively, in 5-FU–exposed cells, and each domain regulated p53, HIPK2, and ZNF313 in a fairly separate manner (Fig. S8 E and F). Whereas all these mutants manifested substantially reduced activity in stimulation of p53 interaction with apoptotic target promoters, ΔZF6–XAF1 rather increased p53 interaction with the p21WAF1 promoter (Fig. S8G). These results support that XAF1-driven apoptotic switch of p53 signaling occurs through the collaborative interplay of ZF5–7 domains. Based on these findings, we evaluated the p53-regulating function of XAF1C, a representative tumor-expressing variant lacking the ZF6 and ZF7 domains. XAF1C retained the ability to bind to and stabilize p53 but failed to interact with Siah2 and ZNF313 (Fig. 7 A and B and Fig. S8H). Compared with XAF1A, XAF1C displayed considerably low apoptotic and colony formation-inhibiting activities, and these activities were not associated with the p53 status of the cells (Fig. 7 C and D). Mouse xenograft assays also revealed that XAF1A strongly suppresses growth of p53+/+, but not of p53−/−, tumors, whereas XAF1C evokes a mild effect in both tumor types (Fig. 7 E and F). These results support that XAF1 suppresses tumor growth in a highly p53-dependent manner and that preferential loss of XAF1A provides tumor cells with a growth advantage.
Fig. 7.
No p53-activating function of XAF1C. (A) Loss of Siah2- and ZNF313-binding activity of XAF1C. (B) No effect of XAF1C on p53 phosphorylation, HIPK2, and ZNF313. (C and D) Comparison of XAF1A and XAF1C effect on 5-FU–induced apoptosis and colony formation of p53+/+ and p53−/− cells. Data represent the mean ± SD. *P < 0.05; **P < 0.01 (Student t test). (E) Mouse xenograft assay of the XAF1A and XAF1C effect on tumor growth and its dependency of p53 (n = 5 per group). **P < 0.01. (F) Representative photographs of xenograft tumors derived from XAF1A- or XAF1C-expressing HCT116 p53+/+ cells.
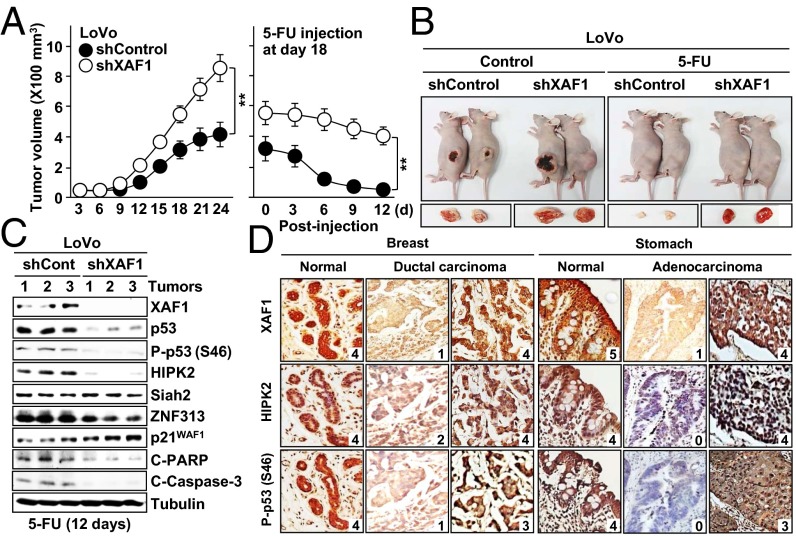
A p53–XAF1 Loop is Commonly Disrupted in Human Cancers.
Next, we examined the XAF1 effect on chemotherapeutic drug-induced tumor regression using xenograft tumors of LoVo colon cancer cells and short hairpin RNA-mediated XAF1 knockdown. Compared with shControls, shXAF1 tumors exhibited markedly reduced regression in response to 5-FU and lower expression of p53, Ser-46–phosphorylated p53, HIPK2, ZNF313, and cleaved PARP and caspase-3, but higher expression of p21WAF1, indicating that XAF1 drives the apoptotic switch of p53 signaling in vivo (Fig. 8 A–C and Fig. S9 A and B). To delineate the integrity of a p53–XAF1 loop in human cancers, we defined its mutational status in 104 cancer cell lines and 66 primary colorectal tumors. XAF1 promoter hypermethylation was detected in 36 of 44 (81.8%) cell lines and 25 of 48 (52.1%) primary tumors harboring WTp53, but only in 13 of 60 (21.7%) cell lines and 3 of 18 (16.7%) tumors carrying MTp53 (Fig. S9B). Consistently, loss or reduction of XAF1 mRNA was highly prevalent in WTp53 vs. p53-deficient cells, showing an exclusive relationship between p53 and XAF1 alterations in many tumors (Fig. S9 C and D). In immunohistochemical study of breast, stomach, lung, liver, and kidney tumor tissues, a strong copositivity (greater than or equal to level 3) of XAF1, HIPK2, and phospho-p53 (Ser-46) was observed in 21 of 50 (42%) WTp53 tumors and 45 of 50 (90%) adjacent normal tissues (Fig. 8D and Fig. S9E). Strong HIPK2 positivity was detected in 21 of 23 (91.3%) high XAF1 tumors but only in 4 of 27 (14.8%) low or no XAF1 tumors (Fig. S9F). Moreover, 21 of 27 (77.1%) tumors with strong phospho-p53 (Ser-46) also showed strong XAF1 positivity, and 21 of 23 (91.3%) tumors with weak phospho-p53 (Ser-46) displayed weak XAF1 positivity, indicating that XAF1, HIPK2, and phospho-p53 (Ser-46) levels correlate closely in both normal and tumor tissues.
Fig. 8.
Role of a p53–XAF1 loop in chemotherapeutic drug-induced tumor regression. (A) Increased resistance of XAF1-depleted tumors to 5-FU–induced regression. Data represent the mean ± SD (n = 5 per group). **P < 0.01. (B) Representative photographs of xenograft tumors at day 21 after inoculation (Left) and at day 9 after 5-FU injection (Right). (C) Comparison of p53, HIPK2, ZNF313, and p21WAF1 levels between in control and XAF1-depleted tumors. C-, cleaved. (D) Expression of XAF1, HIPK2 and phospho-p53 in human tumors. Relative staining levels were classified as levels 0–5.
Discussion
Under cytostatic stress conditions, MDM2 limits p53 growth arrest activity through the negative feedback loop, whereas in response to apoptotic stress, p53 escapes this loop to accumulate rapidly (2, 26). Our present study shows that XAF1 competes with MDM2 for binding to the p53 N-terminal region, generating the physical hindrance in MDM2 interaction with p53. The N-terminal PXXP repeats-containing proline-rich domain of p53 has been known to contribute to the apoptotic response of p53, however the molecular basis for its role is not well understood (27, 28). We found that XAF1 stabilizes p53 via the ZF5 domain-mediated interaction with the proline-rich domain, which is in line with a previous study that found that deletion of the proline-rich domain sensitizes p53 to MDM2-dependent degradation (29). It was also reported that the transcription coactivator p300/CBP stabilizes p53 via the interaction with the PXXP motif and that XAF1 binds to p300/CBP (30, 31). It is thus plausible that the apoptotic function of the p53 proline-rich domain is controlled by the XAF1–p300/CBP interplay in binding and acetylation of p53.
HIPK2 promotes p53 phosphorylation at Ser-46, which allows for p53 interaction with the prolyl-isomerase Pin1, dissociation from the apoptosis inhibitor iASPP, and subsequent induction of apoptotic target genes (6–8, 32). HIPK2 activity is tightly controlled by the ubiquitin–proteasome system, and multiple HIPK2-targeting E3 ligases have been demonstrated (33–35). In this study, we found that XAF1 stabilizes HIPK2 by binding to the RING domain of HIPK2-targeting Siah2 E3 ligase, and this activity of XAF1 is tightly linked to its function to drive the apoptotic switch of p53 signaling. It is therefore conceivable that XAF1 induction during the initial phase of apoptosis allows rapid amplification of the p53-dependent program through the regulation of the Siah2–HIPK2 axis. Given that HIPK2 acts as a tumor suppressor and can induce p53-independent apoptosis through multiple routes—including CtBP degradation and interaction with several proteins containing the high mobility group I domain—our data also suggest that the p53-independent functions of XAF1 might partially stem from its HIPK2-inducing activity (36).
A series of studies demonstrate that p53-induced p21WAF1-mediated cell-cycle arrest attenuates an apoptotic response of p53, suggesting that p21WAF1 may play an important role as a linchpin to govern cell fate (37–39). Our present study provides evidences that XAF1 represses p21WAF1-mediated cell-cycle arrest through induction of ZNF313, a p21WAF1-targeting E3 ligase, and the ZNF313-mediated p21WAF1 ubiquitination is crucial for XAF1 activation of p53’s apoptotic function. This finding indicates that activity of p21WAF1, a key proarrest target of p53, is counteracted by XAF1, supporting the notion that a p53–XAF1 feedback loop plays a key role in the apoptotic switch of p53 signaling. Therefore, XAF1 controls p53-mediated cell-fate decisions by accelerating p53-mediated apoptosis and also by impeding p53-mediated cell-cycle arrest.
XAF1 binds to p53, Siah2, and ZNF313 via the ZF5, ZF6, and ZF7 domains, respectively, and XAF1 regulation of p53 signaling results from the cooperative function of these domains (Fig. 9). Previous studies showed that the full-length XAF1A transcript is preferentially inactivated in cancers, whereas short variants are rather increased (16, 17). Our data show that XAF1C lacking the ZF6 and ZF7 domains is defective in binding to Siah2 and ZNF313 and unable to modulate the Siah2–HIPK2 and ZNF313–p21WAF1 axes. Therefore, XAF1C cannot form the regulatory loop with p53 and has profoundly debilitated growth suppression activity compared with XAF1A. This finding supports that the isoform switch of XAF1 provides tumor cells with selective survival and growth advantages and thus contributes to malignant tumor progression.
Fig. 9.
Schematic representation of the p53–XAF1 loop and its role for p53-mediated cell fate decisions.
Materials and Methods
Details regarding the expression constructs, EMSA, ChIP, ubiquitination, immunofluorescence, in vitro translation, immunoblotting, GST pull-down, yeast two-hybrid, semiquantitative RT-PCR, and single-strand conformational polymorphism analysis are available in SI Materials and Methods. All animal studies were performed with the approval of Korea University Institutional Animal Care and Use Committee and Korea Animal Protection Law.
Human Tissues.
Sixty-six colorectal tumor specimens were obtained by surgical resection in the Kyung Hee University Medical Center (Seoul, Korea) as described (14). Human tissue arrays were obtained from SuperBioChips Laboratory, and immunostaining was performed by using the Vectastain ABC (avidin–biotin–peroxidase) kit (Vector Laboratories).
Cellular Assays.
Annexin-V–FITC (Sigma) and 7-aminoactinomycin D (BD Biosciences) staining was performed as recommended. Cell-cycle profile and sub-G1 fraction were analyzed by using the FACScan flow cytometer (Becton Dickinson) and MultiCycle software (Phoenix Flow Systems).
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein (Johns Hopkins University) for HCT116 p53+/+ and p53−/− cells, Dr. M. L. Smith (Indiana University) for 13X p53RE–Luc, and Dr. K. H. Vousden (Beatson Institute for Cancer Research) for mutant p53 (175Asp) vector. This work was supported in part by National Research Foundation of Korea Grants 2009-0078864 (to S.-G.C.) and 2009-353-C00071 (to M.-G.L.) and a Korea University Intramural Grant-in-Aid (2014).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411746111/-/DCSupplemental.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 3.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9(9):702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 4.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10(4):431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 6.D’Orazi G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4(1):11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann TG, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4(1):1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 8.Mayo LD, et al. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem. 2005;280(28):25953–25959. doi: 10.1074/jbc.M503026200. [DOI] [PubMed] [Google Scholar]
- 9.Harris SL, Levine AJ. The p53 pathway: Positive and negative feedback loops. Oncogene. 2005;24(17):2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 10.Fong WG, et al. Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics. 2000;70(1):113–122. doi: 10.1006/geno.2000.6364. [DOI] [PubMed] [Google Scholar]
- 11.Liston P, et al. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3(2):128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 12.Byun DS, et al. Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res. 2003;63(21):7068–7075. [PubMed] [Google Scholar]
- 13.Lee MG, et al. Promoter CpG hypermethylation and downregulation of XAF1 expression in human urogenital malignancies: Implication for attenuated p53 response to apoptotic stresses. Oncogene. 2006;25(42):5807–5822. doi: 10.1038/sj.onc.1209867. [DOI] [PubMed] [Google Scholar]
- 14.Chung SK, et al. Frequent alteration of XAF1 in human colorectal cancers: Implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132(7):2459–2477. doi: 10.1053/j.gastro.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Lunardi A, et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat Genet. 2013;45(7):747–755. doi: 10.1038/ng.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X, et al. Switch to full-length of XAF1 mRNA expression in prostate cancer cells by the DNA methylation inhibitor. Int J Cancer. 2006;118(10):2485–2489. doi: 10.1002/ijc.21636. [DOI] [PubMed] [Google Scholar]
- 17.Yin W, Cheepala S, Clifford JL. Identification of a novel splice variant of X-linked inhibitor of apoptosis-associated factor 1. Biochem Biophys Res Commun. 2006;339(4):1148–1154. doi: 10.1016/j.bbrc.2005.11.128. [DOI] [PubMed] [Google Scholar]
- 18.Siegelin M, Touzani O, Toutain J, Liston P, Rami A. Induction and redistribution of XAF1, a new antagonist of XIAP in the rat brain after transient focal ischemia. Neurobiol Dis. 2005;20(2):509–518. doi: 10.1016/j.nbd.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Ng KCP, Campos EI, Martinka M, Li G. XAF1 expression is significantly reduced in human melanoma. J Invest Dermatol. 2004;123(6):1127–1134. doi: 10.1111/j.0022-202X.2004.23467.x. [DOI] [PubMed] [Google Scholar]
- 20.Xia Y, Novak R, Lewis J, Duckett CS, Phillips AC. Xaf1 can cooperate with TNFalpha in the induction of apoptosis, independently of interaction with XIAP. Mol Cell Biochem. 2006;286(1-2):67–76. doi: 10.1007/s11010-005-9094-2. [DOI] [PubMed] [Google Scholar]
- 21.Leaman DW, et al. Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J Biol Chem. 2002;277(32):28504–28511. doi: 10.1074/jbc.M204851200. [DOI] [PubMed] [Google Scholar]
- 22.Micali OC, et al. Silencing of the XAF1 gene by promoter hypermethylation in cancer cells and reactivation to TRAIL-sensitization by IFN-beta. BMC Cancer. 2007;7:52–59. doi: 10.1186/1471-2407-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu SP, et al. Tumor suppressor XIAP-Associated factor 1 (XAF1) cooperates with tumor necrosis factor-related apoptosis-inducing ligand to suppress colon cancer growth and trigger tumor regression. Cancer. 2010;116(5):1252–1263. doi: 10.1002/cncr.24814. [DOI] [PubMed] [Google Scholar]
- 24.Zou B, et al. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol Carcinog. 2012;51(5):422–432. doi: 10.1002/mc.20807. [DOI] [PubMed] [Google Scholar]
- 25.Han J, et al. ZNF313 is a novel cell cycle activator with an E3 ligase activity inhibiting cellular senescence by destabilizing p21(WAF1.) Cell Death Differ. 2013;20(8):1055–1067. doi: 10.1038/cdd.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 27.Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93(26):15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baptiste N, Friedlander P, Chen X, Prives C. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene. 2002;21(1):9–21. doi: 10.1038/sj.onc.1205015. [DOI] [PubMed] [Google Scholar]
- 29.Berger M, Vogt Sionov R, Levine AJ, Haupt Y. A role for the polyproline domain of p53 in its regulation by Mdm2. J Biol Chem. 2001;276(6):3785–3790. doi: 10.1074/jbc.M008879200. [DOI] [PubMed] [Google Scholar]
- 30.Dornan D, Shimizu H, Burch L, Smith AJ, Hupp TR. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol Cell Biol. 2003;23(23):8846–8861. doi: 10.1128/MCB.23.23.8846-8861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, et al. Interactions between XIAP associated factor 1 and a nuclear co-activator, CBP, in colon cancer cells. Digestion. 2008;77(2):79–86. doi: 10.1159/000121441. [DOI] [PubMed] [Google Scholar]
- 32.Pistritto G, Puca R, Nardinocchi L, Sacchi A, D’Orazi G. HIPK2-induced p53Ser46 phosphorylation activates the KILLER/DR5-mediated caspase-8 extrinsic apoptotic pathway. Cell Death Differ. 2007;14(10):1837–1839. doi: 10.1038/sj.cdd.4402186. [DOI] [PubMed] [Google Scholar]
- 33.Sombroek D, Hofmann TG. How cells switch HIPK2 on and off. Cell Death Differ. 2009;16(2):187–194. doi: 10.1038/cdd.2008.154. [DOI] [PubMed] [Google Scholar]
- 34.Winter M, et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol. 2008;10(7):812–824. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]
- 35.Calzado MA, de la Vega L, Möller A, Bowtell DDL, Schmitz ML. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol. 2009;11(1):85–91. doi: 10.1038/ncb1816. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115(2):177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 37.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 38.Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol. 2013;25(1):52–58. doi: 10.1097/CCO.0b013e32835b639e. [DOI] [PubMed] [Google Scholar]
- 39.Abbas T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.