Significance
By globally mapping the position of stalled ribosomes in macrolide-treated Staphylococcus aureus, we identified the proteins whose synthesis is affected by this antibiotic. Rather than acting as a nonselective inhibitor, the binding of macrolides to the ribosome tunnel only induces stalling at a few unique sites that are enriched in proline and charged residues. The sequences that flank the stalling sites are highly variable but are essential for efficient stalling. Although it has long been assumed that macrolides inhibit translation after the synthesis of a few residues, we found that stalling could occur at any point during translation. Our results reveal a striking conservation of stalling motifs among all known arrest peptides that strongly suggests a universal ribosome stalling mechanism.
Keywords: antibiotic, ribosome stalling, Staphylococcus aureus
Abstract
The prevailing “plug-in-the-bottle” model suggests that macrolide antibiotics inhibit translation by binding inside the ribosome tunnel and indiscriminately arresting the elongation of every nascent polypeptide after the synthesis of six to eight amino acids. To test this model, we performed a genome-wide analysis of translation in azithromycin-treated Staphylococcus aureus. In contrast to earlier predictions, we found that the macrolide does not preferentially induce ribosome stalling near the 5′ end of mRNAs, but rather acts at specific stalling sites that are scattered throughout the entire coding region. These sites are highly enriched in prolines and charged residues and are strikingly similar to other ligand-independent ribosome stalling motifs. Interestingly, the addition of structurally related macrolides had dramatically different effects on stalling efficiency. Our data suggest that ribosome stalling can occur at a surprisingly large number of low-complexity motifs in a fashion that depends only on a few arrest-inducing residues and the presence of a small molecule inducer.
During translation, nascent polypeptides travel through a ∼100-Å-long aqueous tunnel in the large ribosomal subunit before entering the cytoplasm. The ribosome tunnel is highly irregular in shape and contains numerous cavities and grooves (1). Despite its narrow diameter (∼15 Å on average), recent studies have indicated that at least some nascent chains adopt a more compact conformation (2–4). Furthermore, the detection of specific peptide sequence motifs can arrest elongation or termination in both eukaryotes and prokaryotes (5). The cis-acting translational attenuator serves to regulate the expression of downstream genes in response to physiological signals (6, 7) and small molecules, such as antibiotics or amino acids (8–10). In bacteria, ribosome stalling at the upstream element often promotes the rearrangement of structured mRNA and activates downstream translation. Many resistant pathogens exploit the translational stalling mechanism to up-regulate the synthesis of antibiotic resistance genes (10). Curiously, the ribosome-stalling peptides identified to date vary in length and share no sequence homology.
The ribosome tunnel is the target of many clinically important macrolide antibiotics. Macrolides bind to the tunnel close to the peptidyltransferase center (PTC) and are thought to inhibit translation by physically obstructing the progression of the nascent chain, thereby leading to the premature drop-off of peptides that are only six to eight amino acids long (11). The prevailing view of macrolide action suggests that these antibiotics indiscriminately block the elongation of every protein during the early stage of translation. In contrast to this model, recent studies have shown that several proteins are able to bypass the inhibition, presumably by threading through the drug-bound tunnel or by stimulating dissociation of antibiotic from the ribosome (12–14). Although the precise mechanism is unclear, the identification of a short N-terminal motif that circumvents erythromycin blockage (12) suggests that the extent of macrolide inhibition is highly sequence-specific. The full scope of this selectivity remains unknown.
To identify the protein sequence motifs that are susceptible and resistant to macrolide action, we performed ribosome profiling to examine the Staphylococcus aureus translatome in the presence of the macrolide azithromycin (AZ). In addition to identifying previously unidentified ORFs and noncoding RNAs, we found incomplete down-regulation of the translation in cells exposed to AZ. Of particular interest, we found that AZ-mediated translation arrest selectively occurs at stalling sites that are enriched in proline and charged residues, despite a lack of overall sequence conservation. Surprisingly, many ribosomes do not enrich the N-terminal region as predicted; instead, they undergo arrest at a much later stage of translational elongation depending on the locations of specific stalling sites. Even more unexpectedly, the AZ-bound ribosomes have the ability to overcome the initial AZ inhibition within a multistalling-site-containing sequence, possibly by rerouting the path of the nascent chain.
Results
Ribosome Profiling in Staphylococcus aureus.
Ribosome profiling provides a snapshot of ribosomal movement on its template mRNA (15). A high number of ribosome-protected fragments (RPFs) mapping to the transcriptome (mRNA-seq) is indicative of prolonged ribosome occupancy at a given position, also known as ribosome stalling/pausing. AZ causes ribosome stalling and is one of the most commonly used macrolide antibiotics. The minimum inhibitory concentration (MIC) of AZ for S. aureus NCTC 8325 is ∼0.125 μg/mL (Fig. S1A). We performed growth inhibition and pulse-chase analyses and found that residual protein synthesis persisted for 45 min, even at a dose of AZ ∼200-fold higher than the MIC, whereas bacterial growth was completely halted within 24 h (Fig. S1 B and C). Cell lethality may occur owing to the imbalance of the cellular proteome caused by the partial inhibition of a large proportion of proteins (12). Long-lived protein bands observed in the pulse-chase analysis could in part be due to the slow protein turnover, accumulation of truncated products, or bypassing of AZ-mediated translational inhibition.
To investigate the effects of AZ on the global translatome and whether AZ preferentially stalls synthesis of particular protein sequences, we performed ribosome profiling on AZ-treated and untreated S. aureus cells, with the hypothesis that AZ-induced ribosome stalling (and hence translation arrest) would result in an unusually high ribosome density at unique stalling sites. Exponentially grown S. aureus were treated with and without AZ for 15 min because longer treatment severely impaired the assembly of ribosomal subunits. Before cell harvest, 2-min pretreatment of chloramphenicol was conducted to stabilize the translating ribosomes. Cell lysate was prepared by cryomilling (16). Sucrose density fractionation was performed after micrococcal nuclease (MNase) treatment to isolate ribosome-protected mRNAs from the monosomes. We were unable to fully convert the “disome” peak into the monosomes (Fig. 1A), despite using different MNase sources and digestion conditions, unlike Escherichia coli and Bacillus subtilis, which produce homogenous monosomes after MNase digestion (16, 17). Additional analyses confirmed that the “disome” peak in the MNase-negative sample was indeed a mixture of 100S ribosomes (>85%) and disomes (∼10–15%) (Fig. S1F). The binding of hibernation promoting factor (HPF) to the interface of the 30S and 50S subunits has been shown to stimulate the dimerization of the 70S to form 100S ribosomes (18, 19). In contrast to E. coli 100S ribosomes, which only exist during stationary phase, S. aureus 100S ribosomes persist throughout all growth phases (18). We performed Western blots using an anti-S. aureus HPF (SaHPF) antibody and confirmed that the protein was highly expressed in S. aureus and was ubiquitously present in all ribosomal fractions. SaHPF was also particularly enriched in the MNase-treated “disome” peak (Fig. 1A and Fig. S1 D and E). The “disome” peak diminished dramatically upon the inactivation of the hpf gene in S. aureus (Fig. S1F). Because the residual “disome” peak might consist of a small fraction of stacked “disomes” that protects longer footprints (20), a “disome” library (RPF2) was constructed in parallel with the libraries derived from the monosomes (RPF1).
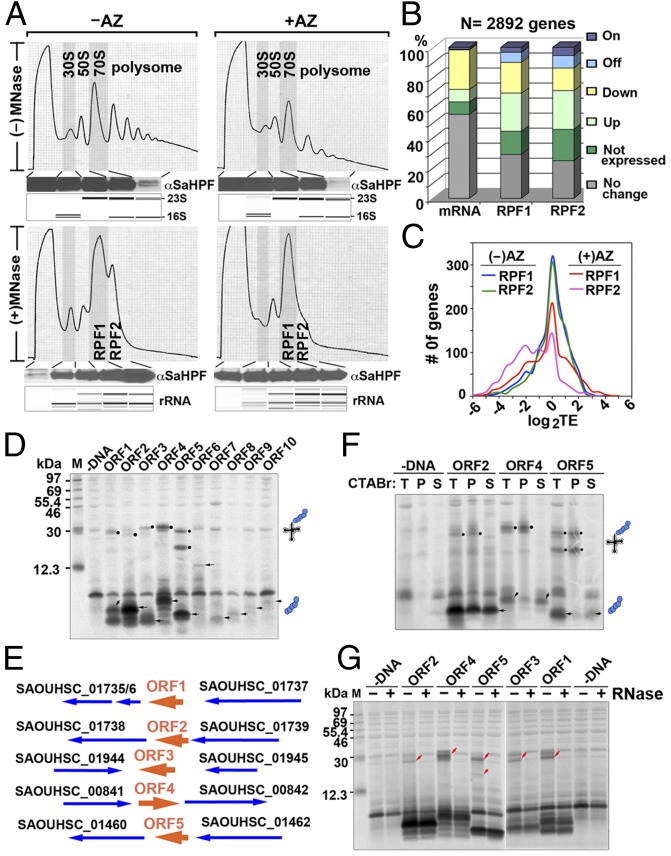
Fig. 1.
Analysis of RPFs and identification of previously unidentified ORFs. (A) Sucrose gradient profiles of S. aureus ribosomes digested and undigested by MNase with or without AZ treatment. The y axis corresponds to the absorbance at 254 nm of the ribosome particles separated on a 10–55% sucrose gradient. The association of SaHPF to the 100S ribosome, which constituted >85% of the RPF2 (Fig. S1F), was detected via immunoblotting. (Lower) Bioanalyzer electropherograms of the RNA species are shown as controls. (B) Global effects of AZ on the transcriptome and the translatome. Differential gene expression is defined as ≥twofold changes (<0.05 false discovery rate) in the RPKM (reads per kilobase of transcript per million mapped reads) ratios between the AZ-treated to -untreated samples. “On” and “Off” are defined as genes that are transcriptionally or translationally inactive (zero read count) in one sample. (C) Translation efficiency (TE) was moderately decreased after AZ exposure. The efficiency was calculated as the log2 ratios of the RPFs to the mRNA fragments that were measured in RPKM. (D) Cell-free coupled transcription–translation of representative short ORFs. Arrowheads indicate the protein products, and solid circles denote stalled peptidyl-tRNAs. (E) Genetic organization of arrest peptide-encoded ORFs (in orange). Genes are depicted by thick arrows, which point in the direction of transcription. ORF1–ORF5 undergo translation arrest without AZ. (F) CTABr precipitation of peptidyl-tRNAs that remain tethered to the stalled ribosomes. Full-length proteins were fractionated into the CTABr supernatant. T, total; P, pellet; S, supernatant. (G) The RNase A susceptibility of translation-arrested peptidyl-tRNAs is indicated by the disappearance of the ∼20- to 30-kDa bands (red arrowheads).
Analysis of RPFs and Global Translational Activity.
On average, we obtained >10 million and >4 million uniquely mapped reads of randomly fragmented total mRNAs and RPFs, respectively. The two independent biological replicates were highly reproducible (Fig. S2A). The total mRNA libraries were enriched in fragments of 29 nt, whereas the RPF1 and RPF2 libraries displayed a multimodal distribution with maximal peaks at 27 nt, 30 nt, and 33 nt (Fig. S2B). The broader distribution of lengths of RPFs could be due to the sequence bias of MNase (21) and an asymmetric 3′-end extension of footprints induced by ribosome pausing (22). In general, AZ treatment indirectly altered the mRNA abundance of 40% of the genes, presumably by changing the expression level of transcription factors central to the metabolic and stress response pathways (23). As a result, ∼60% of the genes were differentially translated, a majority of which was attributable to the changes in the mRNA levels (Fig. 1B). To measure the extent of AZ-mediated translational inhibition, we calculated the translation efficiency (the ratio of the RPF reads to the mRNA reads) after AZ treatment, focusing on the top 1,000 most abundantly expressed mRNAs. Overall, the translation efficiency was only reduced by 35%, suggesting a significant leakiness of translation in the presence of AZ (Fig. 1C).
Identification of Previously Unidentified ORFs Encoding AZ-Independent Ribosome Stalling Peptides.
By closely examining the read coverage across the entire genome, we identified more than a dozen new small RNAs and twenty novel (ORFs) (Figs. S3 and S4 and Table S1). The previously unidentified ORFs were generally less than 70 amino acids in length, except for ORF6 (129 aa) and contained weak Shine-Dalgarno (SD) sequence. Although some start codons were AUG, the vast majority of the newly identified ORFs initiated with the alternative GUG, UUG, or even CUG (Table S1). Using an in vitro translation system with T7 promoter-driven templates carrying the native SD and initiation codons, we detected the protein products of 10 ORFs, 5 of which also yielded a prominent product of ∼30 kDa (Fig. 1 D and E). The migration of the ∼30-kDa product conspicuously matched the molecular weight of an ∼25-kDa tRNA linked to the short peptide. Taking advantage of the ability of cetyl trimethylammonium bromide (CTABr) to precipitate nucleic acids, we performed CTABr fractionation and confirmed that the ∼30-kDa products were peptidyl-tRNAs that were tethered to the stalled ribosomes (Fig. 1F). Furthermore, the tRNA moiety of the ∼30-kDa products could be removed by RNase A treatment (Fig. 1G). These results indicate these ORFs are able to stall the ribosome in the absence of AZ. On the basis of the identity of the downstream genes, the predicted locations of −10 and −35 boxes, and the genetic organization of these ORFs, they likely function as cis-acting regulatory peptides to modulate downstream gene expression (5).
Mid- and Late-Stage Ribosome Stalling Are Unexpectedly Prevalent.
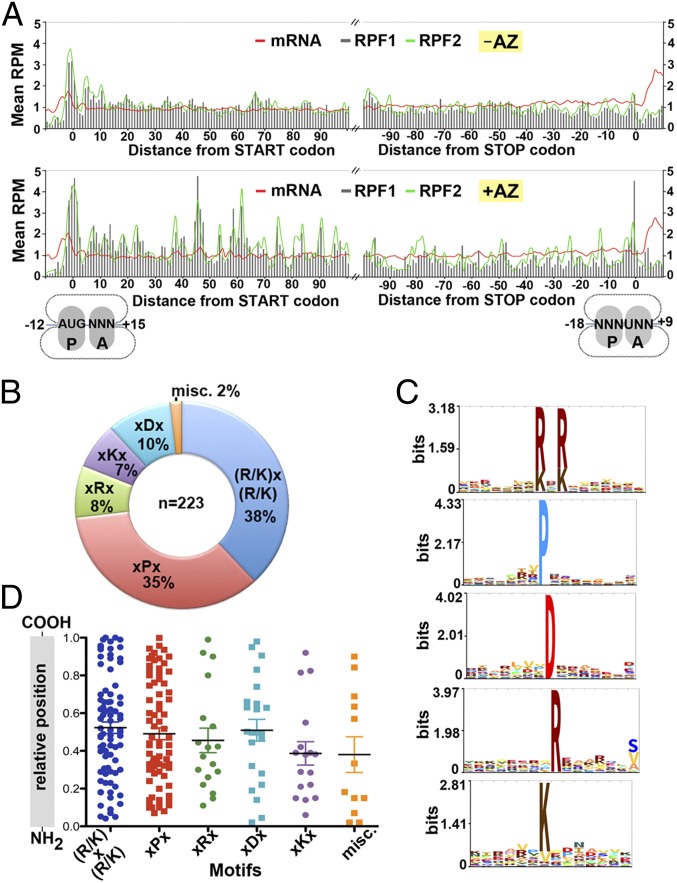
Metagene analysis was undertaken to analyze the average ribosome density across the top 1,200 most abundantly expressed genes. S. aureus ribosomes seemed to protect a footprint of 27–28 nt. As expected, a pronounced accumulation of RPF reads was detected over that of the start codon, whose density was >threefold greater than those within the ORFs and the mRNA reads regardless of the presence of AZ. The elevated ribosome density around the start and stop codons corresponds to the slower kinetics of initiation and termination in comparison with the elongation rate (16). We found that the reads from RPF1 and RPF2 libraries followed the same 3-nt periodicity and displayed no substantial differences in intensity (Fig. 2A). Further investigation revealed that both RPF1 and RPF2 shared >85% of the ORFs in common, and their correlations were 0.8943 (no AZ) and 0.9505 (with AZ). The disome may result from the stacking of the trailing ribosome after the lead ribosome has undergone translational pausing or from the ribosomes stacked on the truncated mRNAs (20). At present we cannot distinguish them owing to a lack of resolution at the 3′-end of RPF2 libraries. Here we will focus on analysis of the RPF1 library. On the basis of the conventional view that macrolide antibiotics stall nascent chain elongation at the very N terminus of the ORFs, we expected that AZ treatment would only lead to an elevation of the RPF density within the first 5–10 codons. In sharp contrast to this prediction, unusually high-density peaks were detected throughout the ORFs, and each of the consecutive elevated peaks was on average ∼10 residues apart (Fig. 2A). To our knowledge, these results provide the first evidence that AZ-mediated translation arrest could occur at later stages of elongation.
Fig. 2.
AZ-specific ribosome stalling sites are scattered along ORFs and are enriched in proline and charged residues. (A) Ribosome density as a function of position. Metagene analysis of read densities in the AZ-treated (Lower) and untreated (Upper) samples. The normalized RPM correspond to the average ribosome density across the most abundantly translated ORFs (>50 counts), which were aligned relative to the start and stop codons. Total mRNA and RPF2 are shown with red and green lines, respectively. RPF1 is depicted by gray bars. (B) The fraction of stalling sequences falling into each indicated class. (C) Hidden Markov model logos showing the frequencies of specific residues at a given position in each motif group. Sequences of miscellaneous class are too variable to build the logo. (D) The relative location of the stalling motifs within their full-length proteins. Error bars indicate mean.
AZ-Targeted Ribosome Selectively Stalls at Specific Residues.
The widespread ribosome stalling locations prompted us to examine whether these results could be corroborated by the distribution of particular stalling-inducing sequences in the ORFs. By subtracting the density between the AZ-treated and -untreated RPF1 reads, we were able to locate the precise ribosome stalling sites. We found that AZ-bound ribosomes had a high propensity to stall at the Pro, Arg, Lys, and Asp residues, as well as any residues that were flanked by positively charged residues (Table S2). The stalling sites could be categorized into six classes, in which (R/K)x(R/K) and xPx represented 72% of the stalling sequences (Fig. 2B). Interestingly, the (R/K)x(R/K) motif closely resembles that of the uncharacterized Erm leader peptide homologs in many macrolide-resistant bacteria (Fig. S5A) (10).
Although the majority of the protein sequences carrying the AZ-specific stalling hotspots are virulence-related or essential genes (Table S2), none of these genes bear any sequence similarity around the stalling sites (Fig. 2C). Furthermore, the locations of the stalling sites are randomly distributed throughout the entire protein sequence, without any apparent clustering at the N terminus or C terminus (Fig. 2D). In fact, most of the stalling sites are not positioned within the first 30 codons, suggesting the ribosomes are able to bypass AZ inhibition during the early phase of translation.
Although the prolonged dwell time of ribosome on its template, in principle, would lead to a lack of ribosome density downstream of the stalling sites, we unexpectedly found that in many instances weaker RPF reads perpetuated toward the C terminus. In 17 cases, two to three strong stalling peaks of equal intensity from the same stalling sequence motif could be detected within a single protein sequence (Table S2). Residual RPF reads could have been obtained from the ongoing translation before the addition of AZ to the cultures. However, results from a synchronized in vitro toe-printing assay argued against this possibility (see below). To determine the distance that ribosomes advance toward the C terminus, we calculated the distance between the first encountered stalling site and the position at which the read density receded completely. On average, the residual read density was found 100–200 nt away from the stalling sites (Fig. S5B). These values are remarkable, given that the average length of an S. aureus ORF is merely 290 nt. The ability of stalled ribosomes to overcome arrest suggests that some AZ-mediated stalling events are inefficient.
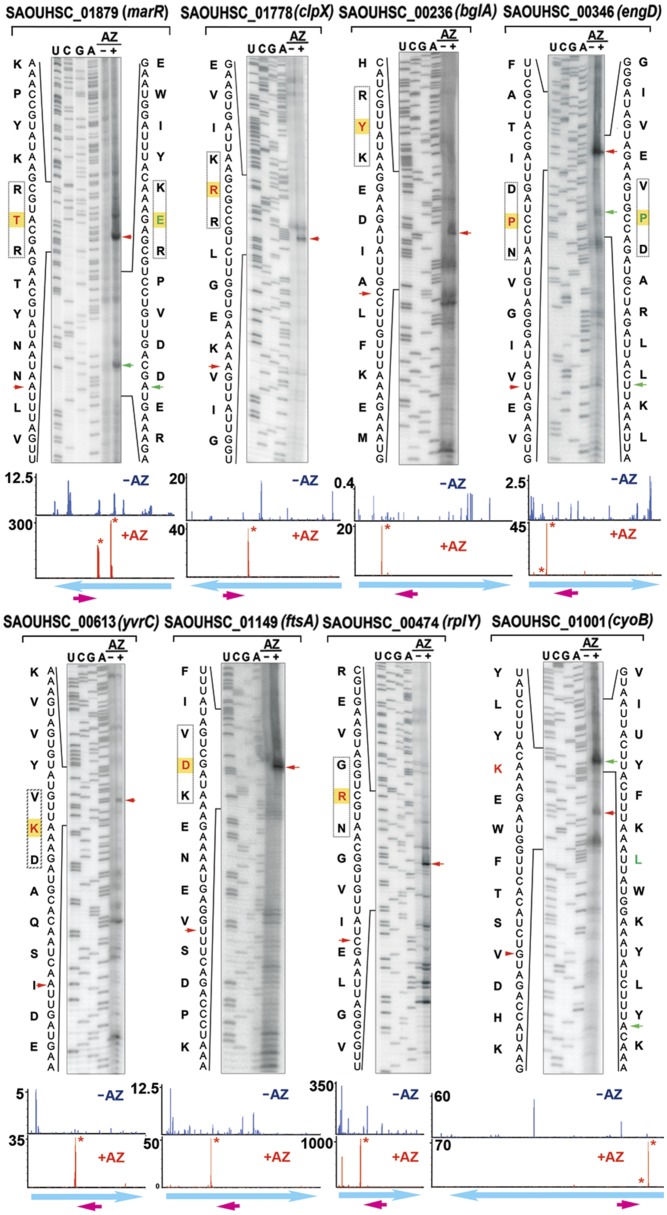
We performed in vitro toe-printing assays (primer extension inhibition) to map the precise ribosome stalling sites of at least one randomly chosen gene for each motif class (Table S2) and were able to faithfully recapitulate in vivo ribosome stalling. In all cases, we found that the ribosomes were arrested in the presence of AZ at the position precisely corresponding to the highest peak in the read density plots (Fig. 3 and Figs. S6 and S7A). Surprisingly, we also detected two consecutive, adjacent stalling sites in a single translation–primer extension reaction (marR [R/K]x[R/K], and engD [xPx]). Although the toe-print signal of the second stall site is weaker than that of the preceding site, these results provide an additional indication that some translation occurs beyond a strong AZ-mediated stalling site.
Fig. 3.
In vitro mapping of AZ-specific ribosome stalling sites by toe-printing analysis. Cell-free translation was programmed with the respective DNA templates in the presence and absence of AZ, followed by primer extension. Tripeptide stalling sequences are boxed. The residues labeled in red and green are positioned in the P site of the stalled ribosome, which are located 16–17 nt upstream of the blocked reverse transcriptase. Red and green arrowheads mark the primary and secondary toe-print signals. The ribosome density maps corresponding to the genes are shown on the bottom panels. The y axis indicates RPM-normalized ribosome density. The magenta arrows indicate the location of the reverse primer. The stalling sites are labeled with an asterisk. Weaker density peaks might not be visible owing to the scale difference.
The Path of the Nascent Peptides Modulates the Stalling Efficiency.
The arrest motifs obtained from this screen contain by far the fewest conserved residues among naturally occurring stalling peptides. This minimalism raises a puzzling question as to how a sequence as simple as one to three amino acids in length is sufficient to trigger translation arrest given that these residues are ubiquitously present in most, if not all proteins. The sequence plasticity of ribosome stalling motifs has been demonstrated in SecM and polyproline-containing proteins, as well as in laboratory-evolved arrest peptides (24–27). In SecM, the flexible flanking residues seem to play a secondary yet important role in positioning the nascent peptide into the proper intratunnel location (24, 28). In addition, the structure of the antibiotic ligands has been implicated to create an optimal tunnel environment for Erm leader peptide-induced stalling (29, 30). Therefore, we posit that beyond the dependency of the one to three conserved residues, the path of the nascent peptide, specified by the flanking sequence, and the type of antibiotic applied also contribute to the inducibility of translational stalling.
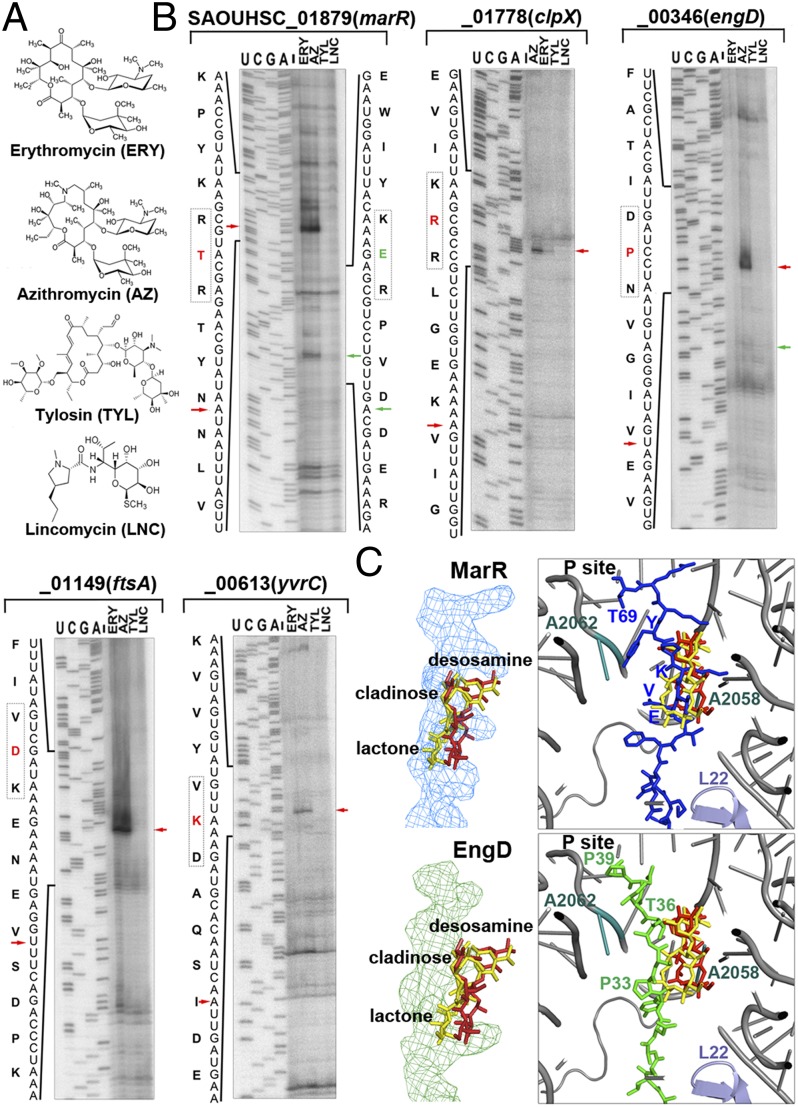
To test this hypothesis, we performed toe-printing assays that were programmed using templates bearing the major motif classes and various antibiotics (Fig. 4A). As expected, tylosin (a 16-membered lactone macrolide) and lincomycin, whose binding sites overlap with the A-site, did not produce strong toe-print signals in all proteins (Fig. 4B). Presumably, early inhibition of peptide bond formation prevented the ribosome from reaching the stalling site. Surprisingly, all but one (EngD) of the arrest sequences underwent ribosome stalling in the presence of the structurally related antibiotics erythromycin (ERY) and AZ, which are 14- and 15-membered lactone macrolides, respectively. Although they differ from one another by an absence of one carbonyl group and the methyl-substituted nitrogen group in AZ, only AZ induced ribosome stalling at P39 of EngD (Fig. 4B). Thus, to our knowledge, EngD represents the first example of macrolide-dependent arrest peptide that differentially responds to ERY and AZ.
Fig. 4.
Distinct dispositions of MarR and EngD peptides inside the tunnel affect stalling efficiency. (A) Chemical structures of macrolides (ERY, AZ, and tylosin) and lincomycin. (B) Toe-printing analysis of representative stalling sequences in the presence of different antibiotics. Labeling schemes are identical to those of Fig. 3. (C) Structural modeling of the MarR and EngD stalling peptides inside the antibiotic-bound tunnel. The relative locations of previously reported ribosome stalling “sensors” (L22, A2062, and A2058) to the superimposed ERY (red) and AZ (yellow) molecules are indicated.
To investigate what might account for the differential induction of ribosome stalling in EngD, the ERY-responsive MarR and ERY-insensitive EngD nascent peptides were modeled into the antibiotic-bound ribosome tunnel (Fig. 4C). The superposition rmsd values among 10 macrolide-bound ribosome crystal structures are almost negligible (0.05–1.59 Å), and the tunnel structures are virtually identical with the ribosome-nascent chain complexes bound with (ErmBL) and without (TnaC) antibiotic. We predicted that the path of the nascent chain would determine the degree of contact between the antibiotic and the peptide. We found that the lactone ring and the cladinose sugar group directly sterically clashed with the MarR peptide, specifically in the 64KVE62 segment, because K64 was “entangled” between the cladinose and the lactone moieties of both ERY and AZ. In contrast, P33 of EngD seemed to swing the nascent peptide chain away from the antibiotics. In this case, the expanded lactone ring in AZ facilitates its interaction with the nascent peptide chain; as a result, the AZ-bound ribosome became stalled at EngD elongation. ERY, however, displayed very minimal contact with EngD at T36, which was not able to elicit stalling (Fig. 4C). Consistent with the model, a P33V mutation abolished AZ-induced stalling, and a T36Y substitution (equivalent of Y66 in MarR) restored the ERY-dependent stalling (Fig. S7B). Our analyses suggest that ribosome stalling efficiency could significantly change if a nascent peptide alters its route along the tunnel. The results help to explain why not every xPx- or (R/K)x(R/K)-containing protein was subjected to ribosome stalling and thus was not captured in the ribosome profiling.
Discussion
Here we describe, to our knowledge, the first genome-wide snapshot of ribosome distribution along mRNAs in S. aureus. This approach enabled us to discover previously unidentified ORFs and to identify hundreds of AZ-specific translation arrest hotspots that were not detectable via conventional proteomics. To our surprise, AZ-mediated translation arrest occurs at specific stalling sites that are located throughout the entire sequence, in contrast to previous prediction that macrolide-induced ribosome stalling occurs predominately at the N terminus. These stalling sites are enriched in charged residues and prolines. The proteins subjected to AZ-mediated stalling do not belong to any particular category but instead seem to perform diverse cellular functions. The sequences flanking the stalling sites are highly degenerate and are not marked by residues of any particular chemical property. Finally, the identity of the stalling site per se does not seem to be the sole determinant of the establishment and maintenance of the arrested state. The context-dependent passage of the nascent peptides and the structure of antibiotic ligand are also essential components of this interactive process.
The isolation of previously unidentified arrest peptides suggests that regulation of gene expression by peptide-mediated ribosome stalling in S. aureus might be more common than in other bacteria (5). Our isolation of AZ-specific translation arrest motifs was by no means exhaustive, in large part because some mRNAs were not transcribed under the experimental conditions used and also because extraribosomal factors could be involved in relieving the elongation stalling. Translation arrest mediated by SecM and MifM could be released upon extracellular secretion and membrane insertion when a pulling force of the translocon acts on the N terminus of the proteins (6, 7). Moreover, elongation factor P alleviates the translation arrest triggered by polyproline sequences (26, 27, 31).
The emerging theme from this analysis was that AZ-bound ribosomes do not simply stall at any random residues, but rather only at specific sites [D, R, K, P or (R/K)x(R/K)]. Intriguingly, these residues seem to be the preferred stalling site or to play critical roles in the translational stalling of other arrest peptides. R163 is essential for SecM peptide recognition (24). Cat and CmlA leader- and ErmBL-dependent stalling occurs when an Asp enters the P site of the antibiotic-bound ribosome (32, 33). The “9VDK11” motif in ErmBL is identical to two of the arrest sequences in S. aureus FtsA and protein A. The (R/K)x(R/K) stalling sites also share a striking resemblance to the “RLR” class of short regulatory peptides upstream of macrolide resistance genes (34). The size of the “RLR” peptides is as short as eight amino acids, whereas the (R/K)x(R/K) motif is embedded in much larger proteins that predominantly perform housekeeping functions. Presumably, if the short RLR regulatory peptides have evolved from the larger housekeeping proteins to counteract macrolide inhibition, then the evolutionary conservation of the stalling sites and the diversity of the overall sequences can be conceived.
Positively charged residues inside the tunnel have a high propensity to induce ribosome pausing (35–38). A-site codons corresponding to the charged residues in ErmAL1 produce the strongest stalling (39). The preferences for Arg and Lys at the PTC cannot be easily explained by their side-chain properties because peptidyl-tRNAs bearing C-terminal Arg or Lys seem to display a faster peptidyl transfer rate (40). Instead, slow reactions may involve the additional relay of the stalling signal from the region close to the tunnel entrance, where the nascent peptide and antibiotic interact, leading to further conformational changes in the PTC (41). TnaC, SecM, and polyproline sequences, as well as the viral UL4 uORF2, all stall the ribosome at a P or A site occupied by a proline moiety (5). Except for TnaC, none of these systems requires a small molecule. The mechanism underlying the translation arrest has been attributed to poor reactivity of proline in peptidyl transfer (42, 43). Coincidentally, the presence of (R/K)x(R/K) and xPx sequences in E. coli fusA and firefly luciferase also induce ribosome stalling in the presence of erythromycin (12). Together, the data suggest that the basic principles of peptide-dependent ribosome stalling are generally similar regardless of the origins of the sequence and the requirements of the small molecule, although effector molecules might alter the tunnel to create ligand-specific sites of peptide discrimination.
In both ligand-independent and -dependent arrest sequences, peptide-mediated ribosome stalling typically requires only a few key residues at discrete locations along the 7- to 17-amino acid sequence from the PTC (5). Nevertheless, the overwhelming heterogeneity of the AZ-specific stalling sequences implies that other elements must be involved in peptide discrimination. Rare codons, internal SD-like sequences, and mRNA secondary structures could account for the slowing of ribosome movement (17, 44, 45). However, our attempts to identify codon use bias and SD-like features at or within the 90-nt upstream and downstream regions of the stalling sites were not successful. The flanking residues also do not seem to fall into any chemical side group. In silico structural analysis revealed that ∼70% of the identified AZ-arrested sequences contains one to two stable mRNA secondary structures 17−30 nt immediately downstream of the stalling sites. The significance of these structured mRNAs is unclear, although they could aid in the stabilization of the arrested ribosomes.
The binding of small molecule ligands to the ribosome tunnel can directly or indirectly alter the tunnel environment and, in turn, restrict or redirect the passage of the peptide chain to initiate translation arrest. Indeed, distinct disposition of the arrest peptide inside the ribosome tunnel and the perturbation of tunnel components can profoundly influence the stalling efficiency (4, 32). In light of these findings, we found that the presumed unique route of the MarR and EngD nascent peptides through the tunnel determines the degree of obstruction by macrolide antibiotics and helps to explain the differential stalling of the EngD peptide by two subtly different macrolides. Therefore, the ability of the nascent chains to detour away from the antibiotics may explain the bypassing of AZ-mediated translation arrest and the continual synthesis of GFP and polyphenylalanine peptides in the presence of erythromycin (13, 14). Alternatively, the growing peptide chain might induce the dissociation or displacement of the antibiotic from the ribosome (46). Finally, the ubiquity and the number of the AZ-specific stalling sites along the ORFs provide an alternative explanation for the mid- and late-stage translational inhibition and the magnitude of this inhibition.
Experimental Procedures
Bacterial Growth.
S. aureus strains RN4220 (a gift from Taeok Bae, Indiana University, Gary, IN) and NCTC 8325 (Network on Antimicrobial Resistance in Staphylococcus aureus Repository) were used in this study. S. aureus were grown in tryptic soy broth (BD Difco) at 37 °C until cultures reached an OD600 of 0.45–0.50. The cells were treated with and without 25 μg/mL of AZ for 15 min, followed by a 2-min pretreatment of 100 μg/mL chloramphenicol. Cultures were fast-cooled by pouring over ice and collected by centrifugation. Cell pellet was flash-frozen in liquid nitrogen and pulverized on a cryomiller and prepared as previously described (16, 17). Details are provided in SI Experimental Procedures.
Library Construction and Data Analysis.
Ribosome footprints were prepared as previously described, with some modifications (15, 17). Raw FastQ sequencing data were processed using a locally installed Galaxy platform. The rRNA-less reads were aligned to the NCTC 8325 reference genome (GenBank CP000253) using Bowtie v.0.12.0. The alignment .map files were used as inputs for the modified Python scripts (21). The normalized read densities were visualized in MochiView. To identify the stalling sites, differences in the read density [in reads per million mapped reads (RPM)] were subtracted between the AZ-treated and -untreated samples using in-house scripts. The gene expression levels were calculated using Cufflinks and Cuffdiff. Sequencing data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession number GSE57175).
Structural Modeling.
Nascent peptide chains were modeled by superimposing the antibiotic-bound 50S subunits of the structures 3OHZ (AZ), 3OFR (ERY), 2WWQ (TnaC), and 3J5L (ErmBL) using the phosphate backbone of the 23S rRNA. The sequences for the MarR and EngD peptides were generated by changing the residues of the leader peptide in TnaC to each of the respective sequences. Bond angles and lengths of the modified residues were assigned their standard values by energy minimization, with restraints placed on the amino acid at the stalling site. Energy minimization on the remaining residues of the peptide was performed while docked within the ribosome tunnel using the standard minimization routines provided with BioLuminate (Schrödinger).
Supplementary Material
Acknowledgments
We thank Harris Bernstein for critical reading of the manuscript and Matthew Sachs for sharing the toe-printing protocols. This work is supported by National Institutes of Health Grant R00GM094212 (to M.-N.F.Y.) and the Saint Louis University President's Research Fund. M.-N.F.Y. is a Pew Scholar in the Biomedical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE57175).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410356111/-/DCSupplemental.
References
- 1.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289(5481):920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 2.Bhushan S, et al. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17(3):313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44(23):8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 4.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006;22(5):587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, Chiba S. Arrest peptides: Cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 6.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28(22):3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108(5):629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19(3):333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci USA. 2004;101(12):4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 11.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330(5):1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 12.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151(3):508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Odom OW, Picking WD, Tsalkova T, Hardesty B. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur J Biochem. 1991;198(3):713–722. doi: 10.1111/j.1432-1033.1991.tb16071.x. [DOI] [PubMed] [Google Scholar]
- 14.Starosta AL, et al. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol. 2010;17(5):504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147(6):1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484(7395):538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueta M, Wada C, Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 2010;15(1):43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 19.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336(6083):915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156(5):950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc. 2013;8(11):2212–2239. doi: 10.1038/nprot.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor PB, Li GW, Weissman JS, Atkins JF, Baranov PV. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013;29(12):1488–1491. doi: 10.1093/bioinformatics/btt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9(5):445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34(2):201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110(10):E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 27.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 28.Bhushan S, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9(1):e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30(2):190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 30.Vázquez-Laslop N, et al. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci USA. 2011;108(26):10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez E, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51(1):35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arenz S, et al. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence MG, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol. 2008;190(17):5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sothiselvam S, et al. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA. 2014;111(27):9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151(5):1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11(3):e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284(16):10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol. 2008;384(1):73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41(3):321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 40.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283(47):32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 41.Vázquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29(18):3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun. 2008;366(4):1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- 43.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106(1):50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, et al. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol. 2013;20(5):582–588. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovmar M, et al. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem. 2006;281(10):6742–6750. doi: 10.1074/jbc.M511918200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.