Abstract
Background
One controversial source of infection for hepatitis C virus (HCV) involves the sharing of contaminated implements, such as straws or spoons, used to nasally inhale cocaine and other powdered drugs. An essential precondition for this mode of transmission is the presence of HCV in the nasal secretions of intranasal drug users.
Methods
Blood and nasal secretion samples were collected from five plasma-positive chronic intranasal drug users and tested for HCV RNA using RT-PCR.
Results
HCV was detected in all five blood samples and in the nasal secretions of the subject with the highest serum viral load.
Conclusions
This study is the first to demonstrate the presence of HCV in nasal secretions. This finding has implications for potential transmission of HCV through contact with contaminated nasal secretions.
Background
Hepatitis C virus (HCV) is a major cause of liver-related morbidity and mortality worldwide, with an estimated global prevalence of 170 million chronic infections. HCV-induced liver disease is the most common indication for liver transplantation and it has emerged as a leading cause of death among hospitalized HIV-infected patients treated during the HAART era [1]. Transmission of HCV is known to occur through contact with contaminated blood, most notably in the context of injection drug use, transfusion of blood products prior to 1992, chronic hemodialysis, occupational exposure to blood, and nosocomial and perinatal exposure. In addition, several studies have reported low levels of suspected sexual and household transmission of HCV [2].
Although much is known about the routes of HCV transmission, nearly 15% of infected individuals report no identifiable source of exposure. Unexplained cases are particularly high among drug-users who have no history of injection risk and no other identifiable risk factors [3]. One hypothesis that might account for the high number of unexplained HCV infections among noninjection drug-users was proposed by researchers at the US National Institutes of Health (NIH), who identified intranasal cocaine use as a significant risk factor for HCV among volunteer blood donors [4]. They reasoned that HCV might be transmitted through contaminated implements, such as straws or spoons, that are commonly used to nasally inhale powdered drugs, including heroin, cocaine, and methamphetamines. Chronic nasal inhalation of these substances (including the adulterants they contain) can cause tissue deterioration and bleeding of nasal membranes. Implements inserted into an eroded nasal cavity may come into contact with HCV-infected mucus or blood, which may then be transmitted to an uninfected individual sharing the same implement. The debate regarding this potential mode of transmission intensified when the National Heart, Lung and Blood Institute (NHLBI) Retrovirus Epidemiology Donor Study (REDS) was unable to confirm intranasal drug inhalation as an independent risk factor for HCV [5]. These conflicting reports prompted the American Association of Blood Banks (AABB) to add, and then shortly thereafter remove, intranasal cocaine use from their list of criteria used to screen potential blood donors. A subsequent review of the literature found serious methodological limitations with both the NIH and NHLBI/REDS studies [6]. Although HCV has been detected in the saliva, semen, and other nonserological fluids of some plasma-positive patients [7], no virological studies have been undertaken to determine whether HCV is present in the nasal secretions of intranasal drug users, a necessary precondition for internasal viral transmission. Here, we report preliminary findings on the detection of HCV RNA in the nasal secretions of plasma-positive chronic drug sniffers.
Methods
Study subjects
Five patients were recruited for the study from the Boriken Neighborhood Health Center in East Harlem, New York City. The subjects were selected from consecutive clinic patients who had previously tested HCV seropositive and who reported a history of intranasal drug use. The first four subjects were HCV antibody reactive and seropositive on quantitative PCR; the fifth subject tested seropositive for HCV antibodies but HCV PCR negative (<600 copies/mL). All subjects were male between 46 to 56 years of age and HIV-1 seropositive. Table 1 presents patient serological indicators for hepatitis C, as well as hepatitis B, HIV-1, ALT levels, and liver biopsy results.
Table 1.
Study subject serology results and indicators
| Subjects | HCV PCR Copies per ml (Date of test) | HepBsAb | HepBsAg | HepBcAb | HIV Viral Load Copies per ml | ALT U/L | Liver Biopsy |
| 1 | 10,800,000 (11/99) |
Neg | Neg | Pos | 633 (12/03) |
32 (12/03) | Activity: Mild Stage 2 of 4 (08/00) |
| 34,500,000 IU (12/03) | 270,000 (01/03) |
||||||
| 2 | 8,360,000 (09/99) |
Neg | Neg | Pos | 180,000 (10/03) |
16 (02/04) |
Not done due to bleeding diathesis |
| 96,000 (02/04) |
16 (10/03) |
||||||
| 3 | 1,320,000 (10/03) |
Pos | Neg | Pos | 16,900 (02/04) |
110 (10/03) |
Grade 2/3 of 4 Stage 2/3 of 4 (11/03) |
| 182 (02/04) |
|||||||
| 4 | 234,000 (12/02) |
Neg | Neg | Neg | 72,031 (12/02) |
37 (12/03) |
Grade 1 of 4 Stage 1 of 4 (11/03) |
| <50 (12/03) |
|||||||
| 5 | <600 | Neg | Neg | Pos | 212,000 (10/03) |
41 (10/03) |
Not done |
| 104 (04/04) |
|||||||
HCV = hepatitis C virus; HepB = hepatitis B; s = surface; c = core; Ab = antibody; Ag = antigen; HIV = human immunodeficiency virus; ALT = alanine aminotransferase enzyme
Biological samples
Blood samples were collected from each subject using standard clinical procedures for quantitative HCV RNA testing. Nasal secretion samples were obtained using a nasal swab technique. Samples from nasal swabs were placed into sterile tubes containing 1 mL of TRIzol reagent (Glibco BRL) and stored at -70°C. Study protocols were approved by an Institutional Review Board and all study participants provided voluntary informed consent.
Isolation and detection of HCV RNA
HCV RNA was isolated from serum samples by QIAamp MinElute column (Qiagen) based on manufacturer's protocol, and from nasal secretions by TRIzol (Gibco BRL) based on established protocols [8]. Briefly, nasal swab samples were subjected to vortexing for 30 s and incubated at 21°C for 5 min to permit the complete dissociation of nucleoprotein complexes. Subsequently, TRIzol solution was transferred into a new Eppendorf tube, mixed with 0.2 mL of chloroform, mixed vigorously for 15 s, and incubated at 21°C for 3 min. RNA extraction and phase separation was obtained by centrifugation at 12 000 × g for 15 min at 4°C. The aqueous phase was transferred into a new Eppendorf tube and mixed with 1 μL of RNase-free glycogen (New England Biolabs). RNA was precipitated with 0.5 mL of isopropyl alcohol for 10 min at 21°C followed by centrifugation at 12 000 × g for 10 min at 4°C and washed in 75% ethanol. RNA pellets were resuspended in 10 μL of RNase-free water.
Detection of HCV RNA was performed by RT-PCR. The first strand cDNA was synthesized by Superscript™ First Strand cDNA Synthesis kit (Invitrogen) using gene-specific downstream primers targeting the HCV p22 core region [9] with minor modification of the upstream primer (406(m)5'-TAGACCGGTGCACCATGAGC-3'). HCV cDNA was amplified by PCR through 40 cycles of denaturation (94°C-1 min), annealing (55°C-1 min) and elongation (72°C-1 min). Subsequently, PCR products were resolved through 1% agarose gel electrophoresis, hybridized to 32P-labeled internal probe (5'-AGGAAGACTTCCGAGCGGTCG CAA-3'), and exposed to Kodak film.
Results and discussion
Previous studies have demonstrated the presence of HCV in a wide variety of nonserological fluids [7,10]; for instance, HCV has been detected in about 50% of saliva samples from plasma-positive individuals [11]. We reasoned that HCV might be present in nasal secretions at a similar prevalence and would therefore be detectable among a relatively small sample of viremic patients. Five consecutive plasma-positive clinic patients were selected for study and each contributed blood and nasal secretion samples for HCV RT-PCR analysis.
The RT-PCR results confirmed the presence of HCV in the blood of all five subjects. The highest serum concentrations of viral RNA were detected in subjects 1 and 2 and the lowest in subject 5 (see Fig. 1); these findings were consistent with patient record viral loads. It is noteworthy that our assay detected HCV RNA in the serum of subject 5 (previously below the level of detection with commercially available assays), indicating low-level viremia, and verifying the high sensitivity of our analysis.
Figure 1.
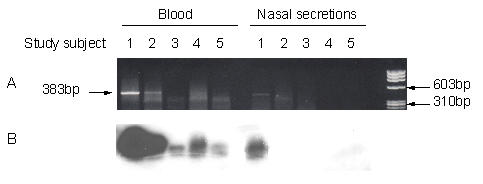
HCV RNA detection from serum and nasal secretions of five intranasal drug users. A: Ethidium bromide staining of DNA fragments; B: Southern blot hybridization
Significantly, HCV particles were also detected from the nasal secretions of subject 1 (Fig. 1). This subject, a 56-year-old African-American male, also exhibited the highest HCV serum viral load (34 500 000 IU/mL). HCV was not detected in the nasal secretions of the other four study subjects. This finding represents the first demonstration of the presence of HCV RNA in nasal secretions.
Great care was taken to avoid sample contamination during all phases of the study. Blood samples of known serology were collected from each of the five subjects and tested for HCV RNA to confirm the validity of our laboratory procedures. The TRIzol assay used on the nasal secretion samples in this study has been shown to be effective for RNA isolation with a variety of other nonserological samples [11,12].
Conclusions
To our knowledge, this is the first study to demonstrate the presence of HCV in the nasal secretions of an intranasal drug-user. While this finding does not confirm internasal viral transmission, it does lend virological support to previous indications that intranasal drug use poses a risk by confirming an important precondition for this route of infection. Additionally, detection of HCV in nasal secretions advances the debate regarding potential iatrogenic and nosocomial transmission of HCV in the context of ENT practices. More research involving larger samples is needed to replicate our results, provide an accurate estimate of HCV prevalence in the nasal secretions of plasma-positive patients, and determine whether intranasal HCV derives from blood in the nasal cavity or directly from nasal secretions. Future research should also address whether the presence HCV in the nasal cavity affects the progression of upper respiratory infections such as influenza, rhinovirus, adenovirus, coronavirus, and severe acute respiratory virus.
Authors' contributions
JM was the principal investigator; he conceived, designed and coordinated the study, and prepared the report. MS performed viral RNA isolation and RT-PCR analysis and interpretation, prepared the graphic, wrote the virology section of the report, and contributed to several revisions of the manuscript. DM recruited and enrolled study subjects from among her clinic patients, administered screening and informed consent, collected blood and nasal secretion samples, and helped with manuscript revisions. MC contributed to the design and coordination of the study and assisted with revisions. All authors reviewed the report and approved the final version.
Acknowledgments
Acknowledgments
This research was supported by the National Institute on Drug Abuse and the National Development and Research Institutes. The authors thank Dr. S. Tortu for initiating this line of investigation and for advising on aspects of the study design. Dr. G. Santos advised on the clinical protocols; L. Torres assisted with the material and procedural aspects of the study; and J. Botta provided clerical and technical assistance. The sponsors of the study had no role in study design, data collection, data analyses, data interpretation, or writing of the report.
Contributor Information
James M McMahon, Email: mcmahon@ndri.org.
Malgorzata Simm, Email: ms130@columbia.edu.
Danielle Milano, Email: dmi3070332@aol.com.
Michael Clatts, Email: clatts@ndri.org.
References
- Monga HK, Rodriguez-Barradas MC, Breaux K, Khattak K, Troisi CL, Velez M, Yoffe B. Hepatitis C virus infection-related morbidity and mortality among patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;332:240–7. doi: 10.1086/321819. [DOI] [PubMed] [Google Scholar]
- Ackerman Z, Ackerman E, Paltiel O. Intrafamilial transmission of hepatitis C virus: a systematic review. J Viral Hepatol. 2000;7:93–103. doi: 10.1046/j.1365-2893.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- Flamm SL, Parker RA, Chopra S. Risk factors associated with chronic hepatitis C virus infection: limited frequency of an unidentified source of transmission. Am J Gastroenterol. 1998;93:597–600. doi: 10.1016/S0002-9270(98)00048-3. [DOI] [PubMed] [Google Scholar]
- Conry-Cantilena C, van Raden M, Gibble J, Melpolder J, Shakil AO, Viladomiu L, Cheung L, DiBisceglie A, Hoofnagle J, Shih JW, Kaslow R, Ness P, Alter HJ. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996;334:1691–96. doi: 10.1056/NEJM199606273342602. [DOI] [PubMed] [Google Scholar]
- Murphy EL, Bryzman SM, Glynn SA, Ameti DI, Thomson RA, Williams AE, Nass CC, Ownby HE, Schreiber GB, Kong F, Neal KR, Nemo GJ. Risk factors for hepatitis C virus infection in United States blood donors. Hepatology. 2000;31:756–62. doi: 10.1002/hep.510310329. [DOI] [PubMed] [Google Scholar]
- McMahon JM, Tortu S. A potential hidden source of hepatitis C infection among noninjecting drug users. J Psychoactive Drugs. 2003;35:455–60. doi: 10.1080/02791072.2003.10400492. [DOI] [PubMed] [Google Scholar]
- Ackerman Z, Paltiel O, Glikberg F, Ackerman E. Hepatitis C virus in various human body fluids: a systematic review. Hepatol Res. 1998;11:26–40. doi: 10.1016/S1386-6346(98)00004-7. [DOI] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chan SW, McOmish F, Holmes EC, Dow B, Peutherer JF, Follett E, Yap PL, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–41. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- Liou TC, Chang TT, Young KC, Lin XZ, Lin CY, Wu HL. Detection of HCV RNA in saliva, urine, seminal fluid, and ascites. J Med Virol. 1992;37:197–202. doi: 10.1002/jmv.1890370309. [DOI] [PubMed] [Google Scholar]
- Hermida M, Ferreiro MC, Barral S, Laredo R, Castro A, Diz Dios P. Detection of HCV RNA in saliva of patients with hepatitis C virus infection by using a highly sensitive test. J Virol Methods. 2002;101:29–35. doi: 10.1016/S0166-0934(01)00417-7. [DOI] [PubMed] [Google Scholar]
- Xiang X, Qiu D, Hegele RD, Tan WC. Comparison of different methods of total RNA extraction for viral detection in sputum. J Virol Methods. 2001;94:129–35. doi: 10.1016/S0166-0934(01)00284-1. [DOI] [PubMed] [Google Scholar]


