Significance
The frequency of intraindividual mitochondrial DNA (mtDNA) polymorphisms—heteroplasmies—can change dramatically from mother to child owing to the mitochondrial bottleneck at oogenesis. For deleterious heteroplasmies such a change may transform alleles that are benign at low frequency in a mother into disease-causing alleles when at a high frequency in her child. Our study estimates the mtDNA germ-line bottleneck to be small (30–35) and documents a positive association between the number of child heteroplasmies and maternal age at fertilization, enabling prediction of transmission of disease-causing variants and informing mtDNA evolution.
Keywords: mitochondria, heteroplasmy
Abstract
The manifestation of mitochondrial DNA (mtDNA) diseases depends on the frequency of heteroplasmy (the presence of several alleles in an individual), yet its transmission across generations cannot be readily predicted owing to a lack of data on the size of the mtDNA bottleneck during oogenesis. For deleterious heteroplasmies, a severe bottleneck may abruptly transform a benign (low) frequency in a mother into a disease-causing (high) frequency in her child. Here we present a high-resolution study of heteroplasmy transmission conducted on blood and buccal mtDNA of 39 healthy mother–child pairs of European ancestry (a total of 156 samples, each sequenced at ∼20,000× per site). On average, each individual carried one heteroplasmy, and one in eight individuals carried a disease-associated heteroplasmy, with minor allele frequency ≥1%. We observed frequent drastic heteroplasmy frequency shifts between generations and estimated the effective size of the germ-line mtDNA bottleneck at only ∼30–35 (interquartile range from 9 to 141). Accounting for heteroplasmies, we estimated the mtDNA germ-line mutation rate at 1.3 × 10−8 (interquartile range from 4.2 × 10−9 to 4.1 × 10−8) mutations per site per year, an order of magnitude higher than for nuclear DNA. Notably, we found a positive association between the number of heteroplasmies in a child and maternal age at fertilization, likely attributable to oocyte aging. This study also took advantage of droplet digital PCR (ddPCR) to validate heteroplasmies and confirm a de novo mutation. Our results can be used to predict the transmission of disease-causing mtDNA variants and illuminate evolutionary dynamics of the mitochondrial genome.
The centerpiece of cellular metabolic machinery—the mitochondrion—harbors a 16.5-kb genome, mitochondrial DNA (mtDNA). Mutations in mtDNA cause over 200 diseases and contribute to diabetes, cancer, male infertility, Parkinson’s and Alzheimer’s diseases (1). In mammals, mtDNA mutates at high rates and is maternally inherited, making it a popular marker in evolutionary genetics (2). Despite its importance, mtDNA has drifted away from the spotlight eclipsed by nuclear DNA studies (3), and there are still gaps in our understanding of the basic aspects of human mtDNA biology. The lack of cures for diseases caused by mtDNA mutations makes it critical to understand how these mutations arise and are transmitted between generations.
Heteroplasmy, the presence of more than one mtDNA variant in a cell or a tissue, is the result of a de novo mtDNA mutation occurring in an individual or inherited through the maternal lineage. Currently there is no consensus about how prevalent mtDNA heteroplasmy is in human populations (4, 5). Such knowledge is crucial for assessing the load of mtDNA pathogenic mutations, formulating prognoses for patients with mtDNA diseases, and preimplantation diagnostics after mtDNA replacement in oocytes. Most mtDNA diseases are heteroplasmic and their phenotype depends on the allele frequency of the pathogenic variant (1).
Heteroplasmy levels can change dramatically between generations owing to genetic drift during the germ-line bottleneck—a reduction in the number of mtDNA segregating units during oogenesis (6–8). The size of the bottleneck for mice has been evaluated to be 185 (9), yet for humans this size is difficult to obtain experimentally. Published estimates of the human bottleneck size are too broad [1–200 (10, 11)] to be useful in predicting the transmission of disease variants. Genetic drift theory predicts that a small bottleneck size will result in drastic shifts in heteroplasmy levels from a mother to her child, potentially reaching nondisease levels or levels with higher disease severity. After fertilization, mtDNA variants are distributed among cells owing to mitotic segregation—the random partitioning of mitochondria during cell divisions (12). We also lack an accurate estimate of the germ-line mtDNA mutation rate in humans, with pedigree and phylogenetic studies producing conflicting results (13, 14).
To conduct a population study of heteroplasmy transmission, we analyzed full-length mtDNA in 39 mother–child pairs using the MiSeq platform. Accounting for PCR and sequencing errors, we were able to accurately score heteroplasmies with allele frequency above 1%. With these data, we addressed (i) how common heteroplasmy is in a human population, (ii) how heteroplasmy frequency changes between tissues of the same individual and between generations, and (iii) whether maternal age at conception influences heteroplasmy occurrence in a child. We also estimated the size of the germ-line mtDNA bottleneck and the germ-line mutation rate via population genetics modeling of heteroplasmies. Focused on heteroplasmy inheritance in healthy individuals, our data serve as a valuable baseline to study disease associations for mtDNA and provide important insights into mtDNA evolution.
Results
Samples, mtDNA Enrichment, and Sequencing.
We studied the prevalence of mtDNA heteroplasmy in blood and buccal cells from 39 mother–child pairs residing in central Pennsylvania, analyzing 156 samples (39 mothers × 2 tissues + 39 children × 2 tissues) grouped in sets of four (two tissues from a mother and two tissues from her child). Total genomic DNA was isolated from each sample. Haplogroup analysis conducted via Sanger sequencing of the D-loop indicated European ancestry for all families (Dataset S1, Table S1). For each sample, we amplified mtDNA from total DNA in two overlapping 9-kb fragments and sequenced them with paired-end 250-bp reads on a MiSeq instrument (Materials and Methods). This enriched for mtDNA and minimized the presence of numts (Fig. S1 and SI Materials and Methods), the majority of which are short (15). Multiplexing 12 samples per run resulted in ∼106 read pairs per sample. We confirmed the efficacy of our approach for mtDNA enrichment by applying it to Rho0 cells not harboring mitochondria (Fig. S2). To minimize potential contamination among samples, we followed previously devised guidelines (16) and used pUC18 and PhiX174 as spike-ins (Materials and Methods).
Heteroplasmy Discovery.
The sequencing read pairs were mapped to human mtDNA and nuclear genomes. On average, 85% of the reads per sample mapped to mtDNA (97% for samples without a spike-in; Fig. S3). We then applied our heteroplasmy discovery pipeline (Materials and Methods and Fig. S4). We required both reads of the pair to map uniquely, and in a proper orientation, to the reference mtDNA (Fig. S5). To compute minor allele frequency (MAF) at each site for each sample, we used bases with sequencing quality ≥30 from reads with mapping quality ≥20 (other thresholds led to almost identical results; Fig. S6). The mean sequencing depth per sample (averaged across sites) was 19,789× ± 770× (mean ± SEM). Tabulating depth on a per-site basis, 90% of sites in the mitochondrial genome were sequenced at ≥7,858× per sample (Fig. S7). The proportion of spike-in reads aligning to their respective references was as anticipated, suggesting absence of contamination among adjacent samples (Fig. S3).
In the search for heteroplasmies, we first identified sites with MAF ≥1% in individual samples. The sequencing depth per site required to detect true heteroplasmies with MAF ≥1% over the base quality error (0.1% for Phred score 30) with 99% power is 839× per site (one-sided power calculation for one-sample proportion test). Conservatively, we rounded up the depth requirement to 1,000×. A detection limit of MAF ≥1% allows detection of inherited and de novo variants that pass through the bottleneck if its size is <100. Mutations with lower frequency are accounted for with population genetics modeling (discussed below). After filtering for potential sequencing artifacts (Dataset S1, Table S2 and Materials and Methods), we identified 174 point heteroplasmies distributed among 100 quartets—groups of site-specific heteroplasmy frequencies from two tissues of a mother and two tissues from her child (Dataset S1, Table S3). These heteroplasmies were found in 31 families (eight families had no heteroplasmies).
Statistical Validation of Point Heteroplasmies.
To validate heteroplasmies, we used a novel statistical method that identifies heteroplasmic sites via a likelihood function accounting for instrument sequencing and mapping errors (SI Materials and Methods). With this method, all 174 point heteroplasmies were significant (P < 0.0003 for each site; Dataset S1, Table S4). Additionally, the allele counts for all 174 heteroplasmies tested were significant (P < 0.0003 for 172 sites, and P < 0.03 for the remaining two sites; Table S4) based on the variability observed for the same position among all samples (17).
Experimental Validation of Point Heteroplasmies.
We used Sanger sequencing to test all point heteroplasmies with MiSeq MAF ≥10% (Sanger method detection limit, Fig. S8A and Dataset S1, Table S5) in at least one sample per family and the corresponding sites from the other samples from the same family (we always sequenced newly amplified fragments). In total, we examined 21 sites × 4 samples = 84 sites, 44 of which had MiSeq MAF ≥10% (Dataset S1, Table S6). The presence of heteroplasmy was successfully validated in all these 44 cases. Thus, our false-positive rate for detecting heteroplasmies with MAF ≥10% is below 0.023 (1/44). The MAFs from the MiSeq and Sanger methods were well correlated (R2 = 75%; Fig. S9A).
A set of point heteroplasmies with MiSeq MAF <10% was analyzed with droplet digital PCR (ddPCR) (18), which can detect heteroplasmies with MAF >0.2% (Fig. S8 B and C and Dataset S1, Table S7). Here we analyzed point heteroplasmies with MiSeq MAF between 1% and 10% in at least one sample per family and the corresponding sites from the other samples of the same family, a total of 10 sites × 4 samples = 40 sites, 18 of which had MiSeq MAF ≥1% (Fig. S9B and Dataset S1, Table S8). When we assayed the original amplicons used for MiSeq sequencing, the presence of heteroplasmy was confirmed in all these 18 instances. However, when we reamplified mtDNA from these 18 samples, in two instances (site 11,616 in M203C5-ch and site 11,825 in M210-bl) ddPCR did not confirm the presence of heteroplasmy. Repeating amplification and ddPCR for a third time again did not detect heteroplasmy (Dataset S1, Table S8), suggesting PCR errors in the amplicons sequenced with MiSeq. Thus, our false-positive rate for detecting heteroplasmies with MAF between 1% and 10% is 0.11 (2/18). Overall, the MAFs from the MiSeq and ddPCR methods were well correlated for the sequenced and newly amplified amplicons (R2 = 95% and 79%, respectively; Fig. S9B).
Distribution of Point Heteroplasmies.
After removing two sites that failed to validate with ddPCR (discussed above), we retained 172 point heteroplasmies in 98 quartets (Dataset S1, Table S3). We assumed that these 98 point mutations arose independently in the families analyzed (or in their maternal ancestors). Point heteroplasmies were found at 87 unique mtDNA positions (Fig. S10). Six positions (185, 189, 214, 215, 16,093, and 16,183) were heteroplasmic in multiple families (four, three, three, two, three, and two families, respectively; Dataset S1, Table S3), likely owing to high mutation rate at the D-loop (13). Each mother on average carried 1.13 ± 0.04 heteroplasmies in her blood. This value was similar for maternal buccal tissue and for buccal and blood tissues of children (Fig. S11). Among the 98 point heteroplasmies 96 were transversions, resulting in a transition-to-transversion ratio of 48 (Fig. S10 and Dataset S1, Table S3).
There were significantly more and significantly fewer heteroplasmies in the D-loop and protein-coding regions, respectively, than expected based on their length and assuming equal propensity to harbor a heteroplasmy along mtDNA (Table 1). A high mutation rate for the D-loop had been documented previously (13). The nonsynonymous-to-synonymous rate ratio (dN/dS) at (concatenated) protein-coding genes was significantly lower than 1 (P = 5 × 10−3; Fisher’s exact test; Dataset S1, Table S9), suggesting purifying selection (19). Most nonsynonymous mutations were predicted to affect protein function (Dataset S1, Table S10).
Table 1.
The distribution of point heteroplasmies among mtDNA regions
| Region | bp | Observed | Random | Neutral |
| D-loop | 1,122 | 34 | 6.6* | 7.9* |
| tRNA | 1,508 | 6 | 8.9 | 10.6 |
| rRNA | 2,513 | 13 | 14.9 | 17.7 |
| Protein S | 2,834 | 20 | 16.8 | 20.0 |
| Protein N | 8,533 | 25 | 50.5* | 60.2* |
| Intergenic | 88 | 0 | 0.5 | 0.6 |
| Total | 16,569† | 98 | N/A | 197 |
The observed numbers of heteroplasmies and the numbers expected under random and neutral (based on the frequency at synonymous sites) expectations are shown. The numbers of synonymous (S) and nonsynonymous (N) sites were calculated with the Nei–Gojobori method. N/A, not applicable.
Significantly different from observed (P < 0.05, test comparing two proportions).
Owing to overlapping annotations and exclusion of stop codons, the sum of base pairs in regions does not sum up to the overall length of mtDNA.
Disease-Associated Mutations and Mutation Burden.
Eight families harbored eight point heteroplasmies (one per family) that can cause disease when present at high allele frequencies (Table 2). Among 39 mothers, 5 (or 1 in 8) were carriers of disease-associated mtDNA mutations in at least one of the two tissues analyzed. Mutations at four of the eight sites are associated with disease when homoplasmic for the mutant allele (20–23); however, in our data these were heteroplasmic (Table 2). For the other four of the eight sites above, disease can develop even when mutant alleles are heteroplasmic—with disease severity depending on the allele frequency. For A1555G, G13708A, and G3242A mutations, the allele frequencies were much lower than disease-associated frequencies (Table 2) (24–26), suggesting lack of symptoms. Mutations at tRNA-Leu sites 3,242 and 3,243 contribute to several mitochondrial diseases (25, 27); notably, allele frequencies observed at site 3243 in the child of family M512 (Table 2) were comparable to those observed in mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes patients (27).
Table 2.
Disease-causing heteroplasmies
| Site | Region | Wild type | Mutant | Amino acid change | Family | Mother cheek | Mother blood | Child cheek | Child blood | Diseases caused by the mutant allele | Disease allele frequency (ref.) |
| 195 | D-loop | T | C | — | M494 | 0.084 | 0.012 | 0.002 | 0.001 | Bipolar disorder | 1.0 (20) |
| 1,391 | 12S | T | C | — | M513 | 0.040 | 0.027 | 0.001 | 0.000 | HCM | 1.0 (21) |
| 1,555 | 12S | A | G | — | M520 | 0.002 | 0.001 | 0.014 | 0.014 | Deafness | >0.52 (24) |
| 2,352 | 16S | C | T | — | SC16 | 0.429 | 0.437 | 0.247 | 0.245 | LVNC | 1.0 (22) |
| 3,242 | Leu | G | A | — | M242 | 0.001 | 0.001 | 0.008 | 0.016 | RTD | >0.49 (25) |
| 3,243 | Leu | A | G | — | M512 | 0.335 | 0.144 | 0.686 | 0.611 | MELAS, MIDD; MERRF; CPEO | >0.5 (27) |
| 12,634 | ND5 | A | G | I to V | M203 | 0.001 | 0.025 | 0.001 | 0.001 | Thyroid cancer (cell line) | 1.0 (23) |
| 13,708 | ND5 | G | A | A to T | SC8 | 0.001 | 0.000 | 0.022 | 0.016 | LHON | >0.92 (26) |
The allele frequencies for the mutant allele in maternal and child tissues are indicated. CPEO, chronic progressive external ophthalmoplegia; HCM, hypertrophic cardiomyopathy; LHON, Leber’s hereditary optic neuropathy; LVNC, left ventricular noncompaction; MELAS, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; MERRF, myoclonic epilepsy with ragged-red fibers; MIDD, maternally inherited diabetes deafness; RTD, renal tubular dysfunction.
Transmission of Heteroplasmies.
Considering heteroplasmy-containing quartets (Dataset S1, Table S3), we used the presence of heteroplasmy with MAF ≥1% in at least one sample from a family as a “prior” to support the existence of heteroplasmy at the same position for other samples of the same family if their MAF was ≥0.2% (greater than or equal to twice the value of the allowed sequencing quality error of 0.1%—Phred score 30). We classified 98 quartets into five categories (Table 3 and Dataset S1, Table S3) based on whether heteroplasmies were present in (i) both tissues of a mother and both tissues of her child (category “all,” which included dramatic shifts in allele frequency from mother to child, suggesting the germ-line bottleneck); (ii) both tissues of a mother, but absent from both tissues of her child (category “mother,” suggesting loss of a variant in the child owing to the germ-line bottleneck); (iii) both tissues of a child, but absent from both tissues of a mother (category “child” with candidate germ-line de novo mutations); (iv) both tissues of a mother and one tissue of a child, or in one tissue of a mother and both tissues of a child (category “somatic loss,” suggestive of a change in MAF in tissues owing to mitotic segregation) (12); and (v) one tissue of one individual of a family (category “somatic gain” with candidate somatic de novo mutations).
Table 3.
Categories of quartets with examples
| Category | Total | Family | Site | mj | mn | M cheek | M blood | C cheek | C blood |
| All | 22 (11,−) | M188 | 5,107 | C | T | 0.152 | 0.165 | 0.228 | 0.225 |
| Child | 16 (2,3) | M500 | 4,191 | A | T | 0.000 | 0.000 | 0.047 | 0.056 |
| Mother | 22 (4,5) | M494 | 9,196 | G | A | 0.032 | 0.030 | 0.000 | 0.000 |
| SG | 13 (−,2*) | M137 | 8,953 | A | G | 0.000 | 0.013 | 0.000 | 0.000 |
| SL | 25 (4,−) | M236 | 6,791 | A | G | 0.002 | 0.000 | 0.015 | 0.015 |
The total number of quartets (with the number validated with Sanger sequencing and ddPCR in parentheses, asterisk indicates failed validations), followed by an example with family, site, major (mj) and minor (mn) alleles, and MAFs in maternal (M) and child (C) tissues. SG, somatic gain; SL, somatic loss.
Site 4191 in family M500 seemed to harbor a de novo mutation in the child. ddPCR confirmed complete absence of the mutant allele in both maternal tissues but presence in both tissues of the child (with MAF of 4.7% and 5.6% in buccal and blood tissues, respectively; Table 3 and Dataset S1, Table S8). Examination of hair from the same individuals indicated homoplasmy in the mother and MAF of 1.2% in the child (Dataset S1, Table S11), confirming emergence of a novel allele.
The changes in allele frequencies between tissues of an individual, or between two generations, tabulated for our 98 quartets (Dataset S1, Table S3) followed an approximately normal distribution with mean zero (Fig. S12), corroborating the action of genetic drift as the major force affecting heteroplasmy allele frequencies (28). A decrease in allele frequency for a variant from mother to child will be indicative of purifying selection (29). When we plotted the relative change in allele frequency between mothers and children (Fig. S13), such a decrease was significant for nonsynonymous sites (P = 9.54 × 10−7, one-tailed nonparametric sign test), suggestive of purifying selection. Consistent with selection operating against transmission of nonsynonymous mutations, we observed a significantly lower proportion of these mutations among transmitted heteroplasmies (5 out of 43, or 12%, in “all” and “somatic loss” categories) compared with untransmitted heteroplasmies (8 out of 22, or 36%, in “mother” category; P = 0.025, Fisher’s exact test).
Comparing MAFs Between Tissues and Generations.
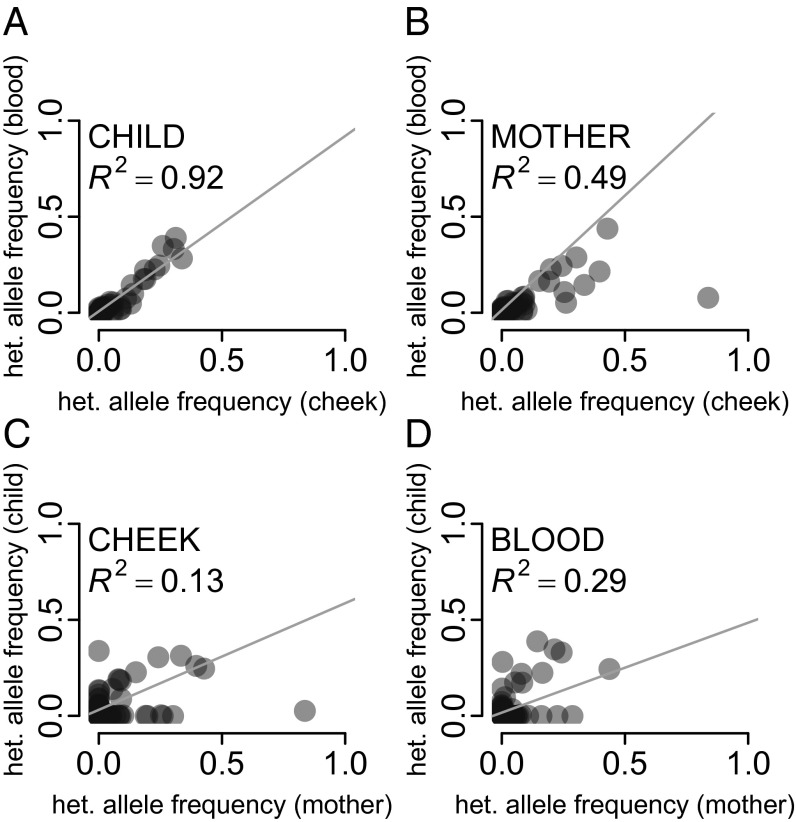
Compared with mtDNA in maternal tissues, mtDNA in child tissues underwent fewer mitotic segregations and replications and was exposed to mutagens for a shorter time. Therefore, we expect heteroplasmy allele frequency at a site to diverge less in the tissues of a child than in those of a mother. Indeed, the allele frequencies for the sites tabulated as quartets (Dataset S1, Table S3) were more strongly correlated between the two tissues for children (R2 = 92%, Fig. 1A) than between the two tissues for mothers (R2 = 49%, Fig. 1B). Stronger correlation for allele frequencies was observed between two tissues of a mother or of a child (discussed above) than between a mother and a child for the same tissue (R2 = 13% for buccal, Fig. 1C; R2 = 29% for blood, Fig. 1D), likely owing to the stronger action of the mtDNA germ-line bottleneck relative to mitotic segregation.
Fig. 1.
Correlation in heteroplasmy allele frequencies between (A) the two tissues of children, (B) the two maternal tissues, (C) buccal tissues of mothers and children, and (D) blood of mothers and children.
Maternal Age Effect.
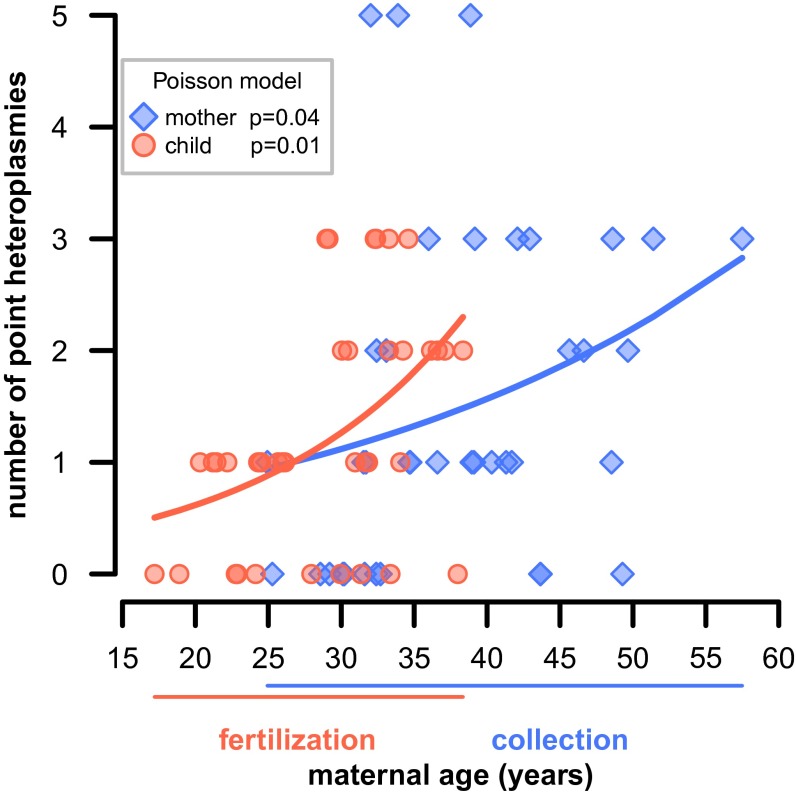
We explored the relationship between age and the total number of point heteroplasmies for each individual. No association was found for children. For mothers we found a significant positive association (P = 0.039, 0.049, and 0.055 for combined, buccal, and blood heteroplasmies, respectively, Poisson regression; Fig. 2 and Fig. S14). Thus, older mothers accumulate more mutations in their somatic tissues, with the number of point heteroplasmies tripling over 30 y of life. Intriguingly, a positive association exists between the number of heteroplasmies in children and maternal age at fertilization (P = 0.010, 0.005, and 0.006, for combined, buccal, and blood heteroplasmies, respectively; Fig. 2 and Fig. S14). This suggests that older mothers accumulate more mutations in their germ-line tissues. In our dataset, there was a correlation between maternal age at fertilization and maternal age at sampling (R2 = 43%, P = 0.002, linear regression), and we found that older mothers, who also had children later, likely transferred a larger number of accumulated mutations to their children (Fig. 2); whereas mothers who conceived under the age of 20 transmitted zero to one heteroplasmies, this number was two to three for mothers conceiving in their late 30s.
Fig. 2.
Maternal age effect. The dependence of the total number of heteroplasmies (blue) in mothers on their age at collection and (red) in children on maternal age at fertilization. Poisson generalized linear model fitted curves are indicated.
Estimating the Size of the Germ-Line mtDNA Bottleneck and Mutation Rate.
Because most heteroplasmy allele frequency changes between the two generations are consistent with genetic drift (Fig. S12), we can estimate the effective size of the germ-line bottleneck, that is, the size of the bottleneck in a traditional population model required to explain the observed amount of genetic drift [the actual number of mtDNA molecules passing through the bottleneck might be different, because they might segregate in units (8)]. Following the method developed by Millar, Hendy, and coworkers (30, 31), we assume that a child samples mutant mtDNA alleles at a given site from a binomial distribution with parameters p, the maternal MAF in the germ line (estimated here from somatic tissues), and N, the germ-line bottleneck size (SI Materials and Methods). Then the variance of the child’s heteroplasmy frequency at conception, or genetic variance, is σ2gen = p(1 − p)/N. Solving for N, we obtain N = p(1 − p)/σ2gen. We estimate the genetic variance as σ2gen = σ2raw − 4σ2measure, where σ2raw is the squared difference between the maternal and the child MAF at the site and σ2measure is the uncertainty in measuring heteroplasmy frequency (includes sampling, PCR, and sequencing errors), which we estimated from sequencing amplified D-loop–containing clones (SI Materials and Methods; σ2measure was multiplied by 4 because we are taking four measurements). This procedure produced an estimate of N in a quartet. To minimize false positives, we applied this approach to quartets where heteroplasmy was present in both maternal tissues (51 quartets in which at least one tissue in the mother had heteroplasmy with MAF ≥1% and the other tissue had MAF ≥0.2%; Dataset S1, Table S3), and thus likely was present in the maternal germ line. The median estimated N across these 51 quartets, when MAFs were averaged between the two maternal tissues and (separately) between the two child tissues, was 32.3 [interquartile range (IQR) 10.5–103.3; Fig. S15]. Similar estimates of bottleneck size were obtained when only blood (median N = 33.5, IQR 14.1–79.6) or only buccal tissues were used (median N = 29.8, IQR 9.5–68.1), and when quartets with nonsynonymous mutations were excluded (median N = 31.9, IQR 8.8–99.2). Accounting for the variance owing to mitotic segregation (SI Materials and Methods) led to median N = 35.0 (IQR 10.0–141.4; Fig. S15). Also, assuming that the single germ-line mutation we observe (at site 4191 in family M500, Table 3) originated in a single mtDNA segregating unit in the maternal germ line, and that its MAF in the child’s zygote was 3.8% (averaged across three tissues), we can estimate N as 1/0.038 = 26.3.
Next, we estimated the mtDNA germ-line mutation rate μ as in Millar, Hendy, and coworkers (30, 31). Assuming that new mutations enter the germ line at rate α, and that they are neutral and have equal probability to be transmitted to the next generation, only 1/N of them will go to fixation, leading to μ = α/N. A heteroplasmy can only be observed when its MAF is above a detection threshold θ. Analytically, it was shown (30, 31) that most heteroplasmies are lost without reaching θ, that most heteroplasmies reaching θ do not go to fixation, and that the rate of observed heteroplasmies can be approximated as μ0 = 2αln(1/θ − 1). Solving for α, one obtains α = μ0/(2 ln(1/θ − 1)). Thus, μ = μ0/(2N ln(1/θ − 1)). Setting θ = 0.01 (our detection threshold) results in μ = 0.109 μ0/N. Having observed 51 germ-line point heteroplasmies among 39 mothers, we estimated μ0 as 51/(39 × 16,569 bp) = 7.9 × 10−5 heteroplasmies per transmission per site. Using N = 32.3, we thus estimated the mutation rate μ as 2.7 × 10−7 mutations per site per generation (IQR 8.3 × 10−8 to 8.2 × 10−7), or, assuming a generation time of 20 y, 1.3 × 10−8 mutations per site per year (IQR 4.2 × 10−9 to 4.1 × 10−8). The mutation rate estimate excluding nonsynonymous sites was 4.4 × 10−7 mutations per site per generation (IQR 1.4 × 10−7 to 1.6 × 10−6), or 2.2 × 10−8 mutations per site per year (IQR 7.0 × 10−9 to 7.9 × 10−8). That for the D-loop was 1.5 × 10−6 mutations per site per generation (IQR 4.8 × 10−7 to 4.7 × 10−6), or 7.7 × 10−8 mutations per site per year (IQR 2.4 × 10−8 to 2.4 × 10−7).
Indel Heteroplasmies.
Using the same thresholds (MAF ≥1% and depth >1,000×), we identified 120 instances of small indels affecting 10 unique mtDNA sites and 28 families (Dataset S1, Table S12 and SI Materials and Methods). All indels occurred in repeats—eight in homopolymer runs, one in a 9-bp tandem repeat, and one in a CA repeat. The latter indel was validated with ddPCR (Dataset S1, Table S13). The MAFs of indels in our samples were above microsatellite sequencing errors for our long-range PCR protocol (SI Materials and Methods). Further experimental validation will allow us to determine indel MAFs more accurately.
Discussion
Prevalence of Heteroplasmy in Humans.
mtDNA heteroplasmy has strong associations with neurodegenerative diseases, aging, and tumorigenesis (1). Our results indicate that an individual carries on average one heteroplasmic variant with allele frequency ≥1% pointing to the ubiquitous occurrence of heteroplasmy (4) and are in remarkable agreement with a recent analysis of the 1,000 Genomes Project data (19) as well as several smaller-scale studies (5, 11, 32–34) (Dataset S1, Table S14).
Maternal Age Effect.
A positive association between an individual’s age and the number of heteroplasmies in postmitotic somatic tissues had already been demonstrated (e.g., refs. 35 and 36). Here, we found evidence for it in the dividing tissues as well. Kennedy et al. (35) found that the frequency of point mutations in brain increases fivefold during 80 y of life. In our data, the number of heteroplasmies in the maternal buccal and blood tissues triples over 30 y. Likewise, with high transition-to-transversion ratio in our data, we do not find transversion-causing oxidative damage (37) to be the major driver of such mutation accumulation.
The positive association we found between maternal age at conception and the number of heteroplasmies in her child has important medical implications. The frequencies of large mitochondrial deletions (38) and the T414G mutation (39) were shown to increase in oocytes as a function of age—consistent with altered mitochondrial cytochemistry and a mutagenic environment with increased glycation and carbonyl stress in aging oocytes (40). However, the number of oocytes with defective mitochondria is significantly reduced during oogenesis (41). Our results suggest that, despite this process, some oocytes with suboptimal mitochondria (e.g., with negatively selected amino acid changes), which are more likely to occur in older women, do proceed to fertilization. This predicts an increase in mtDNA diseases in children born to older mothers—a prediction not evaluated to date—and could be one of the reasons for a lower success rate of assisted reproduction in older women (42).
Germ-Line Bottleneck Size.
Our results support a severe germ-line bottleneck—with effective size of only 30–35—and are particularly striking given ∼100,000 mtDNAs in mature human oocytes (12). Our findings corroborate strong shifts in heteroplasmy frequency observed in Holstein cows (43, 44) but are more robust because they are based on more accurate estimation of MAF changes at many sites, in two tissues, and for a large number of transmissions from multiple families. Moreover, results obtained for other species are not directly applicable to humans, especially for the purposes of genetic counseling. Most previous human studies analyzed one or two sites, some of which were disease-associated, and usually a small number of transmissions (Dataset S1, Table S15). Our estimate is comparable to those in some earlier studies [e.g., 36–180 (45)], higher than in some other studies [e.g., 1–5 (10)], but substantially lower than the recently proposed estimate of 200 (11).
The number 30–35 is obtained by taking medians over effective bottleneck sizes estimated for individual transmission sites, which show a broad variation (Fig. S15). Some of this variation is random, because only one transmission was examined for each site. Selection acting at some sites might have contributed to this variation as well; however, the median bottleneck size remained very similar when nonsynonymous sites were removed. Another contributor to the observed variation in bottleneck size might be the variation among women (46, 47). Future studies examining multiple offspring per mother will allow one to evaluate the differential contribution of these factors to the variability in the bottleneck size in more detail.
Germ-Line Mutation Rate.
The germ-line mutation rate estimated here for mtDNA is an order of magnitude higher than that for the human nuclear genome [1.2 × 10−8 mutations per site per generation (48)], in agreement with previous studies (1). It is similar to estimates obtained in phylogenetic studies (e.g., refs. 13 and 14) and an order of magnitude lower than estimates in most pedigree studies (13, 49–51) (Dataset S1, Table S16). In part this is due to the fact that analyzing two tissues allowed us to identify germ-line (and discard somatic) heteroplasmies (51). However, we also had to perform strict filtering of candidate heteroplasmic sites to minimize sequencing artifacts when estimating the bottleneck size—which may have led to the removal of some real heteroplasmies. Our mtDNA mutation rate estimate should therefore be seen as a “lower bound” (our bottleneck size estimate is not affected by this potential limitation). In agreement with this, our estimate is only two- and fourfold higher than estimates from mutation accumulation cell lines for Caenorhabditis elegans and Drosophila melanogaster mtDNA (9.7 × 10−8 and 6.2 × 10−8 mutations per site per generation, respectively) (52, 53); human mutation rates were shown to be approximately fivefold the rates in these species (54, 55).
Disease-Causing Mutations.
The high prevalence of mtDNA disease-associated mutations found here—with one carrier in eight individuals—is similar to that reported from the 1,000 Genomes Project data (19) and has important practical implications. Indeed, as we demonstrated, the severe germ-line bottleneck can lead to drastic changes in allele frequencies between generations, potentially affecting the manifestation of 200 diseases caused by mtDNA mutations. In one instance we found a disease-associated mutation present at high allele frequencies in child tissues. Genetic background (both mtDNA and nuclear), known to significantly modulate mtDNA disease manifestation (56), may be preventing symptoms in this individual.
Materials and Methods
Sample Collection, DNA Isolation, and Sequencing.
DNA from buccal and blood cells (collected under IRB 30432EP) was isolated as described (32). To determine mtDNA haplogroup, mtDNA was amplified and sequenced using the Sanger method (Dataset S1, Table S17). Before MiSeq sequening, mtDNA was amplified in two ∼9-kb amplicons (16) that were mixed at an equimolar ratio and spiked with 5% (wt/wt) of pUC18 or PhiX174 DNA, or with no spike-in. Sequencing libraries were prepared according to the customized Nextera XT protocol (57).
Experimental Validation of Heteroplasmic Sites.
The primers used for heteroplasmy validation with Sanger sequencing are listed in Dataset S1, Table S17. For ddPCR, we followed the manufacturer’s protocol. TaqMan probes are listed in Dataset S1, Table S18. All experiments were performed in duplicates. To assess the detection limit for ddPCR and Sanger sequencing, we examined artificially mixed variant alleles at predetermined frequencies (SI Materials and Methods).
Preprocessing of Next-Generation Sequencing Data.
Parameters and versions of all tools are listed in Dataset S1, Table S19. The sequencing read pairs were mapped to chrM and hg19 (Fig. S4). For the pair to be retained, we required both reads to (i) map to chrM, (ii) map properly in a pair, (iii) have read length ≥100 bp, and (iv) not form a chimeric alignment.
Identification of Point Heteroplasmic Sites.
The tools Naive Variant Caller and Variant Annotator implemented in Galaxy (16) were used to extract the counts of each nucleotide per position in each strand. We selected sites with MAF ≥1% and depth ≥1,000× . We discarded sites with MAF <1% on one strand or with strand bias >1 (58), low complexity regions as annotated in ref. 32, sites at positions 3106–3107, and sites with the proportion of reads supporting an alternative base within the first and last 25 bp >85%.
Supplementary Material
Acknowledgments
We are grateful to Jessica Beiler, MPH, for coordinating sample collection, to clinical nurses from Penn State College of Medicine Pediatric Clinical Research Office, to Lily Borhan for collecting the samples, and to volunteers for donating the samples. Bonnie Higgins isolated DNA from hair for family M500. Michael DeGiorgio made useful comments on the earlier drafts of the manuscript and Prabhani Kuruppummulage Don provided statistical advice. This work was funded by Battelle Memorial Institute, the Huck Institutes of Life Sciences and Eberly College of Sciences at Pennsylvania State University, and Penn State Clinical and Translational Science Institute. Additional funding was provided, in part, under a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds. The department specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRP047378).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409328111/-/DCSupplemental.
References
- 1.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galtier N, Nabholz B, Glémin S, Hurst GD. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol Ecol. 2009;18(22):4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- 3.Pesole G, et al. The neglected genome. EMBO Rep. 2012;13(6):473–474. doi: 10.1038/embor.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payne BA, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22:384–90. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87(2):237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cree LM, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40(2):249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 7.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40(12):1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 8.Cao L, et al. New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS Genet. 2009;5(12):e1000756. doi: 10.1371/journal.pgen.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14(2):146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 10.Marchington DR, Hartshorne GM, Barlow D, Poulton J. Homopolymeric tract heteroplasmy in mtDNA from tissues and single oocytes: Support for a genetic bottleneck. Am J Hum Genet. 1997;60(2):408–416. [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, et al. Very low-level heteroplasmy mtDNA variations are inherited in humans. J Genet Genomics. 2013;40(12):607–615. doi: 10.1016/j.jgg.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulton J, et al. Transmission of mitochondrial DNA diseases and ways to prevent them. PLoS Genet. 2010;6(8):6. doi: 10.1371/journal.pgen.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons TJ, et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15(4):363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 14.Henn BM, Gignoux CR, Feldman MW, Mountain JL. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol Biol Evol. 2009;26(1):217–230. doi: 10.1093/molbev/msn244. [DOI] [PubMed] [Google Scholar]
- 15.Simone D, Calabrese FM, Lang M, Gasparre G, Attimonelli M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics. 2011;12:517. doi: 10.1186/1471-2164-12-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickins B, et al. Controlling for contamination in re-sequencing studies with a reproducible web-based phylogenetic approach. Biotechniques. 2014;56(3):134–136, 138–141. doi: 10.2144/000114146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13(5):R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci USA. 2014;111(29):10654–10659. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollins B, et al. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PLoS ONE. 2009;4(3):e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prasad GN, et al. Novel mitochondrial DNA mutations in a rare variety of hypertrophic cardiomyopathy. Int J Cardiol. 2006;109(3):432–433. doi: 10.1016/j.ijcard.2005.07.065. [DOI] [PubMed] [Google Scholar]
- 22.Tang S, et al. Left ventricular noncompaction is associated with mutations in the mitochondrial genomes. Mitochondrion. 2010;10(4):350–357. doi: 10.1016/j.mito.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Abu-Amero KK, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene. 2005;24(8):1455–1460. doi: 10.1038/sj.onc.1208292. [DOI] [PubMed] [Google Scholar]
- 24.del Castillo FJ, et al. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40(8):632–636. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wortmann SB, et al. Mitochondrial DNA m.3242G > A mutation, an under diagnosed cause of hypertrophic cardiomyopathy and renal tubular dysfunction? Eur J Med Genet. 2012;55(10):552–556. doi: 10.1016/j.ejmg.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Du W-D, et al. A simple oligonucleotide biochip capable of rapidly detecting known mitochondrial DNA mutations in Chinese patients with Leber’s hereditary optic neuropathy (LHON) Dis Markers. 2011;30(4):181–190. doi: 10.3233/DMA-2011-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, et al. The study of mitochondrial A3243G mutation in different samples. Mitochondrion. 2009;9(2):139–143. doi: 10.1016/j.mito.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Chinnery PF, et al. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16(11):500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 29.Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46(4):389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millar CD, et al. Mutation and evolutionary rates in adélie penguins from the antarctic. PLoS Genet. 2008;4(10):e1000209. doi: 10.1371/journal.pgen.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendy MD, Woodhams MD, Dodd A. Modelling mitochondrial site polymorphisms to infer the number of segregating units and mutation rate. Biol Lett. 2009;5(3):397–400. doi: 10.1098/rsbl.2009.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto H, et al. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biol. 2011;12(6):R59. doi: 10.1186/gb-2011-12-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels DC, et al. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 2013;9(11):e1003929. doi: 10.1371/journal.pgen.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avital G, et al. Mitochondrial DNA heteroplasmy in diabetes and normal adults: Role of acquired and inherited mutational patterns in twins. Hum Mol Genet. 2012;21(19):4214–4224. doi: 10.1093/hmg/dds245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9(9):e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsson N-G. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 37.Itsara LS, et al. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014;10(2):e1003974. doi: 10.1371/journal.pgen.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifer DB, DeJesus V, Hubbard K. Mitochondrial deletions in luteinized granulosa cells as a function of age in women undergoing in vitro fertilization. Fertil Steril. 2002;78(5):1046–1048. doi: 10.1016/s0015-0282(02)04214-0. [DOI] [PubMed] [Google Scholar]
- 39.Barritt JA, Cohen J, Brenner CA. Mitochondrial DNA point mutation in human oocytes is associated with maternal age. Reprod Biomed Online. 2000;1(3):96–100. doi: 10.1016/s1472-6483(10)61946-3. [DOI] [PubMed] [Google Scholar]
- 40.Eichenlaub-Ritter U. Oocyte ageing and its cellular basis. Int J Dev Biol. 2012;56(10-12):841–852. doi: 10.1387/ijdb.120141ue. [DOI] [PubMed] [Google Scholar]
- 41.Barritt JA, Brenner CA, Cohen J, Matt DW. Mitochondrial DNA rearrangements in human oocytes and embryos. Mol Hum Reprod. 1999;5(10):927–933. doi: 10.1093/molehr/5.10.927. [DOI] [PubMed] [Google Scholar]
- 42.Bartmann AK, Romão GS, Ramos EdaS, Ferriani RA. Why do older women have poor implantation rates? A possible role of the mitochondria. J Assist Reprod Genet. 2004;21(3):79–83. doi: 10.1023/B:JARG.0000027018.02425.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivo PD, Van de Walle MJ, Laipis PJ, Hauswirth WW. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 1983;306(5941):400–402. doi: 10.1038/306400a0. [DOI] [PubMed] [Google Scholar]
- 44.Ashley MV, Laipis PJ, Hauswirth WW. Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res. 1989;17(18):7325–7331. doi: 10.1093/nar/17.18.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howell N, et al. Mitochondrial gene segregation in mammals: Is the bottleneck always narrow? Hum Genet. 1992;90(1-2):117–120. doi: 10.1007/BF00210753. [DOI] [PubMed] [Google Scholar]
- 46.Lutz S, Weisser HJ, Heizmann J, Pollak S. Mitochondrial heteroplasmy among maternally related individuals. Int J Legal Med. 2000;113(3):155–161. doi: 10.1007/s004140050288. [DOI] [PubMed] [Google Scholar]
- 47.Monnot S, et al. Segregation of mtDNA throughout human embryofetal development: m.3243A>G as a model system. Hum Mutat. 2011;32(1):116–125. doi: 10.1002/humu.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howell N, et al. The pedigree rate of sequence divergence in the human mitochondrial genome: There is a difference between phylogenetic and pedigree rates. Am J Hum Genet. 2003;72(3):659–670. doi: 10.1086/368264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos C, et al. Understanding differences between phylogenetic and pedigree-derived mtDNA mutation rate: A model using families from the Azores Islands (Portugal) Mol Biol Evol. 2005;22(6):1490–1505. doi: 10.1093/molbev/msi141. [DOI] [PubMed] [Google Scholar]
- 51.Sigurğardóttir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P. The mutation rate in the human mtDNA control region. Am J Hum Genet. 2000;66(5):1599–1609. doi: 10.1086/302902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denver DR, Moris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- 53.Haag-Liautard C, et al. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6(8):e204. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26(8):345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci USA. 2012;109(45):18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kenney MC, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: Implications for population susceptibility to diseases. Biochim Biophys Acta. 2014;1842(2):208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McElhoe JA, et al. Development and assessment of an optimized next-generation DNA sequencing approach for the mtgenome using the Illumina MiSeq. Forensic Sci Int Genet. 2014;13C:20–29. doi: 10.1016/j.fsigen.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo Y, et al. The effect of strand bias in Illumina short-read sequencing data. BMC Genomics. 2012;13:666. doi: 10.1186/1471-2164-13-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.