Significance
Many bacteria produce large periplasmic efflux pumps that impart resistance to excess toxic metal ions. The CusC(F)BA Cu+/Ag+ efflux pump in Escherichia coli is one example. This pump’s mechanism is elusive because it is composed of four distinct proteins, three of which bind metal. By inserting a selenium probe sequentially into the metal-binding sites of the Cus proteins and loading them with Cu+ and Ag+, the bound metal ions could be tracked via X-ray absorption spectroscopy. We found that CusB activates the CusA pump and that CusF is a metallochaperone to CusA. We propose that CusB deactivates CusA once metal ion levels in the cell decrease. These results will aid in combating metal ion resistance by pathogenic bacteria.
Keywords: copper, periplasmic efflux, X-ray absorption spectroscopy, metal ion transport, host–pathogen interaction
Abstract
Copper is an essential nutrient for all aerobic organisms but is toxic in excess. At the host–pathogen interface, macrophages respond to bacterial infection by copper-dependent killing mechanisms, whereas the invading bacteria are thought to counter with an up-regulation of copper transporters and efflux pumps. The tripartite efflux pump CusCBA and its metallochaperone CusF are vital to the detoxification of copper and silver ions in the periplasm of Escherichia coli. However, the mechanism of efflux by this complex, which requires the activation of the inner membrane pump CusA, is poorly understood. Here, we use selenomethionine (SeM) active site labels in a series of biological X-ray absorption studies at the selenium, copper, and silver edges to establish a “switch” role for the membrane fusion protein CusB. We determine that metal-bound CusB is required for activation of cuprous ion transfer from CusF directly to a site in the CusA antiporter, showing for the first time (to our knowledge) the in vitro activation of the Cus efflux pump. This metal-binding site of CusA is unlike that observed in the crystal structures of the CusA protein and is composed of one oxygen and two sulfur ligands. Our results suggest that metal transfer occurs between CusF and apo-CusB, and that, when metal-loaded, CusB plays a role in the regulation of metal ion transfer from CusF to CusA in the periplasm.
As a consequence of its inherent toxicity, unbound copper is not encountered within the living cell (1). Mammals have evolved to exploit this by exposing bacterial pathogens to high concentrations of copper as part of the innate immune response (2–5). Although this will often kill the invading species, copper tolerance pathways have coevolved in many bacteria (2, 6–9). In Escherichia coli, the chromosomally encoded Cu-sensing (CusS) copper-responsive regulator responds to low micromolar concentrations of free periplasmic copper and silver (10–14), and activates the cytoplasmic transcription factor CusR. Activation of CusR results in the production of the periplasmic copper efflux pump CusCFBA, which is specific to cuprous and argentous ions (15) and is essential to copper resistance in E. coli (10, 16) under anaerobic conditions. CusCFBA is composed of the inner membrane-bound proton-substrate antiporter CusA, the periplasmic adapter protein (PAP) CusB, and the outer membrane factor (OMF) CusC (17, 18). The Cus system is distinguished from other RND-type efflux pumps such as AcrB-TolC and its homologs (19), both by its association with the periplasmic metallochaperone CusF (20, 21) and by the active involvement of its periplasmic adapter protein. Unlike other PAPs, which are thought to primarily provide structural support, CusB is conformationally flexible and also possesses a metal-binding site within its N-terminal region (22). Additionally, cusB-knockout strains produce a Cu-sensitive phenotype (22, 23). CusF and CusB have been shown to freely exchange Ag(I) and Cu(I) toward equilibrium in highly specific protein–protein interactions (24, 25).
Crystal structures of CusA (26), CusB (27), and the outer membrane protein CusC (28) have been reported in recent years together with a structure of the CusBA complex (29), revealing a stoichiometry of 2:1:1 for the B:A:C components. Although these structures have provided enormous insight into possible modes of metal efflux (30, 31), the dynamic mechanism of periplasmic metal detoxification by Cus is still undetermined. This is in part due to disorder in the crystal structure of the metal-binding site of CusB (27, 29) and the difficulty of creating an in vitro scenario that includes CusA, CusB, and CusF in biologically relevant conditions. The purpose of our study was to create such a scenario and to determine the method of activation of the CusA pump, thereby defining the exact roles of CusF and CusB in periplasmic Ag and Cu efflux. Recently, a truncated N-terminal CusB fragment (CusB NT; hereafter, CusB) that behaves similarly to native CusB in vivo has been developed, facilitating its study (23).
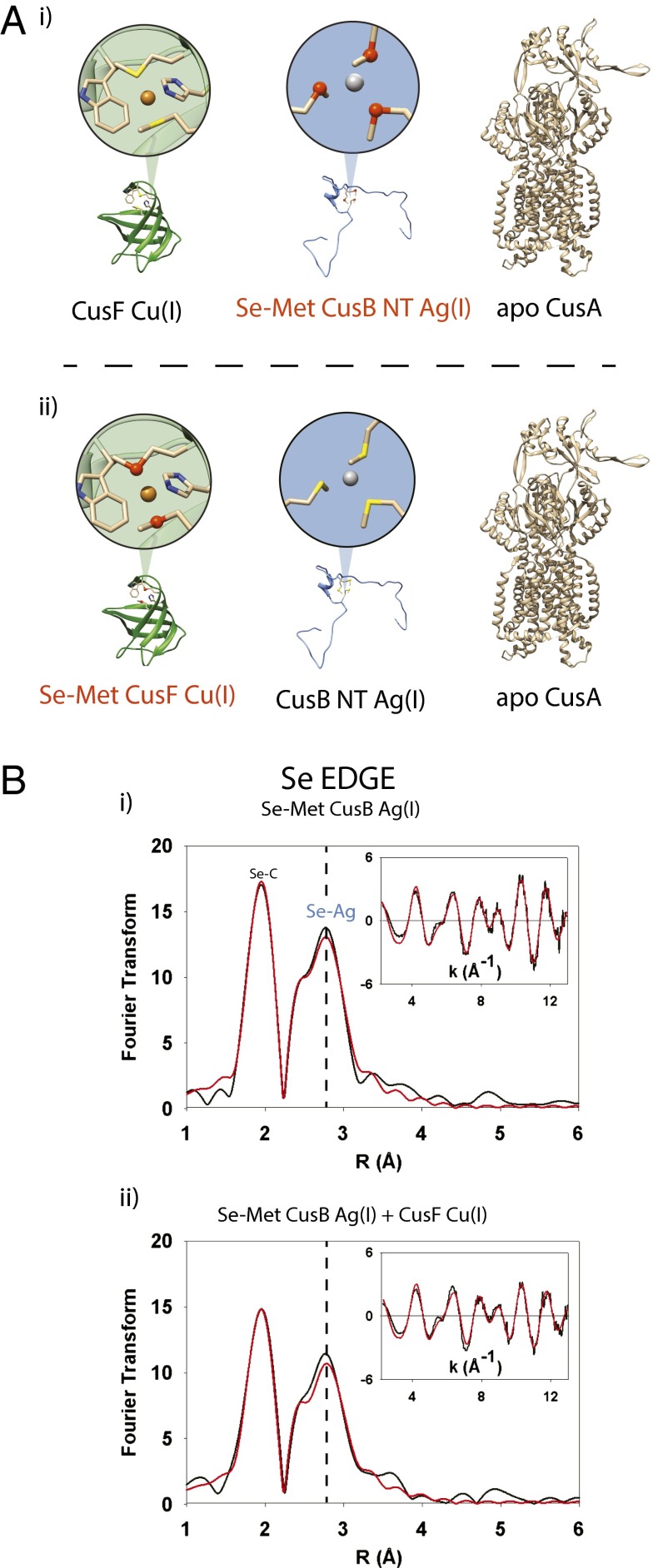
X-ray absorption spectroscopy (XAS) is ideal for the investigation of metalloprotein interactions when combined with selenomethionine (Se-Met) labeling of active site methionine (S-Met) residues (32, 33). By observing the Fourier transform (FT) spectra derived from the Se extended X-ray absorption fine structure (EXAFS), specific holo and apo metallosites within a mixture of proteins can be clearly distinguished. In the Cus efflux system, the metal-binding ligand sets known as of yet are two Mets, one His and a weakly coordinating tryptophan for CusF (20, 21, 24), and three Mets each for CusB (22, 23) and CusA (26), respectively. Therefore, the Cus system is an excellent candidate for Se-Met labeling, because all three of the Cus proteins under investigation contain at least two active-site Met residues. We have used this technique previously to monitor metal transfer between labeled and unlabeled versions of CusF and CusB (24). We now extend this to combine the three key Cus proteins in vitro, in a series of experiments in which either CusF or CusB possess a Se-Met label (Fig. 1A). In this manner, we were able to track the metal gain or loss by any Se-Met–labeled Cus component. Furthermore, we exploited the ability of the Cus system to bind and transport either copper or silver ions. With the ability to incubate copper-loaded CusF, silver-loaded CusB, and apo CusA in the same reaction, and with the Se-Met XAS spectroscopic probes sequentially placed on one or the other of the metal-loaded proteins, we accomplished a full account of metal transfer by a series of trimetal (Cu, Se, and Ag) XAS experiments.
Fig. 1.
(A) Trimetal edge XAS to determine metal transfer in CusF, CusB, and CusA. (Atom legend: sulfur, yellow spheres; selenium, dark orange spheres.) (i) Ag(I)-loaded Se-Met–labeled CusB (red typeface) incubated with unlabeled, Cu(I)-loaded CusF and apo CusA. With the Se-Met label on CusB, any change in Se-Ag coordination could be attributed to metal transfer from CusB to CusF or CusA. (ii) Cu(I)-loaded Se-Met–labeled CusF (red typeface) incubated with unlabeled, Ag(I)-loaded CusB and apo CusA. In these cases, any change in Se-Cu coordination could indicate metal transfer from CusF to CusB or CusA. (B) CusF and CusB do not exchange metal ions when both are fully metal ion loaded. (i) Se K-edge FT of selenomethionine-labeled, Ag(I)-loaded CusB. The CusB Se-Ag interaction is represented in the FT by the peak at ∼2.60 Å. (ii) Se K-edge FT of a stoichiometric mixture of selenomethionine-labeled, Ag(I)-loaded CusB and unlabeled, Cu(I)-loaded CusF. No decrease was observed in the CusB Se-Ag signal intensity, nor did a Se-Cu signal appear. It is of note that the overall intensity of all signals was lowered in the FT of the CusB/CusF mixture; however, this dampening occurred by the same amount in both the Se-C and Se-Ag peaks and is not indicative of metal transfer.
Results
CusF and CusB Only Exchange Metal Ions When One of the Pair Is an Apo-Protein.
Previous work has showed that partially loaded CusF and CusB will freely transfer Ag(I) and Cu(I) toward equilibrium and that these metal-loaded proteins can form a specific protein–protein interaction, but only if one of them is in the apo-state (24, 25). However, no work has yet investigated the extent of metal ion exchange when both are fully loaded with metal. To this end, we incubated Ag(I)-loaded, Se-Met–labeled CusB with Cu(I)-loaded unlabeled CusF and monitored the Se-Met CusB Se-Ag EXAFS for any changes in the Se-Ag signal (Table S1), such as a decrease in the Se-Ag signal or the appearance of a Se-Cu interaction (Fig. 1B). However, we did not observe any evidence of metal ion exchange between the two metal-bound proteins. Any attempt in our models to simulate the presence of Cu in the Se environment as the result of metal exchange between the two proteins resulted in an unrealistic Debye Waller (DW) factor for Cu-Se of 0.040 Å2 despite allowing the amount of Cu to float freely. Although visually the intensity of the overall spectrum did decrease, this appears to be unrelated to metal transfer and may be due to protein–protein interactions between Se-Met CusB and CusF. These data provided an essential control that when Cu-loaded CusF was incubated with Ag-loaded CusB and apo-CusA in subsequent triprotein transfer experiments (see below), any metal exchange that was observed must be between F and A, or B and A and not due to unproductive reversible exchange between F and B.
Metal Transfer Experiments Using Se-Met–Labeled CusB Rule Out a Metal Relay Mechanism.
To define a role for the CusB protein in Cus metal ion efflux, we first incubated apo unlabeled CusA with Se-Met–labeled Cu(I)-loaded CusB to determine whether metal would be transferred from CusB to CusA. The Se-Met CusB Se-Cu interaction at 2.42 Å did not decrease in the Se K-edge FT upon the addition of apo-CusA compared with Se-Met Cu(I) CusB alone (Table S2). We also tested Ag(I)-loaded Se-Met CusB in place of the Cu(I)-loaded protein, and no metal transfer occurred, confirming that CusB alone is not capable of transferring either metal to apo-CusA (Fig. S1).
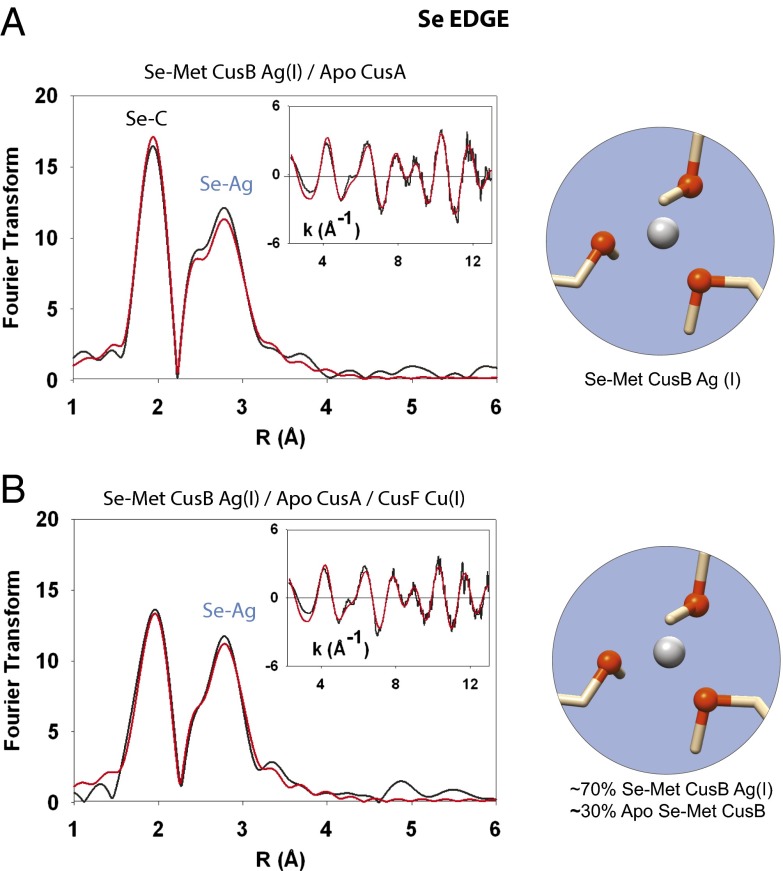
We then sought to determine whether the presence of a metallochaperone activates the transfer of metal from CusB to CusA. We prepared Ag(I)-loaded Se-Met CusB, incubated it with apo CusA, and after 5 min, added Cu(I)-loaded CusF to the reaction. With the Se-Met label on CusB, we looked for any change in Se-Ag coordination at 2.63Å in the FT compared with Se-Met Ag(I) CusB alone, at both the selenium and silver edges (Fig. 2). The addition of CusF resulted in a ∼30% decrease in the intensity of the Se-Ag signal in the Se K-edge FT. However, we did not consider this decrease to be evidence of transfer from CusB to CusA because if CusB had transferred metal to CusA, a Se-Cu signal from Se-Met–labeled, Cu(I)-CusB would appear as apo Se-Met CusB accepted Cu(I) from CusF (see below).
Fig. 2.
Selenium K-edge EXAFS indicates that CusB is not a chaperone to CusA. Black spectra, experimental data; red spectra, simulated fit. (A) Se K-edge spectra of Ag-loaded Se-Met CusB incubated with apo CusA. The Se-Met CusB Se-Ag(I) interaction at 2.63 Å was not decreased in the FT upon the addition of CusA. (B) Se-Met Ag(I) CusB incubated with apo CusA and unlabeled Cu(I)-loaded CusF. The addition of CusF resulted in a small decrease (∼30%) in the Se-Ag signal. However, no Se-Cu interaction could be observed or modeled in the EXAFS of the triprotein incubation, indicating that CusF did not relay metal to CusA via CusB.
Metal Transfer Experiments Using Se-Met–Labeled CusF Indicates an Activator Role for CusB.
These initial results described above indicated to us that metal-loaded CusB does not act as a metal relay to CusA but instead must perform the role of an activator of the pump or “on–off switch,” as has been previously proposed (30). That is to say, CusB turns on the CusA pump when it accepts metal from holo CusF and turns it off when the bound metal is released back to apo CusF. Thus, the extent of metal ion binding to CusB will dictate the open or closed status of the CusA pump, and allows for the subsequent delivery of ions from the periplasm.
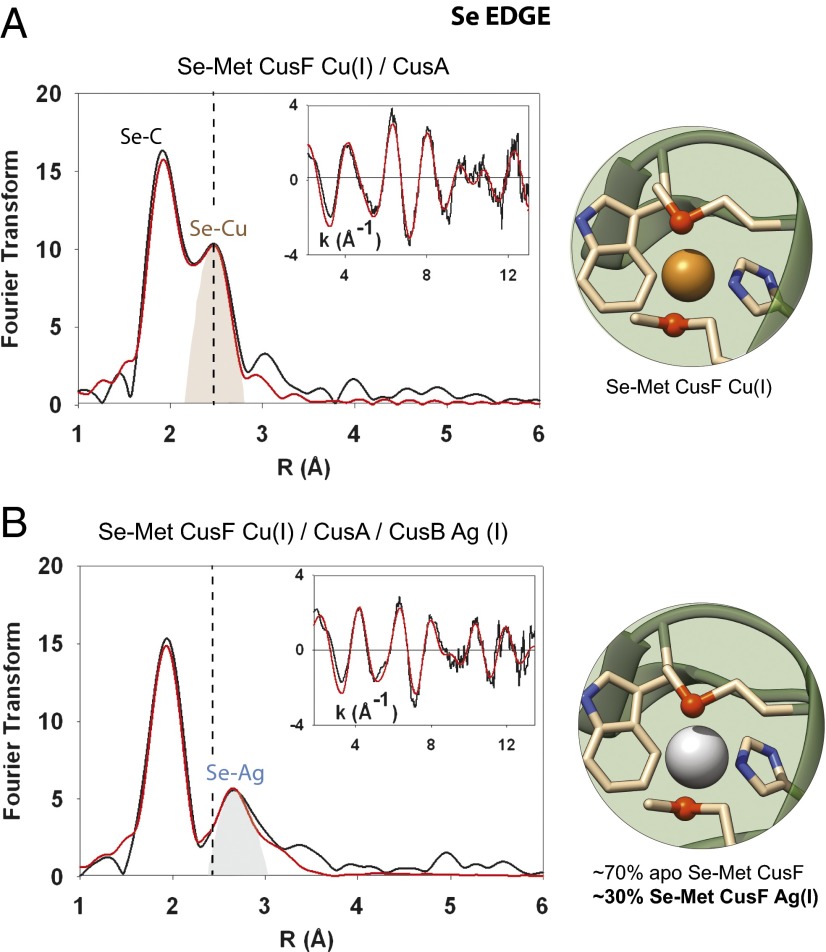
If CusB acts as a switch in the Cus system, then the metallochaperone CusF may be the primary vehicle of metal ion delivery to CusA during metal ion efflux, but CusF should only be able to deliver if it is also in the presence of metal-loaded CusB. To address this, we incubated Se-Met–labeled, Cu(I) CusF with apo CusA and monitored the Se-Met CusF Se-Cu interaction at the canonical bond distance of 2.38 Å. No change in the Se-Cu intensity was detected compared with the spectra of Se-Met Cu(I) CusF alone, indicating that CusF alone was insufficient for the transfer of metal to CusA (Fig. S2). However, when we added unlabeled, Ag(I)-loaded CusB to this reaction, a nearly complete disappearance (∼90%) of the Se-Met CusF Se-Cu interaction occurred in the Se FT within the ∼5-min time frame of mixing and preparation of the XAS sample (Fig. 3). Concurrently, a new peak appeared in the Se FT at 2.63 Å, which we modeled to 0.3 equivalents of Ag(I) (Table S3 and Fig. S3). The appearance of a silver signal was anticipated and served as an important correlate to the previously described Se-Met–labeled CusB data, in which there had been a ∼30% decrease in the Se-Ag signal upon incubation with CusF and CusA (see below). That metal ion is transferred from CusF but only when CusB is in the metal-bound state. These experiments confirm that metal ion is transferred from CusF, but only when CusB is in the metal-bound state. This suggests on–off switch behavior in CusB and may show for the first time (to our knowledge) the in vitro activation of the Cus efflux pump.
Fig. 3.
Metal-loaded CusB activates the CusA pump. (A) Se K-edge spectra of Se-Met–labeled, Cu(I) CusF incubated with apo CusA. No change in the Se-Cu intensity at 2.38 Å was detected. (B) Se K-edge spectra of Se-Met–labeled, Cu(I) CusF incubated with unlabeled, Ag(I)-loaded CusB and apo CusA. A complete disappearance of the Se-Met CusF Se-Cu interaction occurred in the Se FT. Concurrently, a new peak appeared in the Se FT at 2.63 Å, which we fit to 0.3 equivalents of Ag(I). The vertical dashed line illustrates where a Se-Cu signal would have appeared in the spectra in B.
Following the Fate of the Transferred Cuprous Ions by Cu-K Edge XAS Reveals a Putative Acceptor Site on CusA.
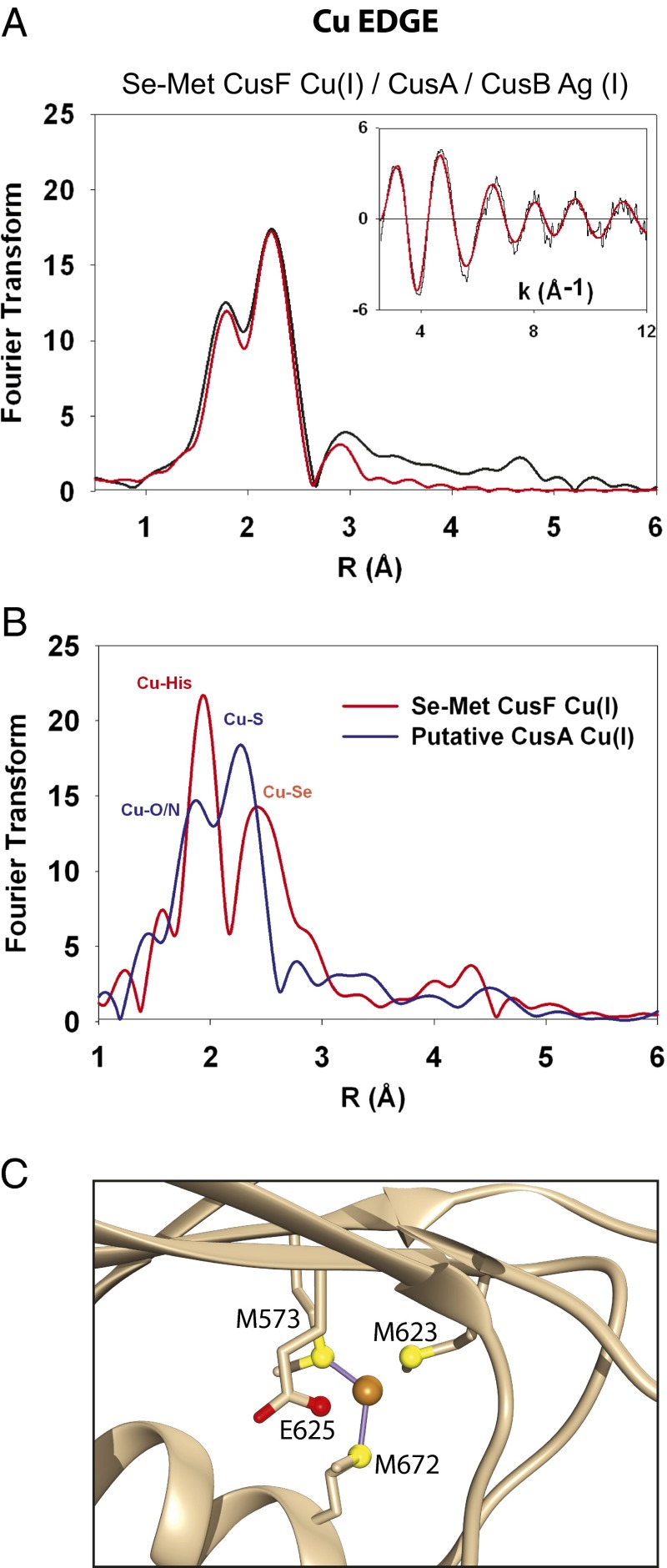
Having determined that CusF released its cuprous ion cargo upon exposure to the other two Cus components, we sought the final destination of the copper. EXAFS data of the Se-Met Cu(I) CusF/Ag(I) CusB/apo CusA mixture was collected at the Cu edge using the same sample as that used previously at the Se edge. The Cu edge FT revealed a metallosite (Fig. 4), which was quite distinct from the copper edge spectra of Cu(I)-loaded Se-Met CusF or unlabeled Cu(I)-loaded CusB. This Cu edge spectrum could be modeled by one oxygen/nitrogen ligand at 1.85 Å and two S ligands (most likely from methionine residues) at 2.23 Å, and in all probability corresponds to the “entry” site for Cu(I) on the CusA pump.
Fig. 4.
Copper edge XAS reveals a putative CusA metallosite. (A) The Cu-K edge XAS spectra of Se-Met CusF Cu(I) incubated with apo CusA and Ag(I)-loaded CusB is distinctly different from the spectra of either Cu-loaded CusF or Cu-loaded CusB, and is best modeled as a Cu-loaded Met-Met-oxygen/nitrogen CusA metallosite. (B) For comparison, Cu K-edge XAS of Se-Met–labeled Cu(I)-loaded CusF (red spectra) overlaid with the spectrum from Fig. 4A (blue spectra). Upon mixing with Ag-loaded CusB and apo CusA, both the Cu-Se and Cu-His signals of Se-Met Cu(I) CusF disappeared and the spectrum in blue was observed, which is best fit to a single oxygen/nitrogen and two sulfur ligands. (C) The crystal structure of Cu(I)-soaked CusA (PDB ID code 3K01) shows that Met623 and Glu625 are nearly equidistant as ligands at ∼3 Å and are therefore equally plausible ligands to Cu(I).
Backtransfer from CusB to Apo-CusF Implies Regulation.
Given that we had previously shown that CusF and CusB can only exchange metal ions when one of them is in the apo form (24), the prediction was that any apo CusF present after Cu(I) transfer to CusA would then accept silver from CusB. When we incubated Se-Met–labeled, Cu(I)-loaded CusF with Ag(I)-loaded CusB and the apo CusA pump, the copper signal of CusF was replaced by a silver signal at the selenium edge, indicating backtransfer from Ag(I) CusB (Fig. 3). Importantly, this appearance of a silver signal in CusF could then be matched to the loss of silver signal in the Se-Met–labeled CusB triprotein incubation at both Se and Ag edges, underscoring the usefulness of monitoring all three metal edges (Fig. S4).
Discussion
CusB Is an Activator or “Switch.”
The detailed mechanism of the CusCBAF efflux system and, in particular, the role played by the periplasmic adaptor protein CusB has been the subject of considerable debate. Two models have been proposed in the literature—the “funnel” model and the “switch” model (23, 30). In the funnel model, excess periplasmic copper is sequestered by CusF, transferred to CusB (24), and then delivered to CusA for extrusion. In the “switch” model, copper loading of CusB induces a conformational change (22), which allows CusB to bind to CusA and open the entry site on the CusA pump, which is known to be located in its periplasmic cleft (26, 34). Although both models predict that either CusB deletion, or metal-site mutagenesis should lead to copper sensitive strains—an expectation that has been experimentally verified (22, 23)—definitive evidence for or against either model has been lacking and is compounded by the fact that the copper-binding N-terminal domain of CusB is disordered in crystal structures of the isolated protein (27), or its complex with CusA (29). Existing evidence, although weak, has leaned more toward the switch model, based mainly on projections of the distance of the CusB metal-binding site from the CusA periplasmic cleft (30), and analogy to structural data on other metal resistance RND adaptor proteins such as ZneB of Cupriavidus metallodurans CH34. The predicted location of the N-terminal copper-binding site of CusB close to the membrane would seem to preclude direct copper transfer to the CusA Met3 acceptor site identified by crystallography (26, 34).
In the present work, we developed an approach to distinguishing between funnel or switch. We took advantage of the fact that the RND efflux pump can extrude either copper or silver and labeled CusF and CusB with copper and silver, respectively, whose coordination environment could then be uniquely determined by XAS. In a further refinement of the method, we substituted the Met ligands on either CusF or CusB with selenomethionine, providing a third XAS-detectable label (Se), which would report on the fate of each metal as it interacted with and was transferred through the RND pump. For example, if CusB acted as a funnel, Ag-loaded SeM-substituted CusB should transfer its Ag(I) ion to apo-CusA, resulting in the loss of the prominent Se-Ag signal in the Se EXAFS. On the other hand, if CusB acted as a switch, the Ag ion should remain bound throughout the transfer reaction, and no change in Se-Ag intensity should be observed. Our experimental data show that the latter scenario was indeed observed, thus providing the first definitive evidence (to our knowledge) for the mechanistic role of CusB as an activator or switch, which opens the entry site on CusA, thereby allowing copper access to the membrane extrusion channel.
CusF and CusB May Exchange Metal as Part of a Regulatory Strategy for Detoxification.
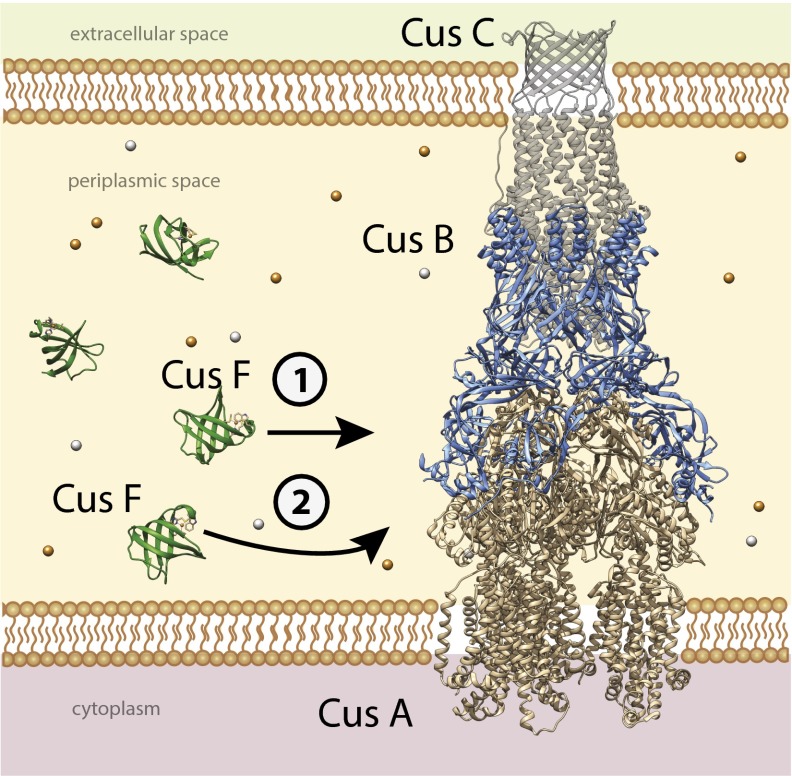
Our previous studies of copper transfer from CusF to CusB have demonstrated that the transfer reaction is rapid and reversible (23, 24), suggesting that the degree of metallation of CusB is dictated by the overall copper flux within the periplasm as sensed by the CusF chaperone. The present observation of backtransfer from CusB to apo CusF after the latter has transferred its copper cargo to CusA strongly supports a mechanism for the Cus efflux system that is finely tuned and responsive to rapidly changing metal ion and environmental conditions. We posit that, as copper and silver fill the periplasmic space under anaerobic conditions, the CusF chaperone acts as a scavenger of metal ions and rapidly fills all available CusB metal-binding sites. Once activated in this manner, CusB affects a structural change, opening the CusA pump to the periplasm and allowing CusF to dock and release its cargo (Fig. 5). As metal ion concentration in the periplasm begins to return to basal levels, CusF retrieves bound metal from CusB, closing CusA to the periplasm and inactivating efflux.
Fig. 5.
Proposed mechanism of the CusCFBA Cu(I)/Ag(I) efflux system. The CusCBA complex spans the periplasmic space of Gram-negative bacteria during copper or silver stress and acts to pump out excess metal ions. CusF is believed to act as a metallochaperone to the complex, first delivering Cu(I) or Ag(I) to CusB metal-binding sites (1), which activates the CusA pump. This in turn facilitates delivery of metal ions by CusF to a previously unobserved binding motif in CusA (2). This mechanism would allow for the pump to be regulated by copper or silver: when excess metal in the periplasm falls to basal levels, CusF will backtransfer Cu or Ag from the CusB metal-binding sites, deactivating the pump and maintaining copper homeostasis.
A Proposed Binding Motif in CusA.
Our data also show that metal-loaded CusB activates the pump to accept copper from CusF, as evidenced from the complete loss of Se-Cu signal in a mixture of Cu-loaded SeM-labeled CusF, Ag-loaded CusB and apo-CusA. The Cu EXAFS of this preparation then reports on the nature of the entry site on CusA when the system is actively transferring metal. The description of the CusA entry site revealed in these experiments contrasts with the (Met)3 ligation determined previously from crystals of CusA soaked with excess cuprous ion (26). Furthermore, we have not been able to replicate the (Met)3 ligand set in our own laboratory by incubation of the detergent-solubilized apo-CusA protein with inorganic cuprous ion (Fig. S5). Rather, our transfer experiment strongly suggests a Met-Met-oxygen/nitrogen–binding site as the product of the triprotein incubation. Close examination of the Cu-incubated CusA crystal structure [Protein Data Bank (PDB) ID code 3K01] reveals that a glutamate residue (E625) is almost equidistant with the longest proposed methionine ligand (M623) at ∼3 Å and appears poised to coordinate with Cu(I) (Fig. 4C) (34). Additionally, mutation of this residue to alanine was shown to negatively impact copper resistance (34). Therefore, it is likely that at least two conformations and possibly more can exist within the copper-binding pocket of CusA. The (Met)2 O/N motif observed in the present work suggests that a dynamic set of Met/oxygen metal-binding sites may coexist and are available to bind the cuprous ion as it moves through the CusA pore and into the large and highly solvated CusC channel.
Mixed Met + O/N ligation is not expected to be a stable ligand set for Cu(I) because the harder (more electronegative) O-donors of amide carbonyls, carboxylates, or –OH groups of serine, threonine, or tyrosine will decrease the reduction potential via stabilization of the cupric state and thus promote oxidation. The preference of Cu(I) for soft bases (His, Cys, and Met) is manifest in the coordination environments of cytoplasmic metallochaperones (35–37) and the Met-rich sites found in periplasmic copper-binding proteins (38–41). However, in the context of Cu(I) transport through membrane channels, kinetic lability may be more important than thermodynamic stability, and indeed high binding affinity is likely to inhibit transport, unless accompanied by energy input that can toggle high- and low-affinity states via conformational change, as has been proposed as a mechanism for intramembrane transport in P1B-type ATPases (42). Of significance to the present study, the two intramembrane Cu(I)-binding sites of CopA from Archaeoglobus fulgidus were identified as being comprised of sulfur plus oxygen-coordination, with Cys-Cys-Tyr and Met-Asn-Ser ligand sets (43). Furthermore, an “entry” site comprised of Met-Asp-Glu triad was identified close to the locus where the N-terminal metal-binding domain was proposed to dock onto the transmembrane domain of Legionella pneumoniae CopA (42) and later shown to be important for ATP-driven transmembrane transport from the chaperone CopZ in the A. fulgidus protein (44).
The preponderance of these mixed sulfur/oxygen ligand environments is an emerging theme in intramembrane metal transport, which may function to ensure that copper is progressively oxidized to the less toxic cupric state as it transits from CusF through the pump and eventually into the large-diameter, negatively charged outer membrane channel CusC (28). Indeed, it is possible that the CusC channel also contains dioxygen molecules, which would oxidize the Cu(I), and extrude it as the nontoxic Cu(II) species. It has been similarly proposed for the CopA Cu+-ATPase in Archaeoglobus fulgidus that the change in architecture of its two transmembrane sites toward increasing oxygen ligands aids in vectorial copper(I) transport from the cytoplasm and likely prohibits backward movement into the cytoplasm (43). Further experiments are planned to test these concepts.
The complexity of the Cus Ag(I)/Cu(I) efflux pump, as well as the prevalence of copper detoxification systems in Gram-negative bacteria, emphasizes our need to fully explore the underlying bioinorganic chemistry. Studying metal transfer spectroscopically and combining this knowledge with the study of the microbiology of the organism should reveal valuable antimicrobial targets. Our future work entails the extensive kinetic profiling of metal transfer between the three key Cus components and will enrich the structural information provided in the current work.
Summary of Materials and Methods
The CusB NT protein was purified as previously described (20). The CusA protein was overexpressed in E. coli and harvested using sucrose and Cymal-6 detergent for membrane solubilization and a streptavidin tag for isolation. The CusF protein was purified via Ni-NTA affinity chromatography of the His6-tagged construct followed by TeV protease cleavage to remove the tag. All Cu(I)-loaded proteins were prepared anaerobically and by slow addition of Cu[(CH3CN)4]PF6, followed by exhaustive stepwise buffer exchange to remove any excess Cu(I). All Ag(I)-loaded proteins were prepared in the dark by slow addition of AgNO3 in Hepes buffer and followed by exhaustive buffer exchange. All protein and metal concentrations were verified by the Bradford assay, the bicinchoninic acid assay, and inductively coupled plasma optical emission spectroscopy, respectively. Proteins were prepared as frozen glasses with 20% (vol/vol) ethylene glycol and analyzed at the Cu, Se, and Ag K-edges by XAS on beamlines 7-3 and 9-3 at the Stanford Synchrotron Radiation Lightsource. Experiments were performed in duplicate, and each of these replicates used newly expressed and purified Cus proteins, to ensure that results were comparable across protein preparations. Full Methods are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to M. Mayfield for optimizing the CusB and CusF plasmid constructs, and to C. Butterfield and Dr. P. Moënne-Loccoz for scientific discussions. We also gratefully acknowledge the use of facilities at the Stanford Synchrotron Radiation Lightsource, which is supported by the National Institutes of Health Biomedical Research and Technology Program Division of Research Resources, and by the US Department of Energy Office of Biological and Environmental Research. The work was supported by the National Science Foundation Graduate Research Fellowship DGE-0925180 (to K.N.C.) and by National Institutes of Health Grants R01 GM054803 (to N.J.B.) and R01 079192 (to M.M.M.). Molecular graphics images of CusA, CusF, CusC, and CusB cocrystallized with CusA were produced using the PDB files 3K01, 3NE5, 3PIK, and 2QCP, and Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a Prearranged Editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411475111/-/DCSupplemental.
References
- 1.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science. 1999;284(5415):805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 2.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284(49):33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolschendorf F, et al. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2011;108(4):1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr Opin Chem Biol. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: Meddling with the metal. Cell Host Microbe. 2012;11(2):106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi KS, Henderson JP. Pathogenic adaptations to host-derived antibacterial copper. Front Cell Infect Microbiol. 2014;4:3. doi: 10.3389/fcimb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood MI, Skaar EP. Nutritional immunity: Transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10(8):525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botella H, Stadthagen G, Lugo-Villarino G, de Chastellier C, Neyrolles O. Metallobiology of host-pathogen interactions: An intoxicating new insight. Trends Microbiol. 2012;20(3):106–112. doi: 10.1016/j.tim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Hernández-Montes G, Argüello JM, Valderrama B. Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol. 2012;12:249. doi: 10.1186/1471-2180-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185(13):3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Outten FW, Huffman DL, Hale JA, O’Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276(33):30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 12.Lok C-N, et al. Proteomic identification of the Cus system as a major determinant of constitutive Escherichia coli silver resistance of chromosomal origin. J Proteome Res. 2008;7(6):2351–2356. doi: 10.1021/pr700646b. [DOI] [PubMed] [Google Scholar]
- 13.Gudipaty SA, Larsen AS, Rensing C, McEvoy MM. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett. 2012;330(1):30–37. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudipaty SA, McEvoy MM. The histidine kinase CusS senses silver ions through direct binding by its sensor domain. Biochim Biophys Acta. 2014;1844(9):1656–1661. doi: 10.1016/j.bbapap.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conroy O, Kim E-H, McEvoy MM, Rensing C. Differing ability to transport nonmetal substrates by two RND-type metal exporters. FEMS Microbiol Lett. 2010;308(2):115–122. doi: 10.1111/j.1574-6968.2010.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass G, Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183(6):2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mealman TD, Blackburn NJ, McEvoy MM. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr Top Membr. 2012;69:163–196. doi: 10.1016/B978-0-12-394390-3.00007-0. [DOI] [PubMed] [Google Scholar]
- 18.Argüello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng TT, et al. The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–125. [PubMed] [Google Scholar]
- 20.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Protein Sci. 2007;16(10):2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Y, et al. Cu(I) recognition via cation-pi and methionine interactions in CusF. Nat Chem Biol. 2008;4(2):107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282(49):35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 23.Mealman TD, et al. N-terminal region of CusB is sufficient for metal binding and metal transfer with the metallochaperone CusF. Biochemistry. 2012;51(34):6767–6775. doi: 10.1021/bi300596a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47(44):11408–11414. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mealman TD, et al. Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry. 2011;50(13):2559–2566. doi: 10.1021/bi102012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long F, et al. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467(7314):484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su C-C, et al. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J Mol Biol. 2009;393(2):342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PLoS One. 2011;6(1):e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su C-C, et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011;470(7335):558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E-H, Nies DH, McEvoy MM, Rensing C. Switch or funnel: How RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011;193(10):2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long F, et al. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos Trans R Soc Lond B Biol Sci. 2012;367(1592):1047–1058. doi: 10.1098/rstb.2011.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackburn NJ, et al. Selenomethionine-substituted Thermus thermophilus cytochrome ba3: Characterization of the CuA site by Se and Cu K-EXAFS. Biochemistry. 1999;38(22):7075–7084. doi: 10.1021/bi982500z. [DOI] [PubMed] [Google Scholar]
- 33.Chacón KN, Blackburn NJ. Stable Cu(II) and Cu(I) mononuclear intermediates in the assembly of the CuA center of Thermus thermophilus cytochrome oxidase. J Am Chem Soc. 2012;134(39):16401–16412. doi: 10.1021/ja307276z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su C-C, et al. Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. J Mol Biol. 2012;422(3):429–441. doi: 10.1016/j.jmb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109(10):4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, et al. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat Chem Biol. 2013;9(3):177–183. doi: 10.1038/nchembio.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnesano F, Banci L, Bertini I, Mangani S, Thompsett AR. A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci USA. 2003;100(7):3814–3819. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Koay M, Maher MJ, Xiao Z, Wedd AG. Intermolecular transfer of copper ions from the CopC protein of Pseudomonas syringae. Crystal structures of fully loaded Cu(I)Cu(II) forms. J Am Chem Soc. 2006;128(17):5834–5850. doi: 10.1021/ja058528x. [DOI] [PubMed] [Google Scholar]
- 40.Chong LX, et al. Unprecedented binding cooperativity between Cu(I) and Cu(II) in the copper resistance protein CopK from Cupriavidus metallidurans CH34: Implications from structural studies by NMR spectroscopy and X-ray crystallography. J Am Chem Soc. 2009;131(10):3549–3564. doi: 10.1021/ja807354z. [DOI] [PubMed] [Google Scholar]
- 41.Sarret G, et al. CopK from Cupriavidus metallidurans CH34 binds Cu(I) in a tetrathioether site: Characterization by X-ray absorption and NMR spectroscopy. J Am Chem Soc. 2010;132(11):3770–3777. doi: 10.1021/ja9083896. [DOI] [PubMed] [Google Scholar]
- 42.Gourdon P, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475(7354):59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 43.González-Guerrero M, Eren E, Rawat S, Stemmler TL, Argüello JM. Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J Biol Chem. 2008;283(44):29753–29759. doi: 10.1074/jbc.M803248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Padilla-Benavides T, McCann CJ, Argüello JM. The mechanism of Cu+ transport ATPases: Interaction with Cu+ chaperones and the role of transient metal-binding sites. J Biol Chem. 2013;288(1):69–78. doi: 10.1074/jbc.M112.420810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.