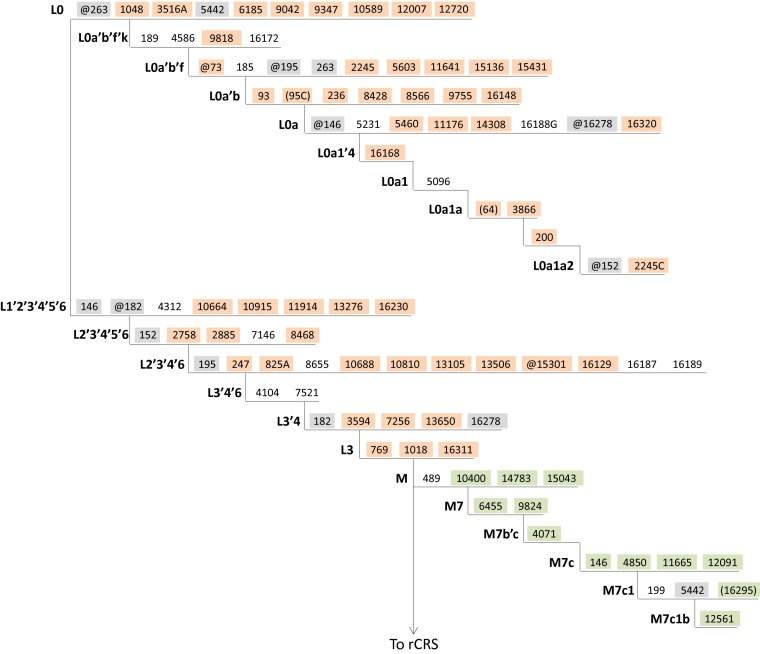
In their analysis of massively parallel sequencing data (MPS) from the 1000 Genomes Project, Ye et al. (1) report a very high rate of human mitochondrial DNA (mtDNA) heteroplasmy (89.68% of individuals), including up to 71 point heteroplasmies within a single individual, when using an ∼1% minor allele frequency (MAF) threshold. Inspection of the heteroplasmy data detailed in dataset S1 of ref. 1 revealed that contamination, not intraindividual variation, is the source of at least some of the reported heteroplasmy. For example, among the 15 samples with 20 or more heteroplasmies, all appear to be a mixture of at least two distinct individuals, and a minimum of 80.7% of the 584 heteroplasmies occurred at positions diagnostic for the mtDNA haplogroups represented in each mixture. To cite specific examples: for sample HG00740, nearly all (90%) of the 71 heteroplasmies can be ascribed to one of two distinct mtDNA haplogroups (L1b1a7a, of sub-Saharan African ancestry, and B2b3a, a Native American lineage); and for sample HG01108, 50 and 12 of 69 total heteroplasmies are diagnostic for haplogroups L0a1a2 (sub-Saharan African) and M7c1b (East Asian), respectively [according to Build 16 of PhyloTree (2)] (Fig. 1). Even among the heteroplasmies reported for those samples that do not match an mtDNA haplogroup motif, some are likely a result of private mutations in either individual represented in each mixture, rather than true intraindividual variation.
Fig. 1.
The figure displays a simplified representation of the revised Cambridge Reference Sequence (rCRS)-oriented version of the currently accepted human mtDNA phylogeny (data from PhyloTree Build 16, ref. 2) that includes only the branches relevant for sample HG01108. Symbols and nomenclature are consistent with PhyloTree, where the “@” symbol signifies mutation toward the rCRS state, nucleotide positions in parentheses represent mutations that may or may not be present, and inclusion of a nucleotide after the base position indicates a transversion. The presence or absence of the mutations highlighted in this figure among the heteroplasmies reported for sample HG01108 provide evidence for a mixture of two distinct individuals. Sample HG01108 heteroplasmies that can be ascribed to haplogroup L0a1a2 are highlighted in orange and sample HG01108 heteroplasmies at positions diagnostic for haplogroup M7c1b are highlighted in green. Branch positions highlighted in gray represent a mutation and reversion combination on the path between the L0a1a2 and M7c1b lineages (positions 146, 152, 182, 195, 263, and 16278), or homoplasy (position 5442), and thus would not be expected to be observed as variant in a mixture of individuals of these two ancestries (and were not reported as heteroplasmic for sample HG01108). L0a1a2 and M7c1b haplogroup diagnostic positions that were not reported as heteroplasmic in sample HG01108 (positions not highlighted) may be accounted for by (i) the 2,930 mtDNA positions that failed quality control standards and thus were not examined by Ye et al. (1) for any sample, and (ii) additional potentially variant positions on a by-sample basis that did not meet the authors’ criteria for heteroplasmy designation (in addition to less-likely explanations, such as reversion as a private mutation). For the seven heteroplasmies reported for sample HG01108 that were not ascribed to either haplogroup (at positions 5112, 12616, 12684, 13095, 15891, 16362, and 16519), these may be due to either (i) private mutations in either individual represented in the mixture or (ii) true mtDNA heteroplasmy.
Absent an in-depth analysis of all 4,342 heteroplasmies reported by Ye et al. (1), it is unclear what MAF threshold would be needed to eliminate all of the variant positions that are the result of mixtures. When we applied a 15% MAF cutoff, no haplogroup diagnostic positions remained for sample HG00740, but a 25% threshold would be required to achieve the same result for sample HG01108. Regardless, it is evident that the conclusions drawn by the authors should be revisited if the data themselves are flawed. For example, in contrast to the positive correlation between substitution rates and heteroplasmy rates reported by Ye et al. (1), no correlation was observed (R2 = 0.003979, P = 0.23) when only the coding region heteroplasmies with a MAF greater than 15% were analyzed using the same substitution rate data used by the authors.
Although use of a higher MAF threshold will undoubtedly exclude authentic heteroplasmies present at lower frequencies, the heteroplasmy detection threshold applied to MPS data must be high enough to both eliminate false positives resulting from chemistry, template, or bioinformatic method limitations, as well as overcome any sample mixtures present in the data (due to contamination resulting from the processing environment, or jumping PCR when indexed samples are pooled during library preparation), even when other quality control measures (such as quality score filtering and double-strand validation) are implemented (3). Given errors identified in another recent study in which extensive human mtDNA heteroplasmy within individuals was claimed (4, 5), the question of whether mtDNA heteroplasmy present at low frequency (less than 5–10%) within an individual can be reliably detected using current MPS technologies and bioinformatic approaches—even when coverage depths are very high—remains unanswered.
Acknowledgments
We thank Melissa Scheible (American Registry of Pathology, and Armed Forces DNA Identification Laboratory) for data review and Eric Pokorak (Federal Bureau of Investigation) for valuable feedback on the manuscript.
Footnotes
The authors declare no conflict of interest.
References
- 1.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci USA. 2014;111(29):10654–10659. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30(2):E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 3.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87(2):237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandelt HJ, Salas A. Current next generation sequencing technology may not meet forensic standards. Forensic Sci Int Genet. 2012;6(1):143–145. doi: 10.1016/j.fsigen.2011.04.004. [DOI] [PubMed] [Google Scholar]