Significance
Protein kinases represent a critically important family of regulatory enzymes. Their activity can be altered by mutations and binding events distant from the active site. To understand the nature of these long-distance effects, we used microsecond-timescale molecular dynamic simulation to subdivide a prototypical kinase, protein kinase A, into contiguous communities that exhibit internally correlated motions. Surprisingly, most of these unconventional structural entities were centered around known protein kinase functions. We thus propose a new framework for analysis of protein kinase structure and function that differs from traditional representations based simply on sequence motifs and secondary structure elements. These results extend our view on the dynamic nature of protein kinases and open a door to understanding of allosteric signaling in these enzymes.
Keywords: phosphorylation, community analysis, molecular dynamics
Abstract
Protein kinases are dynamically regulated signaling proteins that act as switches in the cell by phosphorylating target proteins. To establish a framework for analyzing linkages between structure, function, dynamics, and allostery in protein kinases, we carried out multiple microsecond-scale molecular-dynamics simulations of protein kinase A (PKA), an exemplar active kinase. We identified residue–residue correlated motions based on the concept of mutual information and used the Girvan–Newman method to partition PKA into structurally contiguous “communities.” Most of these communities included 40–60 residues and were associated with a particular protein kinase function or a regulatory mechanism, and well-known motifs based on sequence and secondary structure were often split into different communities. The observed community maps were sensitive to the presence of different ligands and provide a new framework for interpreting long-distance allosteric coupling. Communication between different communities was also in agreement with the previously defined architecture of the protein kinase core based on the “hydrophobic spine” network. This finding gives us confidence in suggesting that community analyses can be used for other protein kinases and will provide an efficient tool for structural biologists. The communities also allow us to think about allosteric consequences of mutations that are linked to disease.
Eukaryote protein kinases (EPKs) are highly dynamic and flexible proteins (1, 2). These kinases developed this property during their evolution from eukaryote-like kinases as they acquired a role of molecular switches, which undergo transformations between “on” and “off” states (3, 4). To understand the mechanisms that govern EPK function and regulation requires an integration of sequence, structure, and dynamics. Early sequence-based analyses showed that the catalytic domain partitioned naturally into conserved, linear sequence motifs (5). The first protein kinase structure provided a 3D context for these motifs and preliminary predictions of function (6).
The conserved protein kinase core consists of two lobes: the N-lobe, featuring a β-sheet with five strands, and the C-lobe, consisting mostly of α-helices and loops. Local spatial pattern (LSP) alignment of EPK structures led to the discovery of two hydrophobic “spines” connecting the N-lobe with the C-lobe (7). The regulatory spine (R-spine) is the hallmark signature of an active kinase and is dynamically assembled as part of regulation (8). In contrast, the catalytic spine (C-spine) is completed by the binding of the adenine ring of ATP. Both spines are anchored to the hydrophobic αF-helix (9). Although these analyses helped to define important structural elements and organizing principles for EPKs, many long-distance allosteric effects of mutations and of ligand and protein binding remained poorly understood.
Dynamic data have also been used as a basis for defining important structural elements in the EPKs (10). An early analysis of rigid bodies in the PKA C-subunit led to the identification of two large, semirigid domains within the protein kinase core, which correspond roughly to the N- and C-lobes (11). An initial molecular dynamics (MD) study of full-length autoinhibited c-Abl kinase showed strongly correlated motions between the Src homology (SH)3 and SH2 domains, and between the SH2 domain and a part of the N-lobe (12). A subsequent coarse-grained model of the protein kinase core, based on normal mode analysis (13), led to the identification of 32–45 semirigid blocks, depending on the specific enzyme, where each block comprised five to eight amino acids. However, the functional context of these blocks was obscure. There is obviously potential value in delineating larger dynamic units that may afford a higher-level description of functional motions, such as large-scale conformational changes upon ligand binding or posttranslational modification.
Fast, effective algorithms to detect communities in large social and biological networks (14–16) have contributed to an explosion of network studies in the social sciences and systems biology (17). Community analysis can also be applied to correlated motions in proteins and nucleic acids (18, 19). In such studies, MD simulations are performed and correlations among residues are evaluated. A graph that then represents a dynamic “portrait” of the molecule is created, where residues are the vertices of the graph and correlation values are the weighted edges between them. Community analysis of such correlation networks identifies relatively independent communities of residues that behave as semirigid bodies. Initial applications of this method to study protein dynamics provided insight into the propagation of allosteric signals in bacterial tRNA-synthetases (18) and in a sortase enzyme from Staphylococcus aureus (19).
Here, we use protein kinase A (PKA) as an exemplar of an active protein kinase, because its multiple structures reflect different stages of phosphotransfer, and wild-type and mutant forms have been extensively studied by biochemical and biophysical methods. PKA belongs to the AGC family of kinases, which are typically characterized by a cis-regulatory C-terminal tail (20). Recently multiple-microsecond timescale simulations have been performed on several tyrosine kinases (21–24), and a Markov model was constructed for activation of Src kinase that captured microsecond–millisecond motions (25). Here we present multiple-microsecond simulations of a Ser/Thr kinase, PKA.
We applied a dynamic community analysis to simulations of PKA and discovered that major communities do not necessarily overlap with the structural elements that were previously perceived as rigid bodies (e.g., the five-stranded β-sheet in the N-lobe, or the αF-helix in the C-lobe). Instead, they center on main functional and regulatory regions, in many cases splitting seemingly stable secondary structural elements into different communities. The size and the number of these communities, as well as the strength of dynamic coupling between them, are sensitive to metal and ligand binding. For example, removal of a single magnesium ion from the active site leads to considerable changes in the community map. Such sensitivity is consistent with the magnesium-dependent large-scale structural changes observed in NMR studies of PKA (26) and also with earlier biophysical measurements (27), MD simulations (28), and recent crystal structures (29). We therefore propose that segregation of the protein kinase molecule into dynamic communities is a more efficient way to analyze the functional state of PKA than traditional representations based on sequence and secondary structure elements. Furthermore, the general architecture of the community map in the fully active form of PKA is consistent with the previously proposed hydrophobic “spines” framework (8, 9). Because the “spines” have been shown to be a conserved feature of all EPKs, we hypothesize that the community maps for all fully active EPKs will have similar patterns.
This analysis offers a novel perspective for the interpretation of experimental and naturally occurring mutations, where one can look not only at which individual residues are directly affected, but also at which communities are likely to be functionally linked. One should no longer be surprised by long-range allosteric effects of perturbations if they occur within the same community, or between communities that are closely linked. The functional annotations associated with these communities allow this signaling enzyme to be viewed as a dynamic switch composed of parts with reasonably well-defined functions. The present work builds upon established methods, such as mutual information and community analysis, to analyze kinase dynamics and identify structurally contiguous sets of residues exhibiting correlated motions and similar functions. Our work sets the stage for a broader study of multiple kinases aimed at identifying common communities that are shared between many kinases and auxiliary communities that are distinct to particular subfamilies.
Results and Discussion
We simulated PKA in four different ligand and conformational states: closed with ATP and two Mg2+ ions bound (5.3 μs), closed with ATP and one Mg2+ ion bound (5.7 μs), open with ATP and one Mg2+ bound (7.8 μs), and open with neither ATP nor Mg2+ bound (apo; 3.3 μs). For each simulation, generalized correlations among residues, each partitioned into side-chain and backbone parts, were quantified in terms of mutual information with the program MutInf (30), and the resulting matrix of correlations was analyzed with the Girvan–Newman algorithm (14) to identify the community structure. We first provide a general overview of the MD simulation results. We then analyze the structure and functions of the communities identified for the closed conformation of PKA with ATP and two Mg2+ ions. This construct is particularly relevant physiologically, because it corresponds to the active kinase, uninhibited by PKI or by regulatory subunits, in an in vivo environment with excess Mg2+. We then highlight how structural elements such as the C-tail, which lies outside the core and is unique to AGC kinases, and the activation segment, which is a conserved feature of all EPKs, contribute to different communities. Next, we describe changes in the communities that result from changing the ligands and open vs. closed conformations. Finally, we provide examples where we use the community map and structural data to interpret mutations.
MD Simulations.
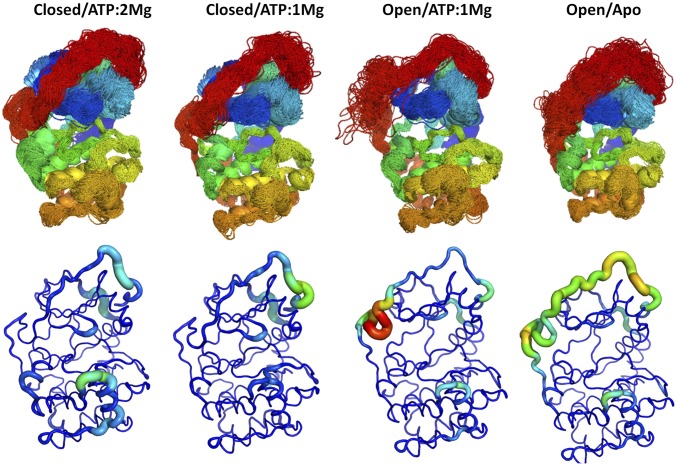
Overlays of sampled conformations and per residue root-mean-square flexibilities (Fig. 1) provide a perspective of the conformational fluctuations sampled in the microsecond-scale simulations analyzed here. The Gly-rich loop opened and closed in this timeframe (SI Appendix, Fig. S1). The segmental flexibility of the C-tail is sensitive to metal binding (one vs. two Mg2+) and to opening and closing of the cleft. The C-tail is most flexible in the apo state, whereas the G-helix in the C-lobe is most mobile in the closed/ATP/two Mg2+ state. Over this time scale the phosphorylated activation loop remains very rigid in all states (Fig. 1 and SI Appendix, Fig. S1).
Fig. 1.
MD simulations of PKA in different ligand and conformational states, as noted, show that the conformation and flexibility of the G-helix and C-terminal tail are particularly sensitive to active site ligands. (Upper) Superimposed MD trajectory snapshots of the PKA backbone illustrate the conformational space sampled in the simulations, including differences among the various ligand and conformational states. The backbone of PKA is colored from blue to red according to position in the primary sequence from the N terminus to the C terminus, so that the C-tail is red. (Lower) Backbone root-mean-squared position fluctuations highlight the most dynamic regions of PKA in various liganded and conformational states. The largest differences in flexibility are seen in the C-terminal tail and the G-helix.
Functional Analysis of PKA’s Dynamic Communities.
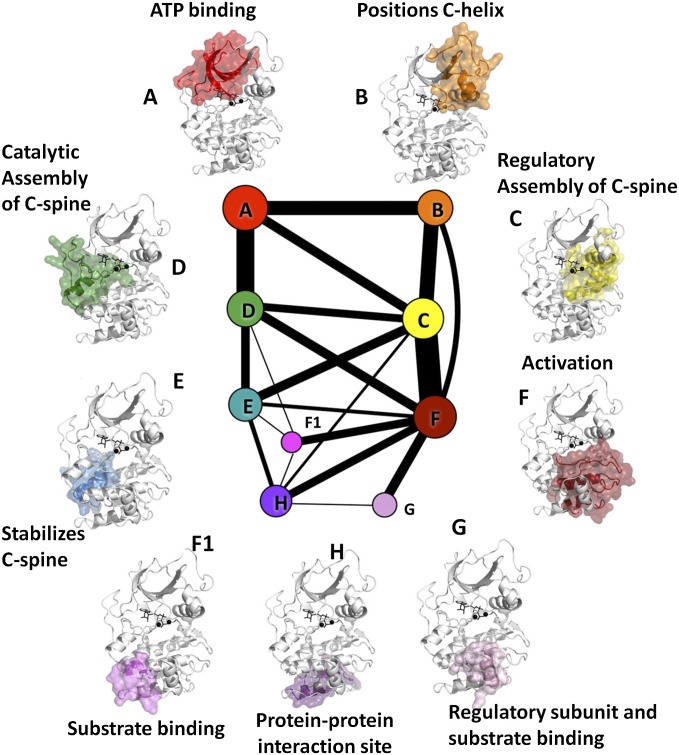
Nine communities (Fig. 2 and Movie S1) emerged from the dynamic analysis of the active form of PKA (closed, with ATP and two Mg2+). We designate them ComA–ComH, in alphabetical order following primary sequence and matching, if possible, community labels to the labels of structurally prominent helices (e.g., the αB-helix for ComB and the αC-helix for ComC). For this analysis, the main-chain and side-chain of each residue are considered independently and sometimes fall into different communities. The smallest community, F1, has 15 main-chain and 15 side-chain members; the largest community, F, has 58 main-chain and 60 side-chain members. The average number of both main-chain and side-chain segments is 37. The thickness of the line segments, also termed edges, joining two communities (Fig. 2) indicates the strength of their dynamic coupling. The edge weights are provided in SI Appendix, Table S1 for reference.
Fig. 2.
Community map of PKA, for the closed conformation with ATP and two Mg bound. The size of each node represents the number of members (residue backbones and side-chains) of each community, and the edge weights are proportional to the thickness of the lines. The full kinase is shown in white, the communities are highlighted in red, and ligands are shown in black. The legend annotates each community by its function.
It is widely accepted that, once the R-spine of an EPK is assembled, its N- and C-lobes perform a “breathing motion,” opening and closing the active site cleft (1, 2, 11, 31–33). This picture might suggest that the kinase lobes behave as semirigid bodies and, consequently, that the borders between communities would lie along the boundaries between the lobes. To our surprise, this is not the case. For example, the five-stranded β-sheet in the N-lobe is divided between ComA and ComB, with ComA covering the ATP binding site, but not the rest of the lobe. ComB includes a part of the Gly-rich loop and portions of β3–β5, and is centered around the hydrophobic FxxF motif in the C-terminal tail, which is a well-recognized regulatory hot spot for multiple kinases (34). ComC is one of the largest communities and the most interconnected. ComC bridges the two lobes and includes α-helix A and α-helix C from the N-lobe, as well as parts of the catalytic loop, the activation segment, β6 and β9, and a part of the αE-helix, all from the C-lobe. Although these results might appear counterintuitive in terms of a traditional perception that the N- and C-lobes represent two largely rigid domains, they are remarkably consistent with the current understanding of the main functions performed by the kinase domain.
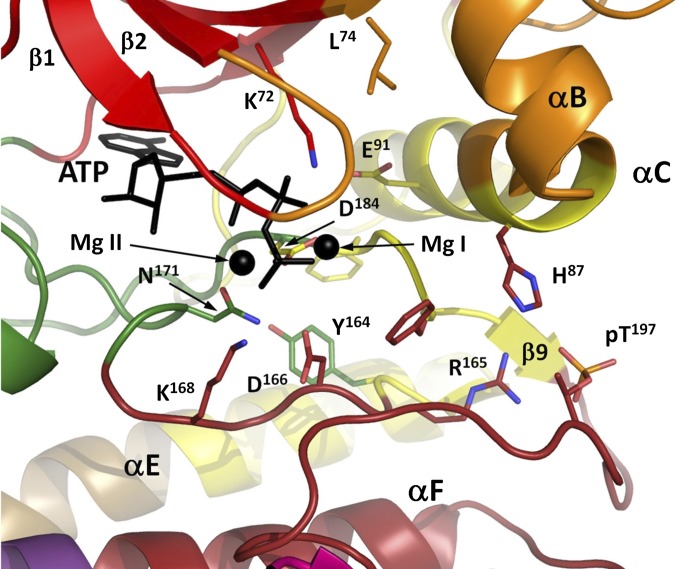
In particular, decomposition of the kinase core into dynamic communities yields arguably a more coherent functional segmentation of the structure (Fig. 2) than does the conventional representation of the protein as a set of secondary structural elements connected by loops, as now detailed. Catalytic machinery (Fig. 3) appears in four different communities (K72 in ComA, D184 in ComC, N171 in ComD, and D166 in ComF) (35). These communities, in turn, are coupled (edges in Fig. 2) to additional communities that lie further from the catalytic machinery, yet harbor sites where substrates or regulatory subunits dock to the kinase (i.e., ComG, which docks to the R-subunit and PKI, and ComH, which also interacts with the R-subunit). A wealth of biochemical and structural data allows us to provide a finer-grained functional annotation of each community, as shown in Table 1.
Fig. 3.
Multiple communities converge around the ATP-binding and catalytic site. ComD provides catalytically critical N171, which coordinates Mg II and D166, a proton acceptor for the substrate’s –OH group in catalysis.
Table 1.
Communities in the active form of PKA
| Name | NMC | NSC | Main function |
| ComA | 60 | 50 | ATP binding |
| ComB | 58 | 58 | Regulates position of the αC-helix |
| ComC | 38 | 41 | Regulatory, assembly of R-Spine |
| ComD | 58 | 62 | Catalytic, ATP binding, and assembly of C-spine |
| ComE | 28 | 29 | Stabilizes C-spine (tentative) |
| ComF | 40 | 44 | Stabilizes active conformation of activation loop |
| ComF1 | 15 | 15 | Substrate binding |
| ComG | 23 | 22 | Regulatory subunit and substrate binding |
| ComH | 16 | 15 | Protein–protein interaction site (tentative) |
Name, name of community; NMC, number of residue backbones in the community; NSC, number of residue side-chains in the community.
We next examine ComA, -B, -C, -D, and -F (Fig. 4–6); analysis of the remaining communities is included in SI Appendix, which provides detailed descriptions of all communities. Partitioning of the N-lobe into three main communities highlights the functional insights that emerge from this analysis. ComA is committed to ATP binding (Fig. 4A). It consists of the N-terminal linker, which connects the αA-helix to β1; the bulk of the β-sheet of the N-lobe (β1–β5); a portion of the C-terminal tail (C-tail), which contributes to ATP binding; and part of the hinge that links the N- and the C-lobes. ComA comprises most of the upper surface of the ATP binding site, along with the F327 that plugs the side of the site.
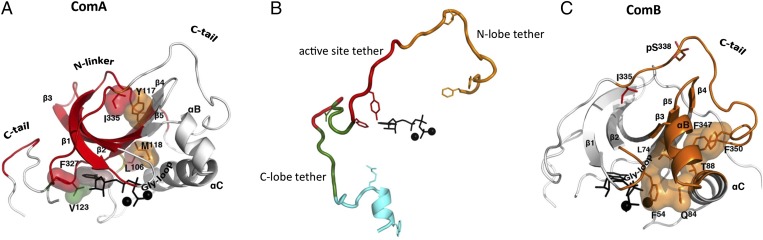
Fig. 4.
The N-lobe is subdivided into three communities that are largely preserved across different ligand/conformational states. (A) In ComA, we show a set of bridging residues’ side-chains in surface representation. (B) In ComB, a chiefly hydrophobic set of residues connects the hydrophobic motif (F347, F350) to the αC-helix and Gly-rich loop. (C) Previously identified subdivisions of the C-terminal tail segregate into different communities, and the entire tail connects communities A, B, D, and E.
Fig. 6.
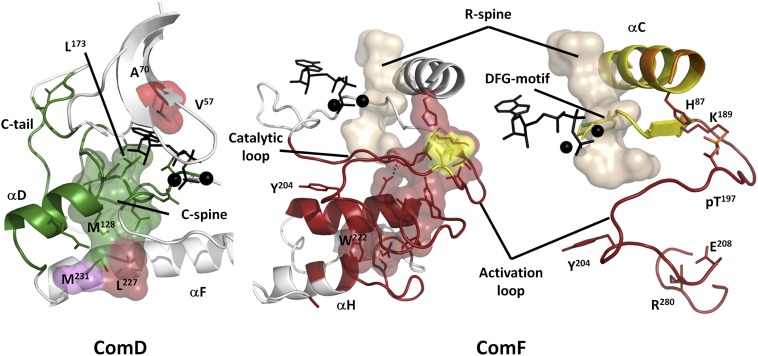
ComD and ComF are C-lobe communities that connect the C-lobe to the N-lobe. ComD (green) encompasses the αD-helix and part of the catalytic loop, coordinating ATP and Mg from below, and includes the majority of the C-spine (surface). Other C-spine residues are colored by their community. ComF (dark red) involves a large section of the C-lobe and includes a set of chiefly polar residues (surfaces) that nucleate around the activation loop phosphate (pT197, yellow) and connect the C-helix to the activation loop, F-helix, and finally AGC insert, involving a conserved salt bridge between E208 and R280. The activation segment is split across two communities and structurally links the C-helix to ATP/Mg, the phosphorylation site, and to the bottom of the C-lobe through the conserved salt bridge between E208 and R280. Additionally, Y204 connects this segment to E230, a critical substrate-binding element in Com-H.
The importance of ComB (Fig. 4C) is emphasized by its incorporation of part of the critical αC-helix, which must be positioned properly for phosphate transfer to occur. ComB also includes the αB-helix, part of the Gly-rich loop (Gly-loop), and the C-terminal part of the C-tail, including its hydrophobic motif. Surprisingly, some of the β-strands belong to two communities (ComA and ComB). Part of the Gly-loop, for example, that lies between β1 and β2 belongs to ComB while β1 and β2 are part of ComA. The partitioning of the C-tail between ComA and ComB is consistent with the sequence-based analysis carried out earlier on the AGC kinases, which identified an active site tether, an N-lobe tether, and a C-lobe tether (Fig. 4B) (20). The N-lobe tether is part of ComB and is anchored to the αC-helix by a conserved hydrophobic motif (FxxF). The active site tether mostly belongs to ComA and F327, which, as noted above, forms part of the ATP binding site (36).
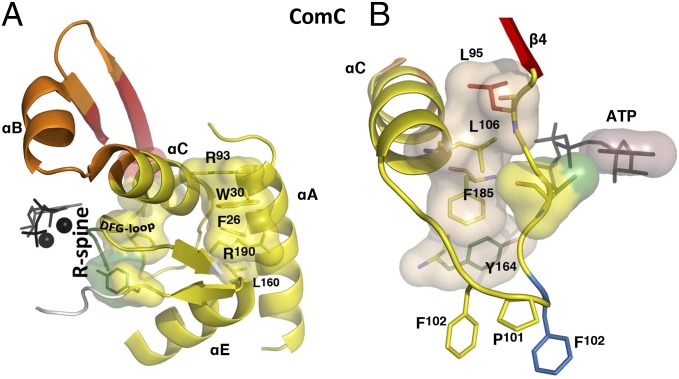
ComC acts as a hub in the community map, having connections to six other communities. It is formed around the αC-helix in the N-lobe (Fig. 5A), which was previously identified as a signal integration motif (1) and emerges here as a major allosteric mediator for regulation of kinase activity. Side-chains from this helix radiate out to several communities (ComA, ComB, ComF). For example, the side-chain of K92 (ComB) helps to anchor the hydrophobic motif in the C-tail, and the side-chain of H87 connects the αC-helix to the activation loop phosphate. R93 forms a cation–π interaction with W30 in the αA-helix, which fills a conserved pocket in kinases that is commonly used to regulate activity (37, 38). Segments adjacent to the αC-helix in ComC include the αA-helix, the DFG-motif, and β9 in the activation segment. These provide important regions whose proper positioning can be regulated by binding at allosteric sites or phosphorylation of the activation loop. β9 and its flanking regions comprise a hotspot for cancer mutations in B-Raf proto-oncogene serine/threonine kinase (39).
Fig. 5.
ComC structurally connects the N-lobe to the C-lobe through the R-spine, DFG-motif, and αA-helix. (A) In ComC, surfaces show the R-spine and a set of cation–π interactions that connect the top of the αE-helix, the activation loop, and the αA- and αC-helices. (B) The αC-β4 region connects communities A, C, D, and E, providing a link between the R spine, ATP, and the C-lobe.
The αC-β4 loop is an anchor that immobilizes the C-terminal part of the αC-helix. Earlier computational work showed that it is the only part of the N-lobe that moves in concert with the C-lobe (11). As seen in Fig. 5B, this loop can also be considered as a signal integration motif; it contributes to many communities, and in some cases the side-chain belongs to one community and the backbone to another (for example F102 at the tip of the loop). Previously, F100 and F102 were identified as AGC family-specific features that are anchored to the C-tail, which in turn helps the C-tail to position both ATP and the αC-helix (40). Another example is V104, whose functional significance was not appreciated until recently (41); its main-chain is in the ComC and its side-chain is in the ComD.
ComD (Fig. 6) consists of the αD-helix and β7 and β8 in the C-lobe, and part of the active site tether of the C-terminal tail (see below). ComD includes the middle part of the C-spine, as well as a critical Asn171 in the catalytic loop that coordinates one of the Mg2+ ions.
ComF (Fig. 6) contains the activation segment, part of the catalytic loop, the N-terminal part of the αF-helix, and the AGC insert. This region is highly regulated. Phosphorylation of the activation loop, for example, can stabilize this region in an active conformation that promotes activity. Also included in ComF is the APE motif, where E208 forms a highly conserved salt bridge with R280. This salt bridge is a defining feature of all EPKs (42). Additionally, this region is commonly involved in substrate binding through the P+1 loop, and the APE-αF loop is often involved in protein–protein interactions with regulatory proteins.
In contrast to our detection of the five main communities with clearly identifiable catalytic and regulatory functions, we discovered that most of the C-lobe is not a single stable conglomerate of hydrophobic residues, as one might expect. Instead the C-lobe contains several smaller communities (ComE, ComF1, ComG, ComH) that are capable of merging with or splitting from the major communities, depending on experimental conditions. The properties of this part of the kinase core remain, in many aspects, obscure. The importance of the C-lobe is emphasized, however, by the universal conservation of the α-helices G, H, and I, (GHI helical subdomain) in all EPKs. This region contains a binding site for regulatory subunits (43–46), substrates (47), phosphatases (48), and as a dimerization site (49). One may speculate that the sensitivity of the C-lobe’s community structure is mechanistically related to long-distance signaling between protein docking sites and the catalytic machinery. This would account, for example, for the previously observed effect of a substrate binding to the GHI subdomain on the activity of yeast isoform of PKA, Tpk1 (50), or, if this generalizes to other kinases, for the mechanism of action of allosteric inhibitors of Bcr-ABL kinase, which bind in a pocket between αE, αF, and αH-helices (51). We theorize that binding of other proteins or ligands to the GHI subdomain may lead to integration of the minor communities in the C-lobe and a subsequent change of the community map for the whole kinase core. Most of the unique EPK features identified by a Bayesian sequence analysis (20, 52) appear in ComF, including the expanded activation loop, regulation of the activation loop by phosphorylation, and the electrostatic linking of the activation segment to a helical domain following the αF-helix through E208 and R280.
Dynamic and Structural Connections Within and Between Communities.
Three key sequence segments of PKA integrate multiple communities and structurally connect the N-lobe to the C-lobe. First, the activation segment (residues 184–208) is split across communities C and F (Fig. 6C). The activation segment is a conserved structural element in all EPKs, which provides an opportunity to regulate kinase activity via phosphorylation of its primary phosphorylation site (T197 in PKA) (3). This segment contacts the N-lobe, goes through the C-lobe, and stabilizes the bottom of the C-lobe with a conserved salt bridge between E208 and R280. Proper positioning of the activation segment is required for full activity in most protein kinases, and its length and surface accessibility allow it to adopt multiple conformations and permit control by phosphorylation or protein–protein interactions. The DFG-β9 motif is partitioned into ComC, which highlights the critical importance of this community because the DFG-aspartate is a key catalytic residue and, therefore, ComC can directly influence the phospho-transfer reaction. The rest of the activation segment belongs to ComF. Second, the αC-β4 loop (Fig. 5B) begins in ComC and then briefly dips into ComE at F100, which is conserved in AGC-kinases and is homologous to the His in the HxN motif, which—in Cdk2 and other kinases—connects the αC-helix to the catalytic core (40). The αC-β4 loop presents V104, part of the R-spine shell (41), into the ATP-binding pocket, and then terminates at L106, an R-spine residue in ComA that connects to ComC. Third, the division of the C-terminal tail (Fig. 4C) across four different communities recapitulates the previously described subdivisions of this linear segment (20).
Earlier we described a general framework for a protein kinase domain internal architecture (7–9). It is based on two hydrophobic spines, the R-spine and the C-spine, which are connected by a long hydrophobic F-helix. The implied function for the spines was a firm but flexible connection between the kinase lobes. Following this model, one can suggest that, in the community map, the spines—in particular the side-chains—should serve as flexible bonds between the contiguous, semirigid, communities: that is, correspond to the edges in the community maps. Indeed, both the R-spine and C-spine are split across multiple communities. Fig. 7 shows that the C-spine connects ComA, ComD, and ComF, which help position ATP, substrate, and key catalytic machinery. The R-spine connects ComA and ComF via the regulatory hub community ComC.
Fig. 7.
Community maps of PKA are similar across different ligand and conformational states and reveal conserved cores. For each of the four ligand and conformational states, the community map is shown with node sizes proportional to the relative number of community members and edge weights proportional to the total mutual information between the communities. To the left of each map, the structure of PKA is colored by community. To the right of each community map, the R-spine and C-spine are shown and colored by community; ATP is shown in sticks, and the C-spine and R-spine are shown in surface representations.
Looking beyond the spines, a number of communities have compact groups of residues that appear to act as stabilizing cores. Although in principle there could be a number of ways to identify such residues, we leveraged a prior LSP alignment to identify structurally conserved residues in the conserved kinase catalytic core (8, 9). This analysis points to the existence of a set of community-forming residues for each community, as detailed in SI Appendix, Fig. S5. Because the community-forming residues were identified by conserved spatial patterns in multiple kinases, we expect that the community-forming residues shown here might also be important in other protein kinases, much as the importance of the spines has already been documented in RAF kinases (53), Bcr-ABL (54), Src (55), EGFR kinase (49), and anaplastic lymphoma kinase (56).
Community Analysis of Other Conformational and Ligand States of PKA.
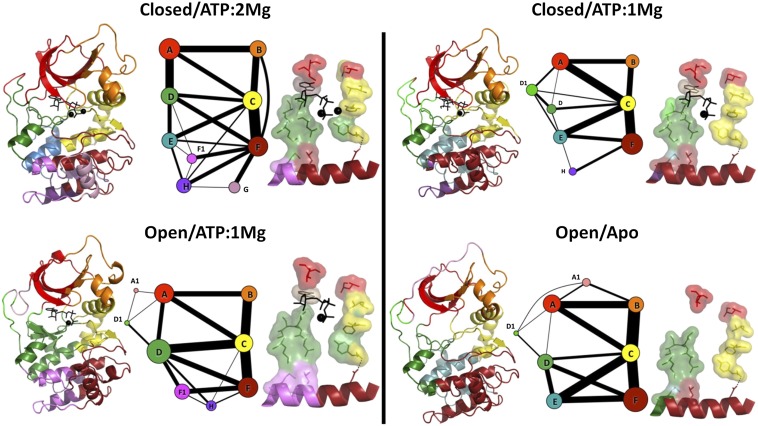
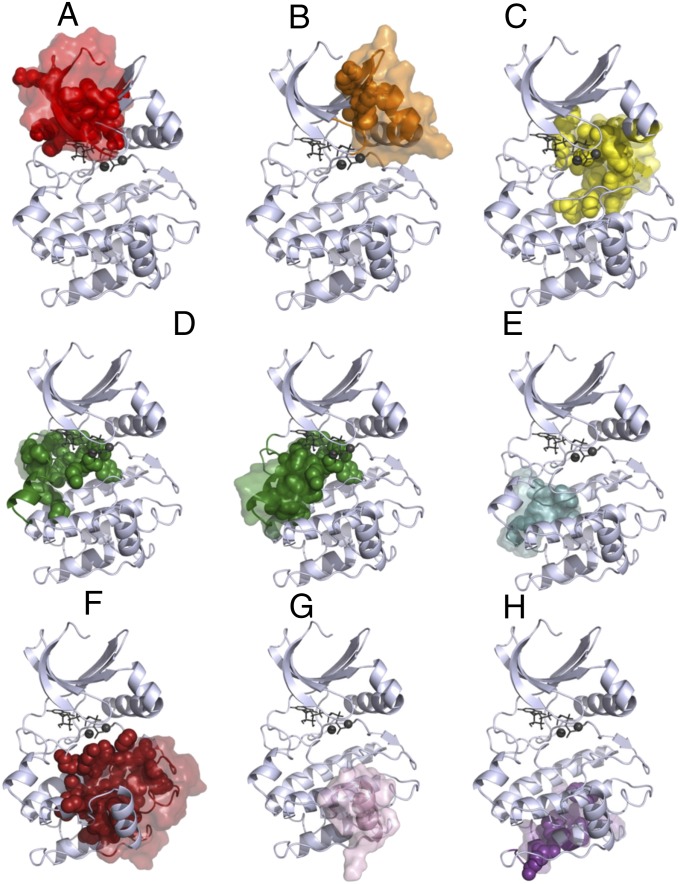
Because PKA’s structure and dynamics depend on the presence of nucleotide and divalent cations, it is important to explore how changes in conformation and ligand status affect the community analysis. Accordingly, Fig. 7 compares the community maps constructed from simulations of PKA in three additional states (closed/ATP/one Mg2+, open/ATP/one Mg2+, and open/apo) to the reference closed/ATP/two Mg2+ state. We noted that some groups of residues in the catalytic core of the kinase (i.e., excluding the N- and C-tails) consistently colocalize in communities with one another in all systems, and refer to these as conserved cores (Fig. 8). These conserved cores form semirigid elements that remain structurally connected in the closed/ATP/two Mg2+ simulation (Movie S2).
Fig. 8.
Community maps (A–H) of PKA in different conformational and ligand states reveal cores (surfaces), members that remain in the same community with each other. These cores contain many of the community-forming residues (spheres). In ComD, because there are two conserved cores, community-forming residues that belong to the other core are not shown.
The majority of the changes in the communities occur in the C-lobe substrate-binding regions and in the C-terminal tail. The N-lobe does not show substantial changes, and the communities that are least affected by changes in conformation and ligand states are ComA, ComB, ComC, and ComF. For all states, the αA-helix and αC-helix remain in ComC, whereas the rest of the N-lobe remains split between ComA and ComB, and the activation segment and part of the F-helix remain in ComF. In the closed/ATP/oneMg2+ state, several residues belonging to the C-tail split from ComA, to form a small additional community termed ComA1. In contrast, the residues belonging to ComD, ComE, ComG, and ComI in the closed/ATP/two Mg2+ structure are grouped into slightly different communities in the other structures. In particular, ComG forms its own community apart from ComF only in closed/ATP/two Mg2+. The regulatory and catalytic spines have remarkably similar community memberships across all states: V57, A70, and the side-chain of L106 remain in ComA; M128 and residues 172–173 remain in ComD; and the backbones of L95, F185, and Y164 consistently remain in ComC, as does the side-chain of L95.
Mutagenesis Validates the Community Maps.
The assignment of function to individual communities suggests that mutations that destabilize a community will tend to disrupt the associated function, and vice versa, even if the key functional site is spatially remote. Indeed, the community map is found to be consistent with a wide range of mutagenesis studies, beginning with the initial Ala scan of the yeast PKA-C-subunit (57) and the more recent Ala scan of the C-tail (58), and extending to individual mutations that have been comprehensively characterized. For example, mutation of the conserved ion pair, E208-R280, in ComF (57, 59) has a profound effect on substrate binding, catalysis, and stability. This observation is consistent with the fact that ComF links ComC and ComD, which form the active site; and that ComF is directly involved in substrate binding. The fact that ComF links the protein docking site with the catalytic communities also is consistent with Herman and colleague’s observation (50) that the docking site on the αH-helix is allosterically coupled to the catalytic site.
The community maps also suggest why the activation loop phosphate is so important. The phosphate provides a multivalent electrostatic link between ComC and ComF, and the community maps suggest that the allosteric range reaches to ComA, ComB, and ComD as well, consistent with the fact that dephosphorylation leads to changes in catalytic activity, stability, global H/D exchange, and conformation, as reflected in a very open crystal structure, where the R-spine is broken and the activation loop and C-tail are disordered (3, 60). The community maps also generate the same partitioning of the C-tail that was previously revealed by deep Bayesian analysis of the evolution of the C-tail (20) and by alanine scanning (61, 62), as well as by detailed characterization of particular sites (36, 62–64). The myristyl binding pocket is in a region of the protein, whose community assignment is particularly sensitive to conditions, such as the presence or absence of bound ATP and Mg, and which comprises residues from multiple other communities. This observation is consistent with recent crystallographic, fluorescence, and computational analyses showing long-ranged effects of myristyl binding (65, 66). Individual residues that have a profound effect on activity and stability [E230Q in ComF1 (67, 68), W30A in ComA (69), and Y204A in ComF (70, 71)] are clearly part of extended allosteric networks defined here by the community maps.
Template for Allosteric Regulation.
In recent years, allostery has reemerged as a major theme for understanding protein structure and function. Here, we provide a new framework for understanding the allosteric integration of structural, dynamic, and functional motifs in kinases. Often, single-site perturbations, such as mutations (W30A, F327A, F350A), or posttranslational modifications (for example adding a myristyl group to the N terminus or a phosphate group to the activation loop) can have significant effects on dynamics and function of distant regions. The community maps presented here provide an estimate of the effective allosteric range of such perturbations: a mutation in one community cannot only alter its own community, but also highly coupled communities. The strength of the couplings are reflected in the width of the edges in the community map. This concept helps explain how perturbations, such as myristyl binding between ComC and ComE, can cause changes in dynamics in not only ComC but also in ComB and ComD, as measured by fluorescence anisotropy (66).
The communities each contain known functional elements and provide a way of understanding how different motifs can be coupled. The two communities in the N-lobe, for example, move together as a semirigid body, with one binding ATP (ComA) and the other positioning the αC-helix (ComB). Because the kinase catalytic core, the spines, and the community-forming residues are all structurally conserved, we also anticipate that the communities will, in general, be conserved between different kinases. We expect that the core residues that are conserved will partition into more or less the same communities, whereas the elements that lie outside the core, such as the C-tail, will be family-specific. Conserved pockets on the core often serve as allosteric docking sites (34). How these pockets are filled will vary for each kinase, but the way in which they integrate with the core and regulate the core might be conserved.
In contrast to powerful evolutionary sequence-based methods, such as protein sector analysis (72) and CHAIN (20), which identify coevolving sets of residues with distinct functions and capture family-level features, our approach focuses on the dynamics of individual family members. Sequence-based approaches cannot account for posttranslational modification or ligand binding, but our approach is quite sensitive to changes in conformation and dynamics upon binding different ligands. The present community map analysis goes beyond primary, secondary, and tertiary structure, providing a new framework for understanding correlated motions and allostery.
Conclusion
Correlation analysis of multiple-microsecond duration MD simulations identifies dynamic communities, structurally contiguous regions of PKA that exhibit correlated motions. Each community has a structurally conserved scaffold provided by community-forming residues, and a largely overlapping group of residues that remain together in all four of the ligand and conformational states of the enzyme examined here. Correlation of mutational analysis with the community structure allows annotation of each community according to key functions. This functional annotation provides arguably a more coherent annotation of structure with function than does the traditional partitioning into secondary structure elements. This structural and functional community analysis complements the previous identification of the C- and R-spines, and offers a more complete picture of the internal architecture of an exemplar protein kinase in an active state. The communities and the spines that connect them provide a conceptual framework for long-distance allosteric communication between different sites on PKA. One may speculate that a similar pattern will emerge from community analyses of other active kinases as well.
Methods
System Preparation.
To prepare the two closed conformations, we began with a high-resolution structure of PKA in the closed conformation with ATP and two Mn2+ ions, (PDB ID code 3FJQ). Residues T197 and S338 were phosphorylated as in the crystal structure. The Protein Local Optimization Program was used to preprocess the PDB file (73), then the structure was imported into Maestro (Schrodinger). The Mn2+ ions were changed to Mg2+ ions and the Protein Preparation Wizard was used to assign bond orders, add hydrogens, and keep all crystal waters. Hydrogen bonds and histidine protonation states were optimized for neutral pH, yielding HIP for H87, HIE for H142, and HID protonation states for the remaining histidines. The crystal waters and PKI peptide were removed. Removal of Mg1 yielded the closed, ATP/Mg2+ starting model. [The protonation state of C199 was modeled as negatively charged based on the very high in vitro reactivity of this cysteine (74, 75).] For PKA in the open state with ATP/one Mg2+ PDB ID code 1CMK (76) was used as a starting point, as it had an ordered C-terminal tail in contrast with other structures that have missing residues in this region. PKI, myristate, and an iodine ion were removed. The Protein Local Optimization Program (77) was used to fix nine side-chain differences between this Sus scrofa structure and the desired Mus musculus sequence (97% sequence identity), and the ATP/Mg2+ was aligned from PDB ID code 3FJQ (34) by the N-lobe, so that the adenine ring would properly hydrogen bond with the hinge. The protonation states were selected to be the same as in the closed state. For comparison with the closed-state structures, residues 1–14 were removed from this model, as these were disordered in the closed state. For the open/apo model, ATP/Mg2+ was removed. Hydrogens were removed from all models before preparation for AMBER 11 (78) and AMBER 12 (79), using the FF99SB-ildn-nmr forcefield (80–82). The proteins were solvated in a cubic box of TIP4P-EW water (83) with 10 Å buffers, with parameters for ATP (84), phosphothreonine (85), and phosphoserine (85) taken from the Bryce group’s AMBER Parameter Database (86). SHAKE (87) was used to constrain all covalent bonds from to hydrogen atoms.
MD Simulations.
The program AMBER12 was used for the initial energy-minimization, heating, and equilibration steps, using the pmemd.cuda module (88, 89). Each system was energy-minimized, first with position restraints on the protein and ATP/Mg2+, and then without restraints. Constant volume simulations using particle mesh Ewald (90) with a 10 Å cut-off for range-limited interactions was used. MD with a 2-fs time-step was performed to heat the system from 0 K to 300 K, linearly over 500 ps, with 10.0 kcal⋅mol⋅Å position restraints on the protein and ATP/Mg2+. Temperature was maintained by the Langevin thermostat, with a collision frequency of 1 ps−1. Then, constant pressure dynamics was performed with isotropic position scaling, first with position restraints for 100 ps and a relaxation time of 2 ps, and then without position restraints for 100 ps. The range-limited cut-off was dropped to 8 Å and the simulation was continued for 50 ns. For closed/ATP/two Mg2+, the system was simulated briefly for 12 ps of Nosé–Hoover constant volume MD (91, 92) using Desmond (DE Shaw Research) at 2-fs time-steps to further prepare for simulation on Anton.
Production runs for closed/ATP/two Mg2+, closed/ATP/one Mg2+, and open/ATP/one Mg2+ were performed on a 512-node Anton supercomputer at constant volume, using time-steps of 2 fs and the Nosé–Hoover thermostat (91, 92), with a relaxation time of 1 ps and center of mass motion removed. For open/apo, three independent equilibrations were performed as above, followed by 1.1-μs production runs using the pmemd.cuda module of AMBER12 and the Langevin thermostat as above, for a total simulation time of 3.3 μs. Trajectories for all systems were saved at 120-ps intervals, because on Anton more frequent output showed decreased performance in this system. Trajectories were aligned by C-lobe residues 128–300, and root-mean-square backbone fluctuations were calculated in GROMACS for all residues after discarding the first 1.2 μs of the Anton datasets.
Mutual Information Calculations.
For our community analysis, we computed Cartesian mutual information values instead of dihedral mutual information values, as the former better capture correlated motions involving semirigid regions, such as secondary structure elements (93), whereas the latter are more suitable for analyzing couplings between surface sites where binding and posttranslational modification often occur (30, 94). We calculated the Cartesian mutual information for backbone Cα atoms, as well as for a representative side-chain atom (SI Appendix, Table S2) of each residue. In the case of glycine, the carbonyl oxygen was used as a second atom because this atom is most distal from the Cα atom. The matrix of mutual information among these atoms was calculated with the MutInf software package (30, 95). To construct the probability densities required to obtain the mutual information values, we used uniformly binned histograms with 24 bins per degree-of-freedom. We then split each trajectory into six blocks of equal size. We used 20 sets of scrambled data to estimate the distribution of mutual information under the null hypothesis of independence, and calculated the excess mutual information by subtracting the average mutual information under the null hypothesis of independence from the observed mutual information, and corrected for undersampling using mutual information between residues in different simulation blocks, as previously described (30).
Community Analysis.
We initially represent the kinase catalytic domain as a graph having a node for each residue’s main chain and side-chain atoms, with edge weights given by the corresponding Cartesian mutual information (96). We then removed edges corresponding to pairs of atoms that were not within a given distance cut-off for 75% of the simulation time (SI Appendix, Fig. S1), and used the Girvan–Newman algorithm (14) to identify the community structure of the resulting pruned graph. This algorithm generates a new graph with one node per community with a size proportional to the number of elements and edges whose weights represent the sum of the weighted betweenness (96) of the edges connecting the communities. The use of a distance cut-off to prune edges in the initial graph allows a focus on direct, rather than indirect correlations (97), as the latter are expected to be more significant at long range. Note that here, the distances are not minimum distances between residues but distances between representative atoms, so longer cut-offs are needed relative to a minimum-distance approach. We chose the cut-off distance between 8 Å and 10 Å that yielded the fewest number of communities, based on the relative insensitivity of the number and shape of the community map in the 8–10 Å range (SI Appendix, Fig. S2). In the case of a tie between two cut-off distances, the longer one was used. In the active state of PKA (closed, two Mg2+), this led to a cut-off of 10 Å. The edge weights (SI Appendix, Table S1) are used to manually redraw the community map in a more accessible format.
LSP Alignment.
To identify community-forming residues, we leveraged our previously described LSP alignment results. Briefly, the LSP alignment method identifies residues with similar physicochemical properties that exhibit similar relative orientations in eight structures of active protein kinases compared with PKA, and calculates the involvement score for each residue as the number of connections to the residue in the similarity graph. We found that an average involvement score of 6.25 (corresponding to a summed involvement score of at least 50 over the eight structures in ref. 7) gave reasonable results, because the community-forming residues in each community formed a structurally contiguous set located near the geometrical center of each community. To identify community-forming residues outside of the structurally conserved catalytic core, we identified residues in the N-tail and C-tail that formed hydrophobic or cation–π interactions and were structurally contiguous with community-forming residues in the conserved kinase catalytic core.
Supplementary Material
Acknowledgments
We thank Mikolai Fajer, whose initial application of community analysis to PKA inspired the present work; Paul Gasper and Jeff Wereszcynski, who provided code for the Girvan–Newman algorithm and community analysis; and Gianluigi Veglia, Fritz Herberg, and laboratory members for valuable discussions. This work was supported in part by National Institutes of Health Grants F32 GM099197 and R01 GM100310.
Footnotes
Conflict of interest statement: M.K.G has an equity interest in, and is a cofounder and scientific advisor of, VeraChem LLC.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418402111/-/DCSupplemental.
References
- 1.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 2.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SS, Keshwani MM, Steichen JM, Kornev AP. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc Lond B Biol Sci. 2012;367(1602):2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AS, Kornev AP, Hu J, Ahuja LG, Taylor SS. Kinases and pseudokinases: Lessons from RAF. Mol Cell Biol. 2014;34(9):1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 6.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 7.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103(47):17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105(38):14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhard S, Noé F. Optimal identification of semi-rigid domains in macromolecules from molecular dynamics simulation. PLoS ONE. 2010;5(5):e10491. doi: 10.1371/journal.pone.0010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsigelny I, et al. 600 ps molecular dynamics reveals stable substructures and flexible hinge points in cAMP dependent protein kinase. Biopolymers. 1999;50(5):513–524. doi: 10.1002/(SICI)1097-0282(19991015)50:5<513::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112(6):859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 13.Shudler M, Niv MY. BlockMaster: Partitioning protein kinase structures using normal-mode analysis. J Phys Chem A. 2009;113(26):7528–7534. doi: 10.1021/jp900885w. [DOI] [PubMed] [Google Scholar]
- 14.Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99(12):7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70(6 Pt 2):066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 16.Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato S. Community detection in graphs. Phys Rep. 2010;486(3-5):75–174. [Google Scholar]
- 18.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci USA. 2009;106(16):6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappel K, Wereszczynski J, Clubb RT, McCammon JA. The binding mechanism, multiple binding modes, and allosteric regulation of Staphylococcus aureus Sortase A probed by molecular dynamics simulations. Protein Sci. 2012;21(12):1858–1871. doi: 10.1002/pro.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA. 2007;104(4):1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci USA. 2009;106(1):139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan Y, et al. How does a drug molecule find its target binding site? J Am Chem Soc. 2011;133(24):9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 24.Shan Y, Arkhipov A, Kim ET, Pan AC, Shaw DE. Transitions to catalytically inactive conformations in EGFR kinase. Proc Natl Acad Sci USA. 2013;110(18):7270–7275. doi: 10.1073/pnas.1220843110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla D, Meng Y, Roux B, Pande VS. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat Commun. 2014;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masterson LR, et al. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108(17):6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herberg FW, Doyle ML, Cox S, Taylor SS. Dissection of the nucleotide and metal-phosphate binding sites in cAMP-dependent protein kinase. Biochemistry. 1999;38(19):6352–6360. doi: 10.1021/bi982672w. [DOI] [PubMed] [Google Scholar]
- 28.Khavrutskii IV, Grant B, Taylor SS, McCammon JA. A transition path ensemble study reveals a linchpin role for Mg(2+) during rate-limiting ADP release from protein kinase A. Biochemistry. 2009;48(48):11532–11545. doi: 10.1021/bi901475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastidas AC, et al. Phosphoryl transfer by protein kinase A is captured in a crystal lattice. J Am Chem Soc. 2013;135(12):4788–4798. doi: 10.1021/ja312237q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClendon CL, Friedland G, Mobley DL, Amirkhani H, Jacobson MP. Quantifying correlations between allosteric sites in thermodynamic ensembles. J Chem Theory Comput. 2009;5(9):2486–2502. doi: 10.1021/ct9001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett CP, Noble ME. Molecular motions of human cyclin-dependent kinase 2. J Biol Chem. 2005;280(14):13993–14005. doi: 10.1074/jbc.M407371200. [DOI] [PubMed] [Google Scholar]
- 32.Lu B, Wong CF, McCammon JA. Release of ADP from the catalytic subunit of protein kinase A: A molecular dynamics simulation study. Protein Sci. 2005;14(1):159–168. doi: 10.1110/ps.04894605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6(11):821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson EE, et al. Comparative surface geometry of the protein kinase family. Protein Sci. 2009;18(10):2016–2026. doi: 10.1002/pro.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, et al. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32(9):2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, et al. Contribution of non-catalytic core residues to activity and regulation in protein kinase A. J Biol Chem. 2009;284(10):6241–6248. doi: 10.1074/jbc.M805862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6(3):569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson EE, et al. Comparative surface geometry of the protein kinase family. Protein Sci. 2009;18(10):2016–2026. doi: 10.1002/pro.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gosal G, Kochut KJ, Kannan N. ProKinO: An ontology for integrative analysis of protein kinases in cancer. PLoS ONE. 2011;6(12):e28782. doi: 10.1371/journal.pone.0028782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kannan N, Neuwald AF, Taylor SS. Analogous regulatory sites within the alphaC-beta4 loop regions of ZAP-70 tyrosine kinase and AGC kinases. Biochim Biophys Acta. 2008;1784(1):27–32. doi: 10.1016/j.bbapap.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meharena HS, et al. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol. 2013;11(10):e1001680. doi: 10.1371/journal.pbio.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351(5):956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 43.Ilouz R, et al. Localization and quaternary structure of the PKA RIβ holoenzyme. Proc Natl Acad Sci USA. 2012;109(31):12443–12448. doi: 10.1073/pnas.1209538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307(5710):690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 45.Wu J, Brown SH, von Daake S, Taylor SS. PKA type IIalpha holoenzyme reveals a combinatorial strategy for isoform diversity. Science. 2007;318(5848):274–279. doi: 10.1126/science.1146447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang P, et al. Structure and allostery of the PKA RIIβ tetrameric holoenzyme. Science. 2012;335(6069):712–716. doi: 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122(6):887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 48.Song H, et al. Phosphoprotein–protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol Cell. 2001;7(3):615–626. doi: 10.1016/s1097-2765(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 49.Jura N, et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deminoff SJ, Ramachandran V, Herman PK. Distal recognition sites in substrates are required for efficient phosphorylation by the cAMP-dependent protein kinase. Genetics. 2009;182(2):529–539. doi: 10.1534/genetics.109.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463(7280):501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5(3):e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, et al. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013;154(5):1036–1046. doi: 10.1016/j.cell.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15(10):1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng Y, Roux B. Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop. J Mol Biol. 2014;426(2):423–435. doi: 10.1016/j.jmb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roskoski R., Jr Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res. 2013;68(1):68–94. doi: 10.1016/j.phrs.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266(14):8923–8931. [PubMed] [Google Scholar]
- 58.Shaltiel S, Cox S, Taylor SS. Conserved water molecules contribute to the extensive network of interactions at the active site of protein kinase A. Proc Natl Acad Sci USA. 1998;95(2):484–491. doi: 10.1073/pnas.95.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, et al. A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold. J Mol Biol. 2012;415(4):666–679. doi: 10.1016/j.jmb.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steichen JM, et al. Global consequences of activation loop phosphorylation on protein kinase A. J Biol Chem. 2010;285(6):3825–3832. doi: 10.1074/jbc.M109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbs CS, Knighton DR, Sowadski JM, Taylor SS, Zoller MJ. Systematic mutational analysis of cAMP-dependent protein kinase identifies unregulated catalytic subunits and defines regions important for the recognition of the regulatory subunit. J Biol Chem. 1992;267(7):4806–4814. [PubMed] [Google Scholar]
- 62.Batkin M, Schvartz I, Shaltiel S. Snapping of the carboxyl terminal tail of the catalytic subunit of PKA onto its core: Characterization of the sites by mutagenesis. Biochemistry. 2000;39(18):5366–5373. doi: 10.1021/bi000153z. [DOI] [PubMed] [Google Scholar]
- 63.Romano RA, Kannan N, Kornev AP, Allison CJ, Taylor SS. A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 2009;18(7):1486–1497. doi: 10.1002/pro.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonemoto W, McGlone ML, Grant B, Taylor SS. Autophosphorylation of the catalytic subunit of cAMP-dependent protein kinase in Escherichia coli. Protein Eng. 1997;10(8):915–925. doi: 10.1093/protein/10.8.915. [DOI] [PubMed] [Google Scholar]
- 65.Bastidas AC, et al. Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase. J Mol Biol. 2012;422(2):215–229. doi: 10.1016/j.jmb.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bastidas AC, Pierce LC, Walker RC, Johnson DA, Taylor SS. Influence of N-myristylation and ligand binding on the flexibility of the catalytic subunit of protein kinase A. Biochemistry. 2013;52(37):6368–6379. doi: 10.1021/bi400575k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J, et al. Crystal structure of the E230Q mutant of cAMP-dependent protein kinase reveals an unexpected apoenzyme conformation and an extended N-terminal A helix. Protein Sci. 2005;14(11):2871–2879. doi: 10.1110/ps.051715205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grant BD, Tsigelny I, Adams JA, Taylor SS. Examination of an active-site electrostatic node in the cAMP-dependent protein kinase catalytic subunit. Protein Sci. 1996;5(7):1316–1324. doi: 10.1002/pro.5560050710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6(3):569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore MJ, Adams JA, Taylor SS. Structural basis for peptide binding in protein kinase A. Role of glutamic acid 203 and tyrosine 204 in the peptide-positioning loop. J Biol Chem. 2003;278(12):10613–10618. doi: 10.1074/jbc.M210807200. [DOI] [PubMed] [Google Scholar]
- 71.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105(2):506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: Evolutionary units of three-dimensional structure. Cell. 2009;138(4):774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacobson MP, et al. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55(2):351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 74.Jiménez JS, Kupfer A, Gani V, Shaltiel S. Salt-induced conformational changes in the catalytic subunit of adenosine cyclic 3′,5′-phosphate dependent protein kinase. Use for establishing a connection between one sulfhydryl group and the gamma-P subsite in the ATP site of this subunit. Biochemistry. 1982;21(7):1623–1630. doi: 10.1021/bi00536a024. [DOI] [PubMed] [Google Scholar]
- 75.Nelson NC, Taylor SS. Differential labeling and identification of the cysteine-containing tryptic peptides of catalytic subunit from porcine heart cAMP-dependent protein kinase. J Biol Chem. 1981;256(8):3743–3750. [PubMed] [Google Scholar]
- 76.Zheng J, et al. Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci. 1993;2(10):1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacobson MP, Kaminski GA, Friesner RA, Rapp CS. Force field validation using protein side chain prediction. J Phys Chem B. 2002;106(44):11673–11680. [Google Scholar]
- 78.Case DA, et al. Amber11. Univ of California; San Francisco: 2011. [Google Scholar]
- 79.Case DA, et al. Amber12. Univ of California; San Francisco: 2012. [Google Scholar]
- 80.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li DW, Brüschweiler R. NMR-based protein potentials. Angew Chem Int Ed Engl. 2010;49(38):6778–6780. doi: 10.1002/anie.201001898. [DOI] [PubMed] [Google Scholar]
- 83.Horn HW, et al. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys. 2004;120(20):9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 84.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24(9):1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 85.Homeyer N, Horn AHC, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12(3):281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 86.Bryce RA. (2014) AMBER parameter Database. Available at www.pharmacy.manchester.ac.uk/bryce/amber. Accessed 22 September 2014.
- 87.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian equations of motion of a system with constraints—Molecular-dynamics of N-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 88.Götz AW, et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput. 2012;8(5):1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Grand S, Gotz AW, Walker RC. SPFP: Speed without compromise-A mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun. 2013;184(2):374–380. [Google Scholar]
- 90.Darden T, York D, Pedersen L. Particle mesh Ewald—An N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 91.Nose S. A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys. 1984;81(1):511–519. [Google Scholar]
- 92.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 93.Morcos F, et al. Modeling conformational ensembles of slow functional motions in Pin1-WW. PLOS Comput Biol. 2010;6(12):e1001015. doi: 10.1371/journal.pcbi.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wan X, Ma Y, McClendon CL, Huang LJ, Huang N. Ab initio modeling and experimental assessment of Janus Kinase 2 (JAK2) kinase-pseudokinase complex structure. PLOS Comput Biol. 2013;9(4):e1003022. doi: 10.1371/journal.pcbi.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McClendon CL, Hua L, Barreiro A, Jacobson MP. Comparing conformational ensembles using the Kullback-Leibler divergence expansion. J Chem Theory Comput. 2012;8(6):2115–2126. doi: 10.1021/ct300008d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Newman MEJ. Analysis of weighted networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70(5 Pt 2):056131. doi: 10.1103/PhysRevE.70.056131. [DOI] [PubMed] [Google Scholar]
- 97.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.