In response to the letter by Campos, “New structural model of adenovirus cement proteins is not yet concrete” (1), we strongly argue for the revised interpretations that we propose for the cement proteins in human adenoviruses (HAdVs) (2).
Fig. 1.
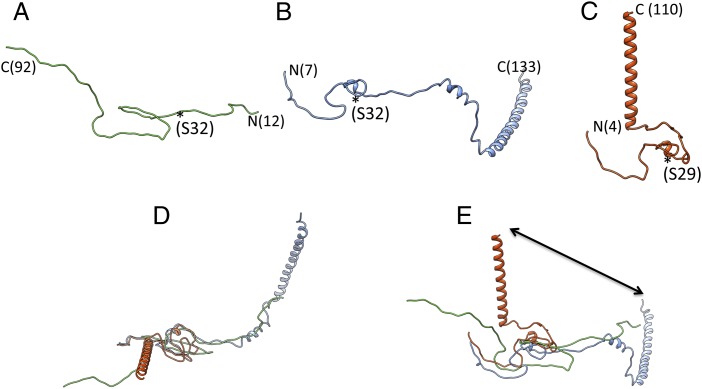
Similarities and differences in the structures of protein IX in HAdV5 and bovine adenovirus 3 (BAdV3). (A) The fold and chain trace of protein IX (green) in HAdV5 (Ad5F35) derived from the crystal structure (2). (B) The structure of IX (blue) from HAdV5 derived from cryo-EM analysis (6). (C) Structure of IX (red) from BAdV3 (7). The asterisk (*) denotes the location of one of the conserved residues in the triskelion region in the respective structures. (D) Approximate structural alignment of IX molecules, a view down a local threefold axis. (E) A 90° rotated view of D. The arrow highlights the dramatic movement of C-terminal helix of IX in HAdV5 (blue; now obsolete) and BAdV3 (red) structures.
Even though Philipson and colleagues observed cross-linking between cement proteins IIIa and VII, these authors favored an external location for IIIa (3). The more recent such studies by Flint and colleagues (4) did not observe any such cross-links between IIIa and VII. We suggest that IIIa is an external protein and can be released in some cases (e.g., ΔIX HAdV) without disrupting the rest of the capsid (5). Taken together with our structural observations, the residual cross-linking observed by Philipson and colleagues (3) between IIIa and VII is perhaps an artifact.
We don’t believe that the reason for the different oligomers of C-terminal coiled-coils (CCs)—trimeric in animal adenoviruses (AAdVs) vs. tetrameric in HAdVs—is because of different size linkers connecting the CCs with the (N-terminal) triskelions. We have shown that the 4-helix bundle (4-HLXB) cannot be comprised of the C termini of IX because of the connections between helices (2). Hence, the chain direction implied by the cryo-EM models of HAdV5 (6) would neither accommodate the remaining ∼75 residues nor allow the surface exposure of C termini. Second, we now know the distinct location of the CCs in AAdVs (7). The oligomeric state of trimeric CCs in AAdVs is consistent with the trimeric triskelions. In our model, the same conserved N-terminal region is involved in forming the triskelions, except that the chain direction is reversed, consistent with the experimental electron density and allowing the surface exposure of the C termini. This chain reversal of IX is also consistent with the formation of both triskelions and trimeric CCs in the bovine and canine adenoviruses. We surmise that the CCs in HAdVs are disordered because of the longer/flexible linkers connecting them to the triskelions.
We also don’t believe that IIIa in AAdVs is at a different place. The density for 4-HLXB (IIIa) may be absent because of its release during virus/sample preparation. Similar loss of IIIa densities were observed in the cryo-EM reconstructions of IX-deleted (thermo-labile) HAdV, even though IIIa can be detected by SDS/PAGE and immunoblotting (5). In this regard, the shorter hypervariable loops (HVRs 2 and 4) in the hexons of AAdVs might facilitate the release of IIIa. This finding is in contrast to wild-type HAdVs, where the longer hypervariable loops closely interact with IIIa and restrict its release.
Even though the Ala-Ser linker of IX mediates interactions between the hexons, their deletion might not totally abrogate hexon–IX associations, because other regions of IX (amino acids 27–53) also closely contact hexon trimers. Taken together, the revised models for cement proteins and the locations that we propose (2) represent a consensus model that is in agreement with majority of the biochemical data. Future structural studies may improve the current X-ray model by revealing the details of missing or poorly resolved regions of the cement proteins.
Footnotes
The authors declare no conflict of interest.
References
- 1.Campos SK. New structural model of adenoviral cement proteins is not yet concrete. Proc Natl Acad Sci USA. 2014;111:E4542–E4543. doi: 10.1073/pnas.1415364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy VS, Nemerow GR. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci USA. 2014;111(32):11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Everitt E, Lutter L, Philipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology. 1975;67(1):197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee PK, Vayda ME, Flint SJ. Interactions among the three adenovirus core proteins. J Virol. 1985;55(2):379–386. doi: 10.1128/jvi.55.2.379-386.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabry CM, et al. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24(9):1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329(5995):1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng L, et al. Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology. 2014;450-451:174–181. doi: 10.1016/j.virol.2013.12.012. [DOI] [PubMed] [Google Scholar]