Abstract
Aims
Cardiac histo-anatomical organization is a major determinant of function. Changes in tissue structure are a relevant factor in normal and disease development, and form targets of therapeutic interventions. The purpose of this study was to test tools aimed to allow quantitative assessment of cell-type distribution from large histology and magnetic resonance imaging- (MRI) based datasets.
Methods and results
Rabbit heart fixation during cardioplegic arrest and MRI were followed by serial sectioning of the whole heart and light-microscopic imaging of trichrome-stained tissue. Segmentation techniques developed specifically for this project were applied to segment myocardial tissue in the MRI and histology datasets. In addition, histology slices were segmented into myocytes, connective tissue, and undefined. A bounding surface, containing the whole heart, was established for both MRI and histology. Volumes contained in the bounding surface (called ‘anatomical volume’), as well as that identified as containing any of the above tissue categories (called ‘morphological volume’), were calculated. The anatomical volume was 7.8 cm3 in MRI, and this reduced to 4.9 cm3 after histological processing, representing an ‘anatomical’ shrinkage by 37.2%. The morphological volume decreased by 48% between MRI and histology, highlighting the presence of additional tissue-level shrinkage (e.g. an increase in interstitial cleft space). The ratio of pixels classified as containing myocytes to pixels identified as non-myocytes was roughly 6:1 (61.6 vs. 9.8%; the remaining fraction of 28.6% was ‘undefined’).
Conclusion
Qualitative and quantitative differentiation between myocytes and connective tissue, using state-of-the-art high-resolution serial histology techniques, allows identification of cell-type distribution in whole-heart datasets. Comparison with MRI illustrates a pronounced reduction in anatomical and morphological volumes during histology processing.
Keywords: Cardiac MRI, Serial histology, Myocytes, Connective tissue, Computational models
What's new?
We report quantitative estimation of cell-type distributions in rabbit heart, based on data integration from magnetic resonance imaging (MRI) and serial histology of the whole heart.
We quantify gross anatomical differences and changes in tissue volume that occur between MRI and histology, due to effects of histological processing.
Introduction
The heart has a complex three-dimensional (3D) structure. Cardiac function at the whole-organ level arises from, and requires, the concerted interplay of intrinsic structure–function gradients at all levels from sub-cellular to whole organ.1 Any change in this well-orchestrated heterogeneity—whether an increase or, indeed, a reduction—may result in detrimental effects on cardiac function.2
Cardiac muscle is composed of multiple cell types, dominated in volume by cardiomyocytes, but in numbers, even in the healthy heart, by fibroblasts.3,4 While much is known about the cellular basis of cardiac electrical activity and the mechanics of cardiac muscle contraction, relating these properties to the pattern of electrical activation and regional mechanics in the whole heart is far from straightforward in view of the heart's complex anatomy and histo-architecture. This task can be aided by computational models, which allow the integration of complex information on organ structure and function.5–10
Early attempts at computational modelling of excitable tissues date back to the 1940s, targeting neural networks,11 which represents work that subsequently gave rise to both biological and artificial intelligence/machine learning tools. Biophysically detailed modelling was introduced in the 1950s, initially to advance the understanding of bioelectric phenomena in neurons.12 In the 1960s, the first biophysically based models of cardiac cell activity were proposed by Noble.13 This was soon followed by computational models for studying bioelectrical activity at the tissue level.14–16 State-of-the-art electrophysiology models now form a well-established area of research and development,17,18 and coupled electromechanical models are also progressing quickly, even though they may not yet have reached quite the same relative degree of maturity and/or accessibility as those focused on electrophysiology alone.19–22 For a recent review of computational modelling of the heart, see Trayanova.23
Continuous improvements of experimental techniques provide researchers with increasingly detailed insight into excitable cell and tissue activity. Similarly, with the rapid development of computer technologies, implementation of 3D computational models of the heart has become more straightforward, incorporating information that allows elucidation of fundamental, and increasingly causal, relationships of biological parameters at various spatial and temporal scales. Computational models allow for the integration of data through which observed phenomena may be assessed, interpreted, and put into context, and to generate hypotheses that can be tested by targeted experimental validation. Ideally, therefore, computational models and experimental research are conducted in close iteration.24,25
Geometrically simple tissue models continue to play important roles in the toolbox of biocomputational analysis.26 Such models may make it easier to identify mechanisms underlying complex responses that would be difficult to spot in high-dimensional simulations. For example, the mechanisms presumed to underlie mechanical induction of reentrant excitation in the heart can be seen plainly in 2D simulations,27 while their confirmation in anatomically structured 3D simulations allows one to assess their plausibility in the presence of important histo-anatomical features of the heart,28 which can guide subsequent experimental designs.29 In terms of clinical utility, patient-specific structure–function models pose an additional challenge. Heterogeneities in structure and function of individual patients contribute to cardiac behaviour in ways that cannot be predicted easily with generic models, and it is often difficult to obtain non-invasively the relevant structural and functional information needed to tailor models.30,31 In spite of these difficulties, patient-specific modelling has made significant strides in the very recent past,10,32 and it is expected to guide clinical decision making in areas such as planning of ablation sites, or resynchronization therapy electrode placement, in the immediate future.
Current state-of-the-art models of the heart largely utilize stylized representations of cardiac anatomy, where finer structural details, such as cell orientation, cell-type distribution, and sheet information, are partially ignored. Recent modelling studies have shown that small structures such as the coronary vasculature, papillary muscles, trabeculations, valves, and extracellular clefts can affect activation and repolarization sequences, and conduction behaviour.33 For instance, fine-scale structures, such as blood vessels, may act as obstacles that could contribute to fractionation of the electrical activation wavefront as it splits into paths of different lengths.34 In addition, trabeculae may provide conduction shortcuts that accelerate propagation, compared with simplified models, and together with papillary muscles they can form virtual electrodes that protect the endocardium from becoming strongly polarized after external electrical shock application.35 Other virtual electrodes are formed at vessels and interstitial cleft spaces, increasing intra-myocardial heterogeneity of voltage distribution.35,36 Thus, fine-scale structural heterogeneities may have significant impact upon basic cardiac function and arrhythmia mechanisms, even though these effects may not always be overtly visible in whole tissue,33 and they are one of the target areas for development and application of the next generation of cardiac models. Histo-anatomical modelling relies heavily on information gleaned from advanced 3D imaging techniques.
Such 3D imaging modalities include magnetic resonance imaging (MRI) and computed tomography (CT), which can produce detailed whole-organ anatomical representations, although usually without cell-type discrimination. In the experimental setting, MRI and CT resolution have been pushed into the 10−5 m domain,37,38 but even that is not enough to provide cellular identification in the mammalian heart. Histological techniques, in contrast, combine high resolution (≤10−6 m in-plane, usually 10−5 m inter-slice) with positive cell-type discrimination, but they are innately based on a tissue-destructive approach. Histological 3D datasets are formed by collection of individual 2D slice images, each of which contains non-rigid deformations (compared both to the pre-cutting state, and to its nearest neighbouring slices), which makes organ reconstruction challenging. The combination of MRI microscopy with histological follow-up of partial organ volumes has been used to develop structure–function models of the heart for several species including mice,39,40 rabbit,41 pig,42,43 and human.44,45
In this study, we present a comparative analysis of 3D tissue data from ex vivo MRI and whole-heart histological imaging, including quantification of tissue alterations that are introduced by histological processing, as well as of tissue composition.
Methods
Wet-lab techniques
All investigations reported in this manuscript conformed to the UK Home Office guidance on the Operation of Animals (Scientific Procedures) Act of 1986; the sample presented formed part of a larger study.46,47 For MRI and histological imaging, hearts were isolated from female New Zealand White rabbits (∼1.2 kg) after induction of terminal anaesthesia (100 mg/kg sodium pentobarbitone; Rhône Mérieux), and swiftly connected via the aorta to a constant flow (12–15 mL/min) Langendorff perfusion system. An incision in the pulmonary artery was made to avoid fluid build-up in the right ventricle (RV). Initial perfusion (2 min) with normal Tyrode-style solution (NT) containing heparin (5 IU/mL) was conducted to wash the tissue before exposure to any intervention (for details see Plank et al.46). Hearts were fixed with minimal delay during cardioplegic arrest (induced using high potassium Tyrode solution). Tissue fixation was by coronary perfusion with 50 mL of the fast-acting Karnovsky's fixative (2% formaldehyde, 2.5% glutaraldehyde mix)48 containing 2 mM gadodiamide contrast agent (Omniscan 0.5 mmol/mL; Nycomed Imaging AS). Hearts were stored overnight at 4°C in the fixative containing gadodiamide. After 24 h, hearts were rinsed in cacodylate buffer and embedded bubble-free in 1% low-melting agarose (Cambrex, Nusieve GTG agarose), containing 2 mM gadodiamide. Subsequent to MRI scans, histological follow-up including dehydration in alcohol, xylene treatment, and wax infiltration, was performed (for details on histological methods, see Burton et al.47).
Magnetic resonance imaging
High-resolution MRI data were obtained on an 11.7 T MRI scanner, using the 3D gradient echo imaging technique described by Schneider et al.37 Image reconstruction included zero-filling in the k-space to reduce the voxel size. The ex vivo rabbit dataset voxel size was 26.4 µm × 26.4 µm in-plane, and 24.4 µm inter-plane, the number of voxels was 1024 × 1024 × 2048 (in the X-, Y-, and Z-axes, respectively), resulting in a data file size of 4 GB. The image was cropped to a region of interest of 925 × 848 × 1359 voxels, containing the whole heart, and down-sampled (using a Gaussian low-pass filter and cubic B-spline interpolation) to produce a 231 × 212 × 340 voxel image (voxel size 105.6 µm × 105.6 µm × 97.5 µm; voxel volume of 1.1 × 106 µm3. The down-sampled image was used as it is much faster to process, and was considered by visual inspection to preserve sufficient anatomical details for volume assessments. A visualization of three orthogonal planes of the down-sampled MRI is shown in Figure 1A–C.
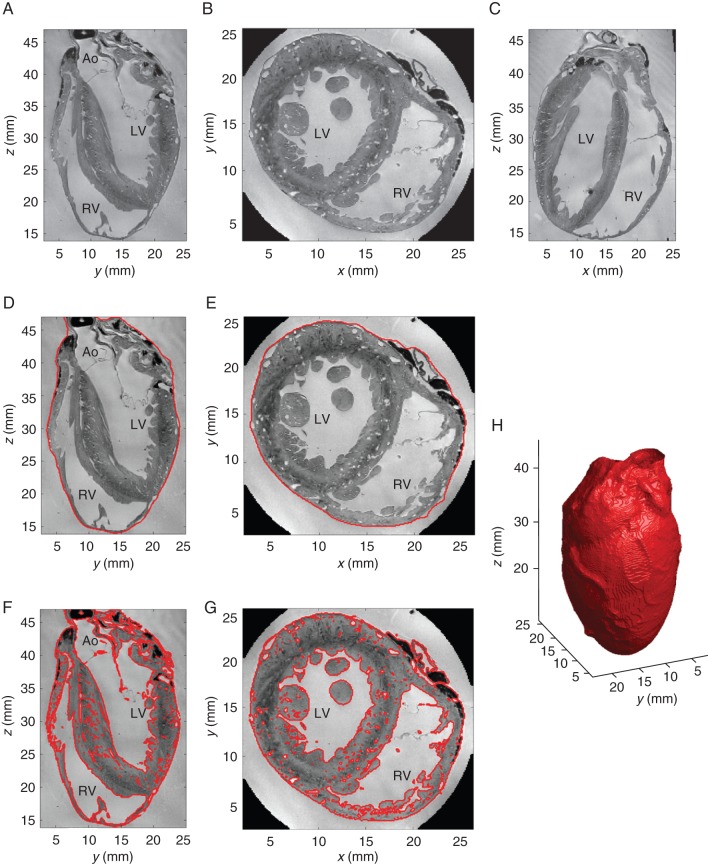
Figure 1.
MRI-based tissue segmentation. (A–C) Down-sampled MRI image in three orthogonal planes. (D, E) Corresponding boundary surface segmentations for anatomical volume determination. (F, G) Detailed tissue segmentations to separate tissue from non-tissue (including clefts, interstitial spaces, blood vessels) for morphological volume calculation. (H) 3D iso-surface visualization of bounding surface segmentation obtained from MRI. LV, left ventricle; RV, right ventricle; LA, left atrium; RA, right atrium; Ao, aorta.
Serial sectioning
Whole hearts were serially sectioned (at 10 µm thickness) using a Leica SM2400 heavy duty sledge-type microtome. The tissue was allowed to relax in a water bath (Leica Microsystems, HI 1210) at 39°C for 2–15 min, depending on the size of the tissue section, and then carefully (aiming for minimal distortion and avoidance of tissue folds) mounted to positively charged slides (SuperFrost, VWR). Slides were air-dried in a laminar airflow hood (overnight), followed by de-waxing and trichrome staining using an automated stainer (Leica AutoStainer XL, ST50-10). Trichrome stain allows one to identify collagen (bluish green), myocytes (pink), cytoplasm (orange, highlighting non-myocytes), and nuclei (blue-black). The stained and mounted sections were imaged at high resolution with Leica Application Suite, operating the LAS Power Mosaic Module, integrated with an automated Leica DM4000B light microscope and a Märzhäuser inverted microscope scanning stage (see methods in Plank et al.46). Sample images are shown in Figure 2A–C.
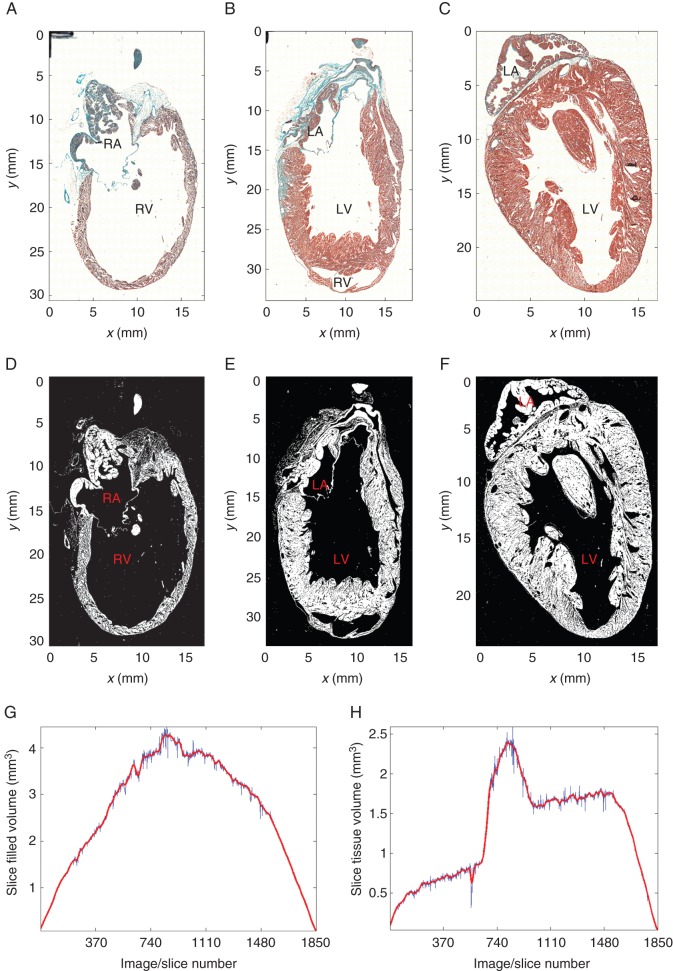
Figure 2.
Histology-based tissue segmentation. (A–C) Histology slices #460 containing the RV cavity), #925 containing LV cavity and part of the septum, and #1387 through the LV cavity (out of 1850 sections in the whole-heart histology stack). (D, F) Corresponding tissue segmentation. Note that the black marker lines at the top left corner in (A–C) have been masked out in the segmentation. (G, H) Volumes measured in slices with valid segmentations (blue curve), and after interpolation to correct for missing sections (red curve). (G) Bounding contour segmentation yielding anatomical volume. (H) Tissue content segmentation yielding morphological volume. LV, left ventricle; RV, right ventricle; LA, left atrium; RA, right atrium.
Each pixel represented an area of 1.1 µm × 1.1 µm. Taking into account the section thickness (10 µm), each pixel in the histology dataset represented a volume of 12.1 µm3. The size of the histology dataset (1853 slices; up to 38 000 × 17 500 pixels per slice; maximum file size per single slice ∼2 GB; 1.6 TB of data for the whole heart) created a computational challenge. As histology images have a higher resolution than necessary for tissue volume quantification, each slice was down-sampled by a factor of 8 in the X- and Y-directions, which facilitated further image processing. The resulting slices have a pixel size of 8.8 µm × 8.8 µm × 10 µm, representing a voxel volume of 774.4 µm3.
Processing of magnetic resonance imaging scans
The bounding surface segmentation of the heart was obtained by computing a local convex hull of the tissue segmentation using an alpha shape49 with radius of 969 µm (∼10 voxels). Each voxel was then labelled as internal or external to the local convex hull; the result is illustrated in Figure 1D and E. The ‘anatomical tissue volume’ was calculated as the content of this bounding surface.
Myocardial tissue in the MRI volume was segmented using an adaptive method to account for spatial intensity inhomogeneities, with the bias field being the foremost contributor. First, the image was split into 10 × 12 × 17 voxel blocks extended by 30 × 32 × 22 voxels on each side to overlap with neighbouring blocks. This large overlap ensured a smooth transition in the estimated typical background/foreground intensities between adjacent blocks, and the extended block size was large enough to contain a significant number of background voxels, whether tissue voxels were also present or not. Intensity histograms were estimated, and iteratively smoothed with an approximating cubic spline until the histogram presented only one or two peaks. The two cases correspond to a block with only background voxels (one histogram peak), or background and tissue voxels (two histogram peaks). The typical intensity value for tissue (foreground, darker) and background voxels was computed as the mode of the peaks. The typical background and foreground intensity values of each block were then fed as input parameters to a classification algorithm using Markov Random Fields [as implemented in the ITK library (Insight Toolkit itk::MRFImageFilter: http://www.itk.org/Doxygen/html/classitk_1_1MRFImageFilter.html)], which classified each voxel as either background or tissue (see Figure 1F and G). The ‘morphological tissue volume’ was computed by multiplying the number of tissue-containing voxels by voxel volume.
Processing of histology slice stacks
To compare to the MRI-based anatomical volume, bounding contour segmentations for histology were computed with morphological operators in the following way. Tissue segmentations were dilated with a disc element with a radius of 10 pixels, holes in the segmentation were filled with a flood-fill operation, and the result eroded with the same disc element. Although this approach fails in part of the cases (9.6% of slices) to fill-up the ventricular or atrial cavities (e.g. if the myocardium has tears, particularly large cleavage planes, and/or is very thin as in parts of the RV), defective slice segmentations can be visually identified with ease and removed.
Tissue segmentation was performed on the full-resolution version of each slice by converting the red-green-blue (RGB) image to greyscale, followed by thresholding. Pixels with intensity <0.9 × Imax, where Imax is the maximum intensity in the slice, were labelled as tissue. The results were revised and easily correctable segmentation artefacts (such as smudges or black marker lines to identify the top left corner of each slide) were manually removed. 12.3% of the slides had complicating artefacts (such as bubbles, tears, folds, or missing tissue fragments) that required removal from the analysed stack. The results are illustrated in Figure 2D–F.
Morphological tissue volume was computed in each slide by multiplying the number of tissue-labelled voxels by the voxel volume. For this, we assumed that (i) in-plane non-rigid tissue deformation during slice sectioning affects tissue shape locally, but not its overall area on the slide; and (ii) that tissue shrinkage in the z-axis occurred prior to sectioning, so that tissue thickness of stained sections is equal to the microtome slicing thickness.
Missing volume values for tissue sections that were removed due to invalid segmentations, as well as for those lost in histological processing (total number of slices analysed: 1653; total number of slices with missing information: 197), were interpolated by averaging the values of the nearest available neighbouring slices. Volume values, interpolated for all invalid slices, were then smoothed with an approximating cubic B-spline (smoothing constant 1.25 × 10−3), both for bounding contour (anatomical) and tissue (morphological) segmentations, as displayed in Figure 2G and H, respectively. Total volumes were computed as the sum of all interpolated and smoothed slice volumes.
Colour classification in the RGB space was used to generate segmentation masks corresponding to three tissue categories: myocytes (red), connective tissue (blue-green), and undefined. The amount of tissue in each of these classes was then quantified. The process is illustrated on a sample slice in Figure 3.
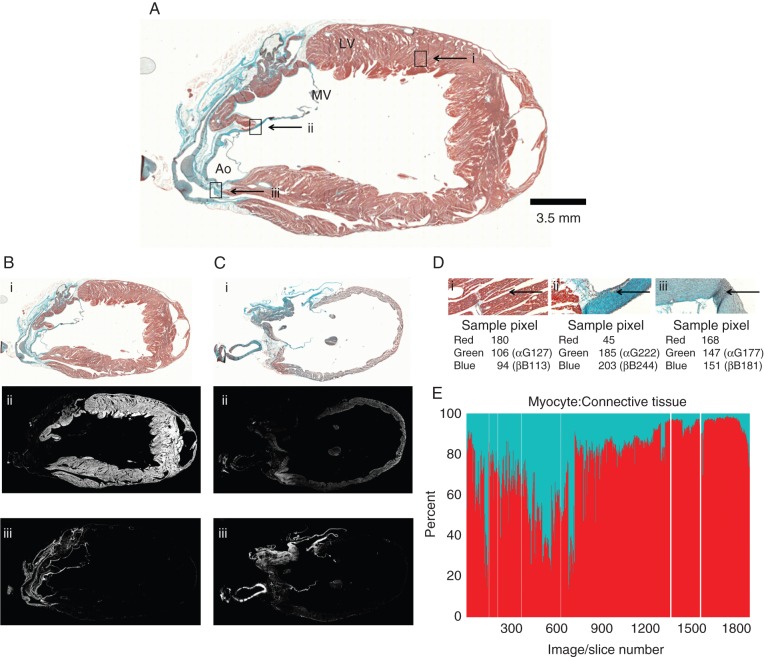
Figure 3.
Histology-based cell-type segmentation. (A) Trichrome-stained rabbit heart section #925, containing. Ao, aorta; LV, left ventricle; MV, mitral valve; scale bar 3.5 mm. Rectangles i, ii, and iii correspond to locations of tissue type segmentation in panel (D). (B, C) Two sample histology images that are shown as trichrome-stained sections (B-i and C-i); after myocyte segmentation (B-ii, C-ii), and after connective tissue segmentation (B-iii, C-iii). (D) Enlarged aspects of the trichrome-stained section in panel (A). Image area dominated by myocytes (D-i), connective tissue (D-ii), or cells that are ‘undefined’ by the algorithm (D-iii). Arrows point to location of sample pixels analysed. (E) Summary data showing the ratio of myocyte: connective tissue pixels in the entire stack of slices, ordered in the same sequence as in Figure 2G and H (red: myocyte pixels, blue: connective tissue pixels). White lines: missing sections in the histology stack.
Figure 3A shows a tissue slice, and Figure 3D illustrates examples of the different distributions of the various colour-coded cell types. Differences in colour are visually apparent and reflected in the numerical values shown for sample pixels.
For colour separation, the relative magnitudes of the colour channels R, G, and B were used. Pixels where the intensities of the red channel were clearly higher than those of blue and green were assigned to the myocyte class (Figure 3D-i). The following condition was used:
Values for α and β (α = β = 1.2) were determined empirically using validation by an expert histologist. In a similar fashion, pixels with blue and green intensities clearly larger than that of the red channel were assigned to the connective tissue group (refer to Figure 3D-ii), using the following formula:
with γ = δ = 1.2, also established empirically.
Pixels that did not meet any of the two conditions could not be unequivocally assigned by the automated procedure to one of the two tissue types, and were thus labelled as ‘undefined’ (Figure 3D-iii).
Results
Magnetic resonance imaging
The contrast between heart tissue and agar was sufficient for effective differentiation between tissue and non-tissue, using the process described in the Processing of magnetic resonance imaging scans section, as illustrated in Figure 1.
Anatomical volume (magnetic resonance imaging)
The anatomical volume, i.e. the region contained inside the bounding epicardial surface, was 7.8 cm3 for the MRI dataset. A 3D reconstruction of the surface is shown in Figure 1H.
Morphological volume (magnetic resonance imaging)
The volume of voxels classified as containing tissue in the MRI scan was 4.5 cm3. This represents a ‘fill factor’ of 57.7% of the anatomical volume inside the bounding surface, and most of this difference is attributable to the volumes contained in the cardiac cavities.
Histology
Figure 3B-i and C-i show two sample sections after trichrome staining. Figure 3B-ii and C-ii are the corresponding segmented monochrome representations for myocytes, and Figure 3B-iii and C-iii are the corresponding monochrome images for connective tissue, generated by our segmentation approach.
The results of the segmentation described in the Processing of histology slice stacks section, corrected for missing tissue slices, were used to calculate the total volume of the histologically processed myocardium, as well as sub-volumes occupied by myocytes or connective tissue.
The ratio of myocyte pixels (61.6% of total) to connective tissue pixels (9.8% of total) for each tissue slice, represented as a percentage, is shown in Figure 3E. Note that this gives a rough indication of relative cell-type volumes, but not cell numbers, as myocytes are significantly larger than non-muscle cells in the heart, and as section sizes and their tissue fill-factor differs (Figure 2G and H). The ratio of pixels classified as either myocytes or connective tissue (71.4%), compared with pixels with undefined tissue content (28.6%), was ∼2.5 : 1, suggesting that there is a significant margin for error in the processing approach we used for digitized images from histological sections.
Anatomical volume (Histology)
The volumes inside the bounding contours outlined on each of the histology slices were added up resulting in a total anatomical volume of 4.9 cm3. The volumes included in the bounding contours for each slice are represented in Figure 2G.
Morphological volume (Histology)
The tissue volume in the histology stack, calculated from voxels in all three groups (myocytes, non-myocytes, undefined; Figure 3D) was 2.3 cm3, filling 47% of the volume inside the bounding contour. Tissue volumes for each slice are represented in Figure 2H. As the heart was sectioned in a plane that was near-parallel to the septum, it is unsurprising that the fill factor (morphological volume of each slice divided by the volume inside its outer contour, i.e. ‘Figure 2H divided by Figure 2G’) was largest in the RV free-wall, septum, and left ventricle (LV) free-wall containing regions (roughly #1–#200, #700–#900, and #1500–#1850; respectively), while it plateaued at relatively low (RV) or medium (LV) levels in sections containing ventricular cavities (roughly #200–#700, #900–#1500; respectively).
Comparison of magnetic resonance imaging and histology data
The volumes, established by histology, were considerably smaller than those of the MRI data from the same heart. The anatomical (bounding surface) volumes were 7.8 and 4.9 cm3 for MRI and histology, respectively. This corresponds to gross organ shrinkage by 37.2% from fixed tissue studied in MRI, to dehydrated/wax embedded/cut tissue in the histology data. Removing intra-cavity volumes, and taking into account further morphological changes (such as enlargement of interstitial cleft spaces), the morphological volumes actually occupied by tissue were 4.5 and 2.3 cm3 for MRI and histology, respectively, representing shrinkage by 48.9%.
The whole-heart ‘fill factor’ of morphological tissue volume inside the epicardial surface (i.e. inside the anatomical volume), was smaller for histology slices (47.0%), compared with MRI data (57.7%), reconfirming the presence of intra-myocardial shrinkage effects.
Discussion
Previous studies have demonstrated that myocytes occupy the majority of cardiac tissue volume, while fibroblasts dominate in terms of cell numbers. Relative cell content, distribution, and inter-cellular mechanical and electrical coupling change during development, and in physiological or pathological remodelling of the myocardium.50–52 The roles of cardiac fibroblasts, in particular, have seen a tremendous increase in experimental, computational, and conceptual research.53–56 This makes the identification and quantification of individual cell types in the heart an important target for fundamental study and clinical application. Here, we describe our methodology, from sample preparation to image acquisition and image analysis, and present the results of our analysis on a sample rabbit heart. Our approach allows one to compare gross anatomical volumes of a heart (based on MRI and histology datasets; Appendix Figure 1), to morphologically characterize the organ fraction occupied by tissue, and to discriminate between cell types within the tissue.
Various factors affect the accuracy of histological data gathering, including the consistency of the staining protocol (here automated, with regular changes in stains, supporting relatively well-sustained quality), the thickness of the sections (here 10 µm), and the preservation, mounting, and analysis of consecutive sections (here including 89% of the entire organ). There is a possibility that tissue processing affects the relative size of various cell populations differentially, which is something we could not control for in the present study. We observe a ratio of 6 : 1 in terms of volume occupied by myocytes relative to non-myocytes. However, this is in the presence of a significant proportion of histology voxels that could not be attributed to either of the two cell populations, raising the level of uncertainty (the ratio of myocytes: non-specified: non-myocytes is 6 : 3 : 1).
Nonetheless, the above data provides an indication of relative volume fractions, as the trichrome method employed allows for distinction between ‘red’ myocytes and ‘green’ connective tissue areas. This is not indicative of cell numbers, though. The nuclei of the different cell types are also stained by the trichrome method, and could be taken into account, using more powerful analysis methods that remain to be developed, to refine and complete the measurements with a more accurate identification of cell types (i.e. nuclei inside collagen-stained areas), and corresponding cell-specific nuclei counts (a task that is not made easier by the presence of multi-nucleated myocytes).57 During paraffin embedding, the content of fat cells is depleted, leaving only their outer membranes intact. Structurally, they can often be identified by large void spaces surrounded by membrane with a thin nucleus near the membrane (Appendix Figure 2). Sub-epicardial adipose tissue in the heart may constitute up to 50% of cardiac weight in excessive adiposis.58 Although visual inspection did not reveal anywhere near these levels of fat cell presence in the young rabbit heart studied, an effect on histological tissue identification cannot be excluded. Our current protocol does not account for the volume occupied by fat cells in the whole-heart histology, as it treats voids as ‘no tissue’. This may have contributed to an overestimation of tissue shrinkage in the histology data, as well as to the fraction of the ‘unidentified’ tissue.
Among the problems in relating MRI and histology samples are tissue deformation and shrinkage. Following the MRI scan, the heart underwent a dehydration process to allow introduction of wax, then sectioning, relaxation on a water bath, mounting, de-waxing/rehydration (although this term is misleading, as re-exposure of tissue sections to H2O does not recover pre-processing cell dimensions), staining, and once again dehydration and cover slipping. As a result of this histological processing pipeline, multiple rigid and non-rigid deformations are introduced at various stages, and a ‘true’ match between MRI and histology data is therefore not possible.
We used two different volume indicators to compare MRI and histology datasets. The anatomical volume was defined as the region contained within the ‘epicardial’ boundary outline. This was 7.8 cm3 for MRI and 4.9 cm3 for histology. While dominated by cardiac tissue, this measure includes also the intra-cardiac chamber volumes, as well as the a-cellular space in the tissue, such as inside blood vessels. The actual volume occupied by tissue, called morphological volume, was identified by voxel-wise segmentation and measured 4.5 cm3 for MRI, and 2.3 cm3 for histology.
The comparatively larger reduction in morphological volumes in histology (compared with MRI) is presumably caused—in addition to the shrinkage mechanisms mentioned in the context of anatomical volume reduction—by an enlargement of interstitial spaces/inter-laminar clefts in the histology samples, chiefly as a result of dehydration. The enlargement of cleavage planes after histological processing is illustrated in Appendix Figure 1A/A1’.
An additional contributor to volume differences could be the inherently lower resolution of MRI source data (i.e. ∼25 µm isotropic voxel dimensions, compared with 1.1 µm × 1.1 µm × 10 µm in histology). Although the MRI segmentation algorithm treats interstitial spaces, clefts, and blood vessel lumens as non-tissue, the difference in resolution is likely to give rise to an under-detection of these areas: as interstitial clefts often are below MRI but within histology resolution (further helped, of course, by the fact that those clefts are widened by histological processing), they may fail to be detected in MRI and thus be counted as part of tissue in the MRI data. On the other hand, MRI might fail to detect small strands of tissue, which would then result in a reduction of detected volume, but this is likely to occur only in thinly trabeculated parts of the endocardium and, hence, will have less of an effect on overall morphological volume identification.
Study limitations
This study reconfirms the presence of tissue shrinkage, caused by histological processing. The observed 37.2% reduction in the anatomical volume inside the epicardial surface is in line with gross histological shrinkage reported in the literature (between 30 and 40% post-histological processing).59
Our comparison of tissue volumes is based on tissue that had been fixed prior to imaging (including MRI). Chemical fixation may have altered tissue volume, compared with the live organ, as the standard Karnovsky's fixative is very strongly hyperosmotic (103 mM). An isotonic glutaraldehyde/cacodylate formulation may be better at preserving overall tissue architecture, though in exchange for weaker fixation. Further work is required to assess shrinkage and distortion caused by different osmolality fixatives. In addition, MRI studies would ideally be performed first on live (i.e. Langendorff-perfused) hearts, before higher-resolution scans are repeated on the fixed sample.60
Morphological volume reduction, based on pixel-wise analysis, is even more pronounced. The observed 48.9% of shrinkage between MRI and histology exceeds expected cell reduction. This may, in addition to the above-mentioned causes, be a consequence of resolution differences, and of a failure to appropriately identify pixels representing the cytosol of fat cells.
The presence of undefined pixels may be attributed to partial volume effects: with a slice thickness of 10 µm, pixels may be occupied partially by overlapping, in the z-direction, heterotypic cells. The majority of heart mass is composed of cardiac myocytes, but significant numbers of other cell types are present, including endothelial cells, fibroblasts, and leucocytes. These non-myocytes are all smaller than cardiac muscle cells, which may bias them for an increased relative contribution to ‘unclassified’ pixels, even though the analysis was conducted on the full-resolution data with voxel volumes in the order of 101 µm3 (for comparison, a myocyte occupies ≥104 µm3, while fibroblasts—even though they form thin sheets that extend over many pixels—have a volume in a pixel-like ball-park). Future studies, using stains specific for some of these other cell types, may help to better understand the presently ‘undefined tissue’ volume.
Finally, staining inconsistencies resulting in colour variations (such as apparent in the images selected for Appendix Figure 1), may also contribute to undefined pixels. The values of α, β, γ, and δ in the segmentation equations were chosen conservatively, aiming at minimizing the number of misclassified pixels, rather than unclassified ones. Choosing smaller values would reduce the number of undefined pixels, but carry an elevated risk of inaccurate cell-type attribution (i.e. false-positives).
Conclusions
The methods and tools used for quantitative analysis reconfirm that cell-type information can be successfully extracted from high-resolution, 3D histology datasets, up to reconstruction of an entire organ such as the rabbit heart, with voxel sizes well below the volume of individual muscle cells. We show that the assessment of tissue volumes using histology techniques is not unproblematic either. In combination with MRI, it may be possible to develop a suitable approach to extracting and correcting volume information, but currently this is limited by restrictions in resolution (hindering segmentation of clefts, interstitial spaces, and small blood vessels) and cell-type identification (relevant for tissue remodelling, in particular if diffuse rather than focal) in MRI data. Combining MRI technology, which preserves histo-anatomical features but provides less microanatomical details, with whole-heart histology allows one to achieve organ representations that have high fidelity, including cell-type identification. The methods presented here can be used for a range of investigations focusing on estimation of cell-types, and their distribution in control conditions (e.g. atria vs. ventricles, left vs. right, base vs. apex) and disease (e.g. diffuse fibrosis, post-ablation or post-infarction scars, surgical lesions). Our approach will be improved by aligning MRI and 3D histological datasets to obtain a detailed geometrically correct anatomical description of the heart,61 using block-face imaging as an intermediate 3D reference volume during the process of histological slice preparation.62 We observed a number of previously underestimated problems with extraction of histology-based information, in particular, related to the large extent of microscopically apparent tissue shrinkage, which can be countered by addition of live-tissue MRI prior to the implementation of this pipeline.
Funding
This research was supported by the BBSRC and the ERC. R.A.B.B. is an EP Abraham Cephalosporin JRF at Linacre College, Oxford. J.E.S. and P.K. are BHF Senior Basic Science Research Fellows (FS/11/50/29038; FS/12/17/29532). R.C., V.C., J.E.S., P.K., and V.G. are supported by the BBSRC BB/I012117/1 and the British Heart Foundation New Horizon Grant NH/13/30238. P.K. holds the ERC Advanced Grant CardioNect. Funding to pay the Open Access publication charges for this article was provided by…
Acknowledgements
We would like to thank Judith Sheldon and Padmini Sarathchandra for technical assistance and advice.
References
- 1.Katz AM, Katz PB. Homogeneity out of heterogeneity. Circulation. 1989;79:712–7. doi: 10.1161/01.cir.79.3.712. [DOI] [PubMed] [Google Scholar]
- 2.Markhasin VS, Solovyova O, Katsnelson LB, Protsenko Y, Kohl P, Noble D. Mechano-electric interactions in heterogeneous myocardium: development of fundamental experimental and theoretical models. Prog Biophys Mol Biol. 2003;82:207–20. doi: 10.1016/s0079-6107(03)00017-8. [DOI] [PubMed] [Google Scholar]
- 3.Adler CP, Ringlage WP, Bohm N. [DNA content and cell number in heart and liver of children. Comparable biochemical, cytophotometric and histological investigations (author's transl)] Pathol Res Pract. 1981;172:25–41. [PubMed] [Google Scholar]
- 4.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Vetter FJ, McCulloch AD. Three-dimensional analysis of regional cardiac function: a model of rabbit ventricular anatomy. Prog Biophys Mol Biol. 1998;69:157–83. doi: 10.1016/s0079-6107(98)00006-6. [DOI] [PubMed] [Google Scholar]
- 6.Peters NS, Wit AL. Myocardial architecture and ventricular arrhythmogenesis. Circulation. 1998;97:1746–54. doi: 10.1161/01.cir.97.17.1746. [DOI] [PubMed] [Google Scholar]
- 7.Hunter PJ, Kohl P, Noble D. Integrative models of the heart: achievements and limitations. Philos Trans R Soc A. 2001;359:1049–54. [Google Scholar]
- 8.Crampin EJ, Halstead M, Hunter P, Nielsen P, Noble D, Smith N, et al. Computational physiology and the Physiome Project. Exp Physiol. 2004;89:1–26. doi: 10.1113/expphysiol.2003.026740. [DOI] [PubMed] [Google Scholar]
- 9.Plank G, Zhou L, Greenstein JL, Cortassa S, Winslow RL, O'Rourke B, et al. From mitochondrial ion channels to arrhythmias in the heart: computational techniques to bridge the spatio-temporal scales. Philos Trans R Soc A. 2008;366:3381–409. doi: 10.1098/rsta.2008.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurthy A, Villongco CT, Chuang J, Frank LR, Nigam V, Belezzuoli E, et al. Patient-specific models of cardiac biomechanics. J Comput Phys. 2013;244:4–21. doi: 10.1016/j.jcp.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCulloch W, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5:115–33. [PubMed] [Google Scholar]
- 12.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble D. Cardiac action and pacemaker potentials based on the Hodgkin-Huxley equations. Nature. 1960;188:495–7. doi: 10.1038/188495b0. [DOI] [PubMed] [Google Scholar]
- 14.Barr L, Dewey MM, Berger W. Propagation of action potentials and the structure of the nexus in cardiac muscle. J Gen Physiol. 1965;48:797–823. doi: 10.1085/jgp.48.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spach MS, Dolber PC, Anderson PA. Multiple regional differences in cellular properties that regulate repolarization and contraction in the right atrium of adult and newborn dogs. Circ Res. 1989;65:1594–611. doi: 10.1161/01.res.65.6.1594. [DOI] [PubMed] [Google Scholar]
- 16.Clayton RH, Bernus O, Cherry EM, Dierckx H, Fenton FH, Mirabella L, et al. Models of cardiac tissue electrophysiology: progress, challenges and open questions. Prog Biophys Mol Biol. 2011;104:22–48. doi: 10.1016/j.pbiomolbio.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Rudy Y. From genome to physiome: integrative models of cardiac excitation. Ann Biomed Eng. 2000;28:945–50. doi: 10.1114/1.1308484. [DOI] [PubMed] [Google Scholar]
- 18.Niederer SA, Kerfoot E, Benson AP, Bernabeu MO, Bernus O, Bradley C, et al. Verification of cardiac tissue electrophysiology simulators using an N-version benchmark. Philos Trans R Soc A. 2011;369:4331–51. doi: 10.1098/rsta.2011.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathmanathan P, Whiteley JP. A numerical method for cardiac mechanoelectric simulations. Ann Biomed Eng. 2009;37:860–73. doi: 10.1007/s10439-009-9663-8. [DOI] [PubMed] [Google Scholar]
- 20.Aguado-Sierra J, Krishnamurthy A, Villongco C, Chuang J, Howard E, Gonzales MJ, et al. Patient-specific modeling of dyssynchronous heart failure: a case study. Prog Biophys Mol Biol. 2011;107:147–55. doi: 10.1016/j.pbiomolbio.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurev V, Lee T, Constantino J, Arevalo H, Trayanova NA. Models of cardiac electromechanics based on individual hearts imaging data: image-based electromechanical models of the heart. Biomech Model Mechanobiol. 2011;10:295–306. doi: 10.1007/s10237-010-0235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang VY, Hoogendoorn C, Engelbrecht G, Frangi AF, Young AA, Hunter PJ, et al. Unsupervised segmentation and personalised FE modelling of in vivo human myocardial mechanics based on an MRI atlas. 9th IEEE International Symposium on Biomedical Imaging (ISBI); 2012. pp. 1360–3. [Google Scholar]
- 23.Trayanova NA. Computational cardiology: the heart of the matter. ISRN Cardiol. 2012;2012:1–15. doi: 10.5402/2012/269680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carusi A, Burrage K, Rodriguez B. Bridging experiments, models and simulations: an integrative approach to validation in computational cardiac electrophysiology. Am J Physiol Heart Circ Physiol. 2012;303:11. doi: 10.1152/ajpheart.01151.2011. [DOI] [PubMed] [Google Scholar]
- 25.Quinn TA, Kohl P. Combining wet and dry research: experience with model development for cardiac mechano-electric structure-function studies. Cardiovasc Res. 2013;97:601–11. doi: 10.1093/cvr/cvt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garny A, Noble D, Kohl P. Dimensionality in cardiac modelling. Prog Biophys Mol Biol. 2005;87:47–66. doi: 10.1016/j.pbiomolbio.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Garny A, Kohl P. Mechanical induction of arrhythmias during ventricular repolarization: modeling cellular mechanisms and their interaction in two dimensions. Ann N Y Acad Sci. 2004;0:133–43. doi: 10.1196/annals.1302.011. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Kohl P, Trayanova N. Induction of ventricular arrhythmias following mechanical impact: a simulation study in 3D. J Mol Histol. 2004;35:679–86. doi: 10.1007/s10735-004-2666-8. [DOI] [PubMed] [Google Scholar]
- 29.Quinn T, Kohl P. Mechanical triggers and facilitators of ventricular tachy-arrhythmias. In: Kohl P, Sachs F, Franz M, editors. Cardiac Mechano-Electric Coupling and Arrhythmias. Oxford: Oxford University Press; 2011. pp. 160–7. [Google Scholar]
- 30.Jacquemet V, Virag N, Kappenberger L. Wavelength and vulnerability to atrial fibrillation: insights from a computer model of human atria. Europace. 2005;2:83–92. doi: 10.1016/j.eupc.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Nickerson D, Smith N, Hunter P. New developments in a strongly coupled cardiac electromechanical model. Europace. 2005;2:118–27. doi: 10.1016/j.eupc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 32.McDowell KS, Vadakkumpadan F, Blake R, Blauer J, Plank G, MacLeod RS, et al. Methodology for patient-specific modeling of atrial fibrosis as a substrate for atrial fibrillation. J Electrocardiol. 2012;45:640–5. doi: 10.1016/j.jelectrocard.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bishop MJ, Plank G. The role of fine-scale anatomical structure in the dynamics of reentry in computational models of the rabbit ventricles. J Physiol. 2012;590:4515–35. doi: 10.1113/jphysiol.2012.229062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibb M, Bishop M, Burton RA, Kohl P, Grau V, Plank G, et al. The role of blood vessels in rabbit propagation dynamics and cardiac arrhythmias. Lect Notes Comput Sci. 2009;5528:268–76. [Google Scholar]
- 35.Bishop MJ, Plank G, Burton RA, Schneider JE, Gavaghan DJ, Grau V, et al. Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function. Am J Physiol Heart Circ Physiol. 2010;298:20. doi: 10.1152/ajpheart.00606.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plank G, Prassl A, Hofer E, Trayanova NA. Evaluating intramural virtual electrodes in the myocardial wedge preparation: simulations of experimental conditions. Biophys J. 2008;94:1904–15. doi: 10.1529/biophysj.107.121343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider J, Böse J, Bamforth S, Gruber A, Broadbent C, Clarke K, et al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol C7–16. 2004;4:1–12. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aslanidi OV, Nikolaidou T, Zhao J, Smaill BH, Gilbert SH, Holden AV, et al. Application of micro-computed tomography with iodine staining to cardiac imaging, segmentation, and computational model development. IEEE Trans Med Imaging. 2013;32:8–17. doi: 10.1109/TMI.2012.2209183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ojha N, Roy S, Radtke J, Simonetti O, Gnyawali S, Zweier JL, et al. Characterization of the structural and functional changes in the myocardium following focal ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2008;294:28. doi: 10.1152/ajpheart.01190.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Linden J, Berr SS, Kron IL, Beller GA, French BA. Timing of adenosine 2A receptor stimulation relative to reperfusion has differential effects on infarct size and cardiac function as assessed in mice by MRI. Am J Physiol Heart Circ Physiol. 2008;295:10. doi: 10.1152/ajpheart.00091.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scollan DF, Holmes A, Winslow R, Forder J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. Am J Physiol. 1998;275:H2308–18. doi: 10.1152/ajpheart.1998.275.6.H2308. [DOI] [PubMed] [Google Scholar]
- 42.Saeed M, Martin A, Ursell P, Do L, Bucknor M, Higgins CB, et al. MR assessment of myocardial perfusion, viability, and function after intramyocardial transfer of VM202, a new plasmid human hepatocyte growth factor in ischemic swine myocardium. Radiology. 2008;249:107–18. doi: 10.1148/radiol.2483071579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malliaras K, Smith RR, Kanazawa H, Yee K, Seinfeld J, Tseliou E, et al. Validation of contrast-enhanced magnetic resonance imaging to monitor regenerative efficacy after cell therapy in a porcine model of convalescent myocardial infarction. Circulation. 2013;128:2764–75. doi: 10.1161/CIRCULATIONAHA.113.002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackowski C, Christe A, Sonnenschein M, Aghayev E, Thali MJ. Postmortem unenhanced magnetic resonance imaging of myocardial infarction in correlation to histological infarction age characterization. Eur Heart J. 2006;27:2459–67. doi: 10.1093/eurheartj/ehl255. [DOI] [PubMed] [Google Scholar]
- 45.Azevedo CF, Nigri M, Higuchi ML, Pomerantzeff PM, Spina GS, Sampaio RO, et al. Prognostic significance of myocardial fibrosis quantification by histopathology and magnetic resonance imaging in patients with severe aortic valve disease. J Am Coll Cardiol. 2010;56:278–87. doi: 10.1016/j.jacc.2009.12.074. [DOI] [PubMed] [Google Scholar]
- 46.Plank G, Burton RA, Hales P, Bishop M, Mansoori T, Bernabeu MO, et al. Generation of histo-anatomically representative models of the individual heart: tools and application. Philos Trans R Soc A. 2009;367:2257–92. doi: 10.1098/rsta.2009.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burton RA, Plank G, Schneider JE, Grau V, Ahammer H, Keeling SL, et al. Three-dimensional models of individual cardiac histoanatomy: tools and challenges. Ann N Y Acad Sci. 2006;1080:301–19. doi: 10.1196/annals.1380.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karnovsky MJ. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J Cell Biol. 1965;27:137A–8A. [Google Scholar]
- 49.Edelsbrunner H, Kirkpatrick D, Seidel R. On the shape of a set of points in the plane. IEEE Trans Inf Theory. 1983;29:551–9. [Google Scholar]
- 50.Lajiness JD, Conway SJ. Origin, development, and differentiation of cardiac fibroblasts. J Mol Cell Cardiol. 2014;70:2–8. doi: 10.1016/j.yjmcc.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shinde AV, Frangogiannis NG. Fibroblasts in myocardial infarction: a role in inflammation and repair. J Mol Cell Cardiol. 2014;70:74–82. doi: 10.1016/j.yjmcc.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohl P, Gourdie RG. Fibroblast-myocyte electrotonic coupling: does it occur in native cardiac tissue? J Mol Cell Cardiol. 2014;70:37–46. doi: 10.1016/j.yjmcc.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohl P, Kamkin AG, Kiseleva IS, Streubel T. Mechanosensitive cells in the atrium of frog heart. Exp Physiol. 1992;77:213–6. doi: 10.1113/expphysiol.1992.sp003576. [DOI] [PubMed] [Google Scholar]
- 54.Xie Y, Garfinkel A, Camelliti P, Kohl P, Weiss JN, Qu Z. Effects of fibroblast-myocyte coupling on cardiac conduction and vulnerability to reentry: a computational study. Heart Rhythm. 2009;6:1641–9. doi: 10.1016/j.hrthm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashihara T, Haraguchi R, Nakazawa K, Namba T, Ikeda T, Nakazawa Y, et al. The role of fibroblasts in complex fractionated electrograms during persistent/permanent atrial fibrillation: implications for electrogram-based catheter ablation. Circ Res. 2012;110:275–84. doi: 10.1161/CIRCRESAHA.111.255026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadoshima J, Weiss JN. Cardiac fibroblasts: the good, the bad, the ugly, the beautiful. J Mol Cell Cardiol. 2014;70:1. doi: 10.1016/j.yjmcc.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bruel A, Nyengaard JR. Design-based stereological estimation of the total number of cardiac myocytes in histological sections. Basic Res Cardiol. 2005;100:311–9. doi: 10.1007/s00395-005-0524-9. [DOI] [PubMed] [Google Scholar]
- 58.Shirani J, Berezowski K, Roberts WC. Quantitative measurement of normal and excessive (cor adiposum) subepicardial adipose tissue, its clinical significance, and its effect on electrocardiographic QRS voltage. Am J Cardiol. 1995;76:414–8. doi: 10.1016/s0002-9149(99)80116-7. [DOI] [PubMed] [Google Scholar]
- 59.Doughty MJ, Bergmanson JP, Blocker Y. Shrinkage and distortion of the rabbit corneal endothelial cell mosaic caused by a high osmolality glutaraldehyde-formaldehyde fixative compared to glutaraldehyde. Tissue Cell. 1997;29:533–47. doi: 10.1016/s0040-8166(97)80054-7. [DOI] [PubMed] [Google Scholar]
- 60.Hales PW, Schneider JE, Burton RA, Wright BJ, Bollensdorff C, Kohl P. Histo-anatomical structure of the living isolated rat heart in two contraction states assessed by diffusion tensor MRI. Prog Biophys Mol Biol. 2012;110:319–30. doi: 10.1016/j.pbiomolbio.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansoori T, Plank G, Burton RAB, Schneider JE, Kohl P, Gavaghan D, et al. An iterative method for registration of high-resolution cardiac histoanatomical and MRI images. IEEE ISBI; 2007. pp. 572–5. [Google Scholar]
- 62.Gibb M, Burton RB, Bollensdorff C, Afonso C, Mansoori T, Schotten U, et al. Resolving the three-dimensional histology of the heart. Computational Methods in Systems Biology; Berlin, Heidelberg: 2012. pp. 2–16. Springer. [Google Scholar]