Abstract
Cancer primarily develops due to somatic alterations in the genome. Advances in sequencing have enabled large-scale sequencing studies across many tumor types, emphasizing discovery of alterations in protein-coding genes. However, the protein-coding exome comprises less than 2% of the human genome. Here, we analyze complete genome sequences of 863 human tumors from The Cancer Genome Atlas and other sources to systematically identify non-coding regions that are recurrently mutated in cancer. We utilize novel frequency and sequence-based approaches to comprehensively scan the genome for non-coding mutations with potential regulatory impact. We identified recurrent mutations in regulatory elements upstream of PLEKHS1, WDR74, and SDHD, as well as previously identified mutations in the TERT promoter. SDHD promoter mutations are frequent in melanoma and associated with reduced gene expression and poor patient prognosis. The non-protein-coding cancer genome remains widely unexplored and our findings represent a step towards targeting the entire genome for clinical purposes.
Introduction
Large-scale cancer genomics projects such as The Cancer Genome Atlas (TCGA)1 and the International Cancer Genome Consortium (ICGC)2 have spent significant effort characterizing the cancer genome. So far, these projects have put their focus on genomic variation in the coding sequences of tumor genomes, and have identified a number of novel alterations, such as recurrent mutations in the exonuclease domain of the DNA Polymerase epsilon3,4. Since most studies rely heavily on targeted exome sequencing, our understanding of somatic variation in coding regions has improved significantly. However, the protein-coding component of the genome accounts for less than 2% of the total sequence and there is very little information on how non-coding variation affects cancer development. Furthermore, even well-studied cancer types such as non-small-cell lung cancer still have significant sub-populations with no observable “driver” mutation5,6.
The Encyclopedia of DNA Elements (ENCODE) project estimates that roughly 80% of the human genome has some sort of biochemical functionality7. It is well known that somatic mutations in non-coding regions are frequent8, but their effect is poorly understood. Recent efforts to understand non-coding variation in the human population have shown that disease-associated genomic variation is commonly located in regulatory elements9,10. Taken together, it is reasonable to expect that a substantial portion of recurrent non-coding somatic mutations observed in cancer could have a regulatory effect. To date, the most notable example is the recent discovery of mutations in the promoter of the TERT gene11,12. Other computational approaches to systematically characterize non-coding variation have primarily focused on nucleotide conservation9,13. Further progress in this area has been hampered by the relatively high cost of whole genome sequencing for large numbers of tumor samples, which are necessary to screen various regulatory regions for significant events. Indeed, previous studies on non-coding mutations in cancer have been limited by sample size12,14. The maturation of sequencing technologies now allows us to systematically sequence whole genomes and so it is only now that we can begin to appreciate the role non-coding mutations might play in the formation and development of cancer.
We performed a comprehensive analysis of somatic mutations from whole-genome sequences (WGS) from 863 cancer patients collected from The Cancer Genome Atlas (TCGA) and other public sources15 (Figure 1a, Supplementary Tables 1, 2). Our approach targets genomic variation in the non-coding part of the genome, which is poorly characterized, and rarely implicated in cancer. We called somatic mutations in tumor-normal pairs across the whole genome and annotated the mutations focusing on those most likely to affect regulatory elements. We then utilized multiple independent approaches to identify functional non-coding alterations.
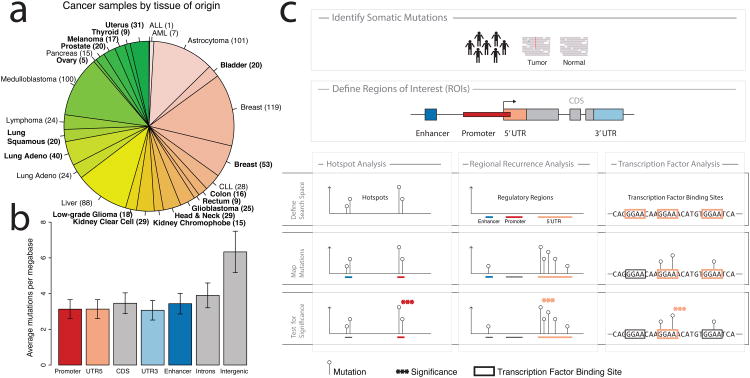
Figure 1.
a) Tumor samples by disease type. Tumor types from TCGA are labeled in boldface; other published samples15 are shown in regular font. b) Mean mutation frequency and 95% confidence interval (n=858) across samples by type of genomic region. c) Workflow for identification of recurrent, non-coding mutations in regulatory regions of interest. Our approach integrates mutation calls from 863 tumor/normal pairs and regulatory regions of interest (ROIs), which are tested for non-coding mutations using three distinct analyses. Hotspot analysis detects recurrent mutations that are often very focal. Regional recurrence analysis identifies annotated regions of interest that are enriched for mutation throughout the entire region. Transcription factor analysis searches for regions that contain recurrent mutations within transcription factor binding sites.
Results
Assessing the genomic landscape of non-coding mutations
The genome-wide mutation burden varied between different cancer types (Supplementary Figure 1) and the trend was generally consistent with previous observations in exome sequencing studies16. In the tumors from TCGA, most genomes had between 1.000 and 50.000 total somatic mutations. Mutations in transcribed regions, including coding sequences (CDS), introns, and 3′ and 5′ UTRs were observed at similar frequencies (Figure 1b). This observation is consistent with previous studies, suggesting a role for transcription-coupled repair8,17. Interestingly, promoter and enhancer regions were mutated at a rate similar to the transcribed genic regions. In contrast, intergenic regions, which by and large should be less implicated in gene regulation and possibly under weaker selective constraint, carry the highest mutational burden across all regions investigated here (Mann-Whitney P < 2.2e-16). Taken together, these observations suggest a functional role for a subset of mutations in annotated regulatory regions.
We used three distinct approaches to identify non-coding mutations that may play a role in tumor development and progression (Figure 1c). First, a “hotspot” analysis identified small regions with frequent mutations by detecting clusters of mutations within 50 bp of each other (see Methods for details). This approach returns very focal regions that are significantly recurrently mutated compared to a random distribution of mutations across the genome. Next, we targeted annotated regulatory regions that are mutated more frequently than expected by chance using a regional recurrence approach. This method takes into account the length and replication timing of different regulatory regions. It computes two measures of significance by comparing the observed frequency of mutations within a region of interest to the mutation frequency in neighboring areas (local P-value), as well as mutation frequencies of similar genomic regions elsewhere in the genome (global P-value). We validated both of these approaches in the TCGA breast cancer exome dataset by re-identifying genes that are frequently mutated in breast cancer (Supplementary Tables 3, 4)18. In the third approach, motivated by the discovery of the TERT promoter mutations, we specifically searched for mutations in ETS-family transcription factor binding sites, which have previously been implicated in cancer11. This approach considers evolutionary conservation of putative binding sites and separately evaluates mutations that disrupt or create ETS binding sites. Overall, we found that many of the most significant events identified by the three methods presented here (Supplementary Tables 5-16) occurred in regulatory regions of known cancer genes19 (P = 3.4e-4 when compared to all Ensembl genes using Fisher's exact one-tailed test), suggesting a possible functional role for these mutations in cancer.
Mutation hotspot in PLEKHS1 promoter
TERT promoter mutations constitute the most significant hotspot in our data (P = 1.1e-127, Figure 2a). The hotspot analysis identified a focal region in the promoter of the TERT gene, the catalytic subunit of telomerase, which is mutated in 56 samples. This hotspot contains two highly recurrent mutations at chr5:1295228 and chr5:1295250, which were found in 38 samples and 15 samples, respectively. Both sites had C->T substitutions, which is consistent with previous reports of TERT promoter mutation11,12. Here we found mutations in these two recurrent sites in 7 cancer types (glioblastoma (16), melanoma (10), bladder (10), low-grade glioma (9), liver (5), medulloblastoma (2), lung (1)), at frequencies similar to other recent reports20,21. We also observe a corresponding increase in TERT gene expression in mutated samples (Supplementary Figure 2). Our results suggest that these may be among the most prevalent functional non-coding mutations across all cancers.
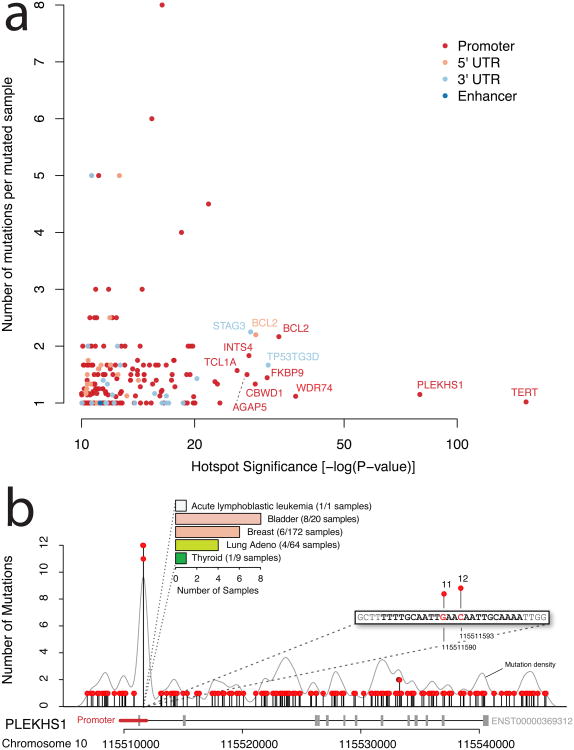
Figure 2. Hotspot Analysis.
a) Significance of mutation hotspots in non-coding regulatory regions. Hotspots are shown according to statistical significance (false discovery rate adjusted p-value on x-axis) and number of mutations per sample (y-axis). Colors represent the types of regulatory regions in which hotspots were found. b) Mutation hotspot in the promoter region of PLEKHS1, including two highly recurrent sites (11 and 12 mutations, respectively) located at the center of a palindromic sequence. Mutation density across the region is shown as grey curve. The bar chart summarizes the frequency of the hotspot mutation in individual cancer types (colors correspond to Figure 1a).
In addition to the hotspot in the TERT promoter, hotspot analysis identified a number of other recurrently mutated hotspots (Figure 2a). The next most significant is a small hotspot in the promoter of PLEKHS1 (P = 4.6e-80). This hotspot contains 23 mutations distributed over 20 samples, with two mutated sites at the far end of the promoter (∼50 bp into the first intron). The two sites were mutated in 11 (chr10:115511590) and 12 samples (chr10:115511593), both of which are predominantly C->T transitions (Figure 2b). Interestingly, these two mutations are flanked by stretches of 10 base pairs on both sides, which are palindromic to each other (Supplementary Figure 3). This hotspot was found in five cancer types and mutated samples appear to have lower expression of PLEKHS1 (Supplementary Figure 4). In bladder cancer, 40% of samples were affected by a mutation in this hotspot (8 out of 20 samples). PLEKHS1 is a largely uncharacterized gene that has not previously been linked to tumorigenesis. The gene contains a pleckstrin homology domain, which suggests a role for the protein in intracellular signaling22.
This analysis identified several other significant hotspots linked to STAG3, BCL2, TCL1A, AGAP5, TRMT10C, TNK2, and WDR74 (Supplementary Tables 5-8). Interestingly, many of these genes have been associated with cancer previously23-26. In contrast to PLEKHS1 and TERT, which mostly have one hotspot mutation per sample, hotspots in the promoter and 5′ UTR of BCL2 are significant, but seem to occur as clusters of several mutations within the same sample (average 2.2 mutations per mutated sample). Closer examination revealed that these are all in B-cell lymphoma samples and are likely a result of targeted somatic hypermutation at hypervariable regions27.
WDR74 promoter is frequently mutated
Protein-coding regions of many tumor suppressor genes display frequent inactivating somatic mutations, not at specific sites, but instead distributed across the entire open reading frame. To identify genes with frequent mutations across an entire regulatory region, we developed a statistical framework that evaluates the mutation rates of annotated regulatory regions in both a local and global genomic context. Briefly, the local approach compares regional mutation rates to the overall mutation frequency in the immediate genomic neighborhood, whereas the global approach compares mutation rates for regions in the same category (e.g. promoter or 3′ UTR) and with similar DNA replication timing (see Methods).
This approach identified larger, more frequently mutated genomic regions, thus complementing the hotspot analysis, which focused on much smaller regions. Apart from frequent promoter mutations in TERT (P < 1.3e-17), we observed a number of regulatory regions that were significantly enriched for non-coding mutations (Figure 3a). In particular, the 5′UTR (P < 5.1e-8) and promoter of WDR74 (P < 3.6e-9) were highly enriched for mutations. In contrast to the hotspot mutations in PLEKHS1, mutations in WDR74 were broadly distributed across numerous positions (Figure 3b) and WDR74 transcript levels were not significantly different in mutated samples (Supplementary Figure 5). While the coding sequence of WDR74 did not contain any mutations, our analysis revealed 35 non-coding mutations in a ∼1kb window near the 5′ end of WDR74 gene, most of which clustered at the start of the untranslated region.
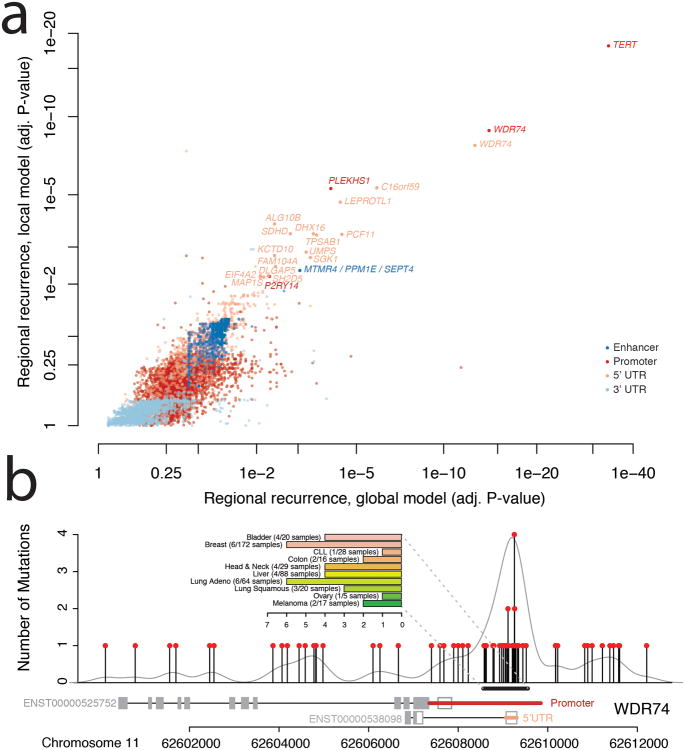
Figure 3. Regional Recurrence Analysis.
a) Significance of recurrent mutations in regulatory regions of interest. Regulatory regions for individual genes are shown according to local (y-axis) and global (x-axis) measures of statistical significance (false discovery rate adjusted p-value). Colors represent types of regulatory region. b) Strong enrichment of mutations in the promoter region of WDR74 in contrast to the remainder of the gene sequence. The bar chart summarizes the frequency of the hotspot mutation in individual cancer types (colors correspond to Figure 1a).
WDR74 contains a WD40 repeat, which has enzymatic activity and has been shown to be involved in a variety of biological processes, including cell cycle control and apoptosis28. The promoter region of WDR74 was previously found to be under purifying selection and likely sensitive to mutation9. Khurana et al. also reported WDR74 promoter mutations in 2/20 analyzed prostate cancer genomes. Here we demonstrate that mutations in this region are more common than previously known. We identified a total of 52 mutations in the promoter region of WDR74 (Figure 3b), including four distinct single nucleotides with recurrent mutations in up to four samples. Overall, 39/863 samples (5%) harbored at least one mutation in the regions.
Other frequently mutated regions were found in non-coding regions of genes such as SGK1, DHX16, and SDHD (Supplementary Tables 9-12). Interestingly, the 5′ end of the SDHD gene contained multiple mutations in putative ETS (E26 transformation-specific) family transcription factor binding sites. We next used transcription factor analysis to specifically assess the significance of mutations in ETS response elements on a genome-wide scale.
Promoter mutations in ETS binding site alter regulation of SDHD
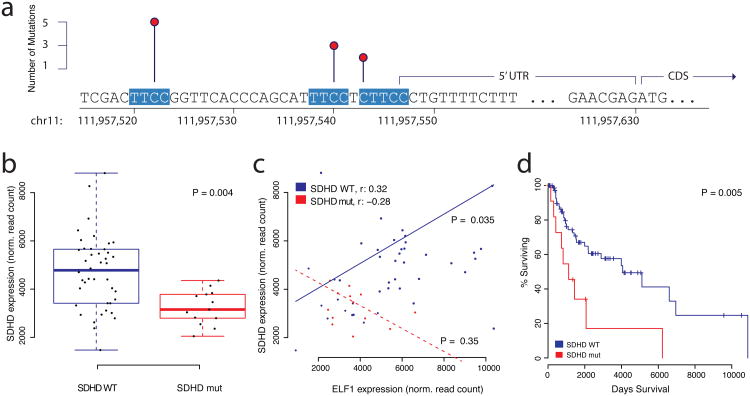
As mentioned above, hotspot analysis identified two known, highly recurrent sites in the promoter of TERT11,12. Both hotspot mutations create novel binding sites for ETS transcription factors by substituting a cytosine nucleotide with a thymine nucleotide (C->T), thereby generating the TTCC response element, which is highly conserved for ETS transcription factors. Both elements are located on the minus strand, which is the coding strand for TERT (CTCC>TTCC at chr5:1295228 and TCCC>TTCC at chr5:1295250). Here, we systematically screened regulatory regions of interest for mutations that either create novel ETS binding sites, or disrupt existing ones (see Methods for details). Apart from TERT, promoter mutations in ANKRD53 were the most significant mutations that created novel ETS binding sites (P < 0.0049). Several regulatory regions contained a significant number of mutations that disrupted ETS binding sites including TAF11, ERLIN2, MEF2C, KRT4, and SDHD, among others (Supplementary Tables 13-16). SDHD, which encodes the succinate dehydrogenase complex subunit D, was also observed in the regional recurrence analysis above. SDHD promoter mutations (C->T) occurred exclusively in melanoma samples and potentially disrupted two separate putative ETS binding sites in a small genomic region upstream of the coding sequence (Figure 4a). The recurrent mutations are located at chr11:111957523 (TTCC>TTTC) and chr11:111957541 (TTCC>TTTC), close enough to the start codon to allow further examination using TCGA melanoma whole-exome data, which exists for a larger number of samples. The exome data revealed a third putative ETS binding site in the SDHD promoter, located at chr11:111957544, which had a mutation just outside of the core response element, converting CTTCC>TTTCC. The mutated base is not conserved in all ETS family transcription factors, but it is highly conserved in ELF1 (Supplementary Figure 6), the only ETS transcription factor that correlated with SDHD gene expression, and also binds the SDHD promoter according to ENCODE data7,29. Out of 128 samples with read-depth 15 or higher, 13 had a mutation in the promoter region (10%), 10 of which had recurrent mutations in ETS binding sites. In contrast to recurrent mutations in the TERT promoter, which create a novel ETS binding site, mutations in the SDHD promoter damage existing ETS binding sites. Since TERT promoter mutations led to increased expression of the TERT gene, we expected expression of SDHD to be lower when compared to a group of ‘wild-type’ melanoma samples without SDHD promoter mutation. Using whole exome sequencing data and gene expression data from the TCGA, we compiled a set of 42 samples that did not present promoter mutations in SDHD (Methods). Analysis of expression data revealed that tumors with SDHD promoter mutations indeed have significantly reduced expression of the SDHD gene (P = 0.004, Figure 4b). Based on CHiP-Seq data from the ENCODE project, we were able to identify three ETS family transcription factors with binding activity in the SDHD promoter (EHF, ELF1, and ETS1). Among these three transcription factors, only ELF1 expression exhibited significant positive correlation with the SDHD expression data in the subset of 42 SDHD proficient samples without promoter mutation (Figure 4c, Supplementary Figure 7, P < 0.0035), indicating that SDHD could be under control of the ELF1 transcription factor under normal circumstances. Interestingly, tumor samples with SDHD promoter mutation do not exhibit a correlation between SDHD and ELF1 mRNA levels (P = 0.35), suggesting a possible adverse effect SDHD promoter mutation has on transcriptional regulation by ELF1 (Figure 4c). In addition to the apparent changes on gene expression, we observed that samples with SDHD mutation had a significantly shorter overall survival compared to a reference group of 88 melanoma samples (P = 0.005, Figure 4d). SDHD, which encodes the subunit D of the succinate dehydrogenase tetramer, is of particular interest since succinate dehydrogenase is the only protein that participates in the citric acid cycle as well as electron transport chain. It has been shown that SDHD mutations can cause paraganglioma30,31, a benign tumor of the head and neck. Previous studies suggest that SDHD acts like a tumor-suppressor30, which is consistent with our observation of reduced mRNA expression in tumor samples with SDHD promoter mutation.
Figure 4. Transcription Factor Analysis.
Mutations in the promoter region of SDHD disrupt ETS transcription factor binding sites in melanoma cancer genomes. a) Three recurrently mutated sites in the promoter region of SDHD, each one altering a separate ETS recognition site, which are highly conserved and highlighted in red. b) SDHD mRNA expression is lower in melanoma samples with SDHD promoter mutations (n = 13, red) compared to ‘wild-type’ tumor samples (n = 42, blue). The box plot displays first and third quartiles (top and bottom of boxes), median (band inside boxes), and lowest/highest point within 1.5 × IQR of the lower/higher quartile (whiskers). c) mRNA expression between ELF1 (ETS transcription factor) and SDHD is positively correlated in samples without SDHD promoter mutations (n = 42, blue) and not in samples with SDHD promoter mutation (n = 13, red). d) Survival analysis shows that overall patient survival is significantly lower in samples with SDHD promoter mutations (n = 12, red) compared to the reference group (n = 88, blue).
Discussion
Here we present a comprehensive analysis of whole genome sequencing data from 863 cancer patients to characterize the landscape of non-coding mutations in cancer. We show that intergenic regions are more often affected by mutation than other transcribed regions in close proximity to the coding sequence, such as introns, promoters, enhancers, and untranslated regions. In addition, our data suggests that regulatory regions at the 5′ end of genes, such as promoters and 5′UTRs, are recurrently mutated more often than 3′ UTRs or distal enhancers (Figure 3a).
We used three complementary types of analysis to identify regions of interest that are significantly affected by mutation: hotspot analysis focused on small regions that frequently contain mutations; regional recurrence analysis identified annotated regions that contained numerous mutations; transcription factor analysis nominated regions with ETS transcription factor binding sites that were disrupted or created by mutation. These three methods used clearly distinct approaches, and as result found different regions of interest, in general. However, the most significant findings, which are highlighted in this study, were identified by multiple methods. Promoter mutations in the TERT gene were found by all three methods. Hotspot analysis identified highly recurrent mutations in PLEKHS1, which contains a pleckstrin homology domain. The mutations occur at the center of a perfectly palindromic sequence. This observation is striking, even though it is not known if this particular palindrome is functional. However, it is also known that transcription factor binding sites can be palindromic32,33. The finding of SDHD promoter mutation was moderately significant in regional recurrence analysis, but was subsequently substantiated by transcription factor binding site analysis. Recurrent mutations in three distinct ETS response elements were associated with loss of correlation with ETS transcription factor (ELF1) on mRNA level, and shorter patient survival. Although our study focused exclusively on ETS transcription factor binding sites, we believe that such an approach will be valuable when carefully applied towards all known conserved binding sites.
It has been shown that large sample sizes are required to accurately detect low frequency cancer mutations34. Here we used data from multiple cancer types across a wide range of studies, most of which had fewer than 50 samples. Our analysis is therefore limited to detecting regions that are mutated at high frequencies in individual tumor types, or across several different tumor types. It is likely that similar analyses on larger sets of samples in individual tumor types will reveal additional insights (Supplementary Figures 8, 9, Supplementary Tables 17, 18). Even with this initial analysis, we observe many mutations at clinically relevant frequencies, which are interesting from a therapeutic perspective. Our results suggest that important tumorigenic mutations occur in non-coding regions, even though large numbers of passenger mutations exist in these regions as well, and the interpretation of such mutations remains a challenge. However, interrogation and interpretation of non-coding mutation will become more accurate and more important as availability of WGS data increases.
URLs
CGHub, https://cghub.ucsc.edu/; Broad GDAC Firehose, http://gdac.broadinstitute.org/; Data from Alexandrov et al.15, ftp://ftp.sanger.ac.uk/pub/cancer/AlexandrovEtAl
Methods
Calling mutations
Whole-genome sequencing data was downloaded from CGHub in the form of tumor and matched normal BAM files. Mutations were called across the whole genome using MuTect35 and Strelka36 with default parameters. The intersection of somatic mutation calls made by both programs was used as the mutation list for each sample.
To investigate promoter mutations in SDHD, whole-exome sequencing data for SKCM was downloaded from CGHub and allele counts were generated for SDHD 5′ coordinates chr11:111957523 and chr11:111957541. Samples without whole-genome sequences but whose whole-exome sequence data exhibited two or more mutant alleles (T instead of the reference C) in these positions were considered to be mutated.
We excluded 5 samples with more than 500,000 mutations each, limiting the data set to 858 samples. All analyses focused on single nucleotide substitutions, and did not consider insertions, deletions, or other structural variants.
Defining non-coding ROIs
We used gene annotation from Ensembl37 (v70) for transcripts of all protein coding genes (having at least one annotated open reading frame). 5′ UTR and 3′ UTR regions were used as defined by Ensembl. Promoter regions were defined as the genomic intervals ranging from 2000 bp upstream to 200 bp downstream of all transcription start sites. We used 66 944 enhancer-region to gene associations (27493 unique regions) from a previous comprehensive study38 in which inferred mid positions of enhancer regions were extended 200 bp up- and downstream. To avoid mutation bias from protein coding regions, we removed ORFs (extended by 5 bp to also account for splice sites) from the collection of ROIs. Furthermore, to avoid bias from immune system coupled somatic hypermutation, we also removed regions of 429 annotated immunoglobulin loci (each region extended by 50 kb, Ensembl v73).
Identification of hotspot mutations
All mutations within 50bp of each other were merged using BEDTools into hotspot “clusters” until no clusters were within 50bp of another39. Clusters with only 1 or 2 mutations were removed from further consideration. A p-value was calculated for each cluster using the negative binomial distribution, taking into account the length of the candidate hotspot, the number of mutations in the cluster, and a “background mutation rate” for the cluster. The cluster “background mutation rate” is calculated as the mean of the background mutation probability for each sample that has a mutation represented in the cluster. The background mutation probability of each sample is calculated as the total number of mutations divided by the genome size. P-values were adjusted for multiple testing with the multtest R package40 using the Benjamini-Hochberg method and hotspot clusters are ranked accordingly.
Testing ROIs for mutation recurrence
All ROIs longer than 50 bp were tested for recurrence of mutations using both a global and local statistical approach. Both approaches assumes that the observed number of mutated samples, k, for a given ROI follows a binomial distribution, Bin(n,pi), where n is the total number of samples with mutation data, and pi is the estimated sample mutation rate for ROI i under the null hypothesis that the region is not recurrently mutated. We can therefore compute the following p-value:
Here we assume that pi depends on the effective length Li of the ROI (with any ORF overlap subtracted, see above) and the estimated nucleotide mutation rate qi for the region under the null hypothesis:
The background mutation frequency qi is not readily available and needs to be estimated before we can compute a p-value using the above equation. We estimate both a local and a global background mutation frequency, which form the basis for the local and global tests, respectively. For the local approach we extracted 10kb flanking regions upstream and downstream of the ROI, excluding ORFs from the flanks to reduce mutation bias from nearby protein coding regions. The local background nucleotide mutation frequency was then estimated by dividing the total number of observed mutations with the effective length of the flanking region. In the global approach, we estimated nucleotide mutation frequencies from other regions of the same ROI category (e.g. promoter, 3′ UTR, etc.). Because DNA replication timing has previously been shown to affect somatic mutation rates in tumors16,41, we further stratified ROIs of the same category by their replication timing in 5 cancer cell lines (HeLa, K562, HEPG2, MCF7, SKNSH, data from the UW ENCODE group7,42). We first computed average replication time values in 100 kb bin sizes for each cell line, and for each ROI we computed a single replication time value (average if spanning >1 bins) for each cell line. For a given ROI in category C, we identified the top 5% ROIs in C with most similar replication timing profiles (Euclidian distance between vectors of replication time values across the 5 cell lines). The global background nucleotide mutation frequency was then estimated by dividing the total number of observed mutations in the top 5% ROIs with the effective length of these regions. P-values were computed using the equation above, and adjusted for multiple testing with the multtest R package using the Benjamini-Hochberg method. For each region/gene we selected the maximum FDR of the individual global and local tests.
Transcription factor analysis
All mutations were annotated if they affected ETS transcription factor binding sites. Mutations were considered to create ETS transcription factor binding sites, if the nucleotide substitution created a novel ETS transcription factor core response element on either strand (e.g. TGCC>TTCC). Mutations were considered to disrupt ETS transcription factor binding sites if they altered an existing ETS core response element (e.g TTCC>TGCC). Using the regions of interest defined above, we then calculated a count statistic for each region of interest by summing up the number of mutations that created or disrupted ETS transcription factor binding sites within each ROI. For each region of interest that contained more than 1 mutation in a ETS binding site, an empirical p-value was computed by comparing the observed count statistic (number of mutations creating/disrupting ETS binding sites within the region of interest) to a reference distribution of count statistics. Reference distributions were generated for each region of interest by iteratively calculating the above count statistic on the same set of mutations after randomizing the ETS transcription factor annotations (i.e. the binary annotation whether of not mutations created novel binding sites were randomized) during each iteration. A p-value was derived by comparing the observed count statistic of a given region of interest to the distribution of count statistics of its corresponding reference distribution (based on 10000 iterations), and was defined as the fraction of count statistics in the reference distribution greater or equal to the observed count statistic. P-values were adjusted for multiple testing using the Benjamini-Hochberg method.
Expression analysis
Expression analysis was performed using RNASeq raw counts from TCGA. P-values are reported using a negative binomial test from the edgeR package43, which is available through Bioconductor44. In-depth analyses of SDHD promoter mutations (Figures 4b-c) were performed on a set of melanoma samples from TCGA (v20130923, Level 3) for which exome-sequencing data was available. The set of reference (‘wild-type’) samples consisted of melanoma samples, which had a read depth of 15 reads or higher in the SDHD promoter region. “Wild-type” samples with putative copy-number alterations at the SDHD locus were excluded. Survival analysis (Figure 4d) was performed on a set of 88 samples with read depth of 15 reads or higher in the SDHD promoter region using the clinical data file for melanoma from TCGA.
Supplementary Material
Acknowledgments
The authors thank The Cancer Genome Atlas as well as numerous other groups who have made their data available for public analysis. The authors additionally thank the CGHub team, members of the MSKCC Bioinformatics Core, Gunnar Rätsch, and John Chodera for setup and administration of high-performance computing resources. This work was supported in part by a TCGA GDAC grant from the NIH for N.W., N.S., C.S., and W.L. (NCI U24-CA143840) and grants from the Danish Research Council and the Carlsberg Foundation for A.J.
Footnotes
Author Contributions: Project planning and design: N.W., A.J., N.S., C.S., W.L., Method design and data analysis: N.W., A.J., W.L., Manuscript writing and figures: N.W., A.J., W.L., Review and editing of the final manuscript: all authors.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church DN, et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Human molecular genetics. 2013;22:2820–8. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. The lancet oncology. 2011;12:175–80. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 9.Khurana E, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann KV, Chen T. Exploring functional variant discovery in non-coding regions with SInBaD. Nucleic acids research. 2013;41:e7. doi: 10.1093/nar/gks800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futreal PA, et al. A census of human cancer genes. Nature reviews Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nature communications. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 21.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer BJ, Ren R, Clark KL, Baltimore D. A putative modular domain present in diverse signaling proteins. Cell. 1993;73:629–30. doi: 10.1016/0092-8674(93)90244-k. [DOI] [PubMed] [Google Scholar]
- 23.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–9. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 24.Virgilio L, et al. Identification of the TCL1 gene involved in T-cell malignancies. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12530–4. doi: 10.1073/pnas.91.26.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan NP, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8438–43. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krol M, et al. Transcriptomic signature of cell lines isolated from canine mammary adenocarcinoma metastases to lungs. Journal of applied genetics. 2010;51:37–50. doi: 10.1007/BF03195709. [DOI] [PubMed] [Google Scholar]
- 27.Schneider C, Pasqualucci L, Dalla-Favera R. Molecular pathogenesis of diffuse large B-cell lymphoma. Seminars in diagnostic pathology. 2011;28:167–77. doi: 10.1053/j.semdp.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends in biochemical sciences. 2010;35:565–74. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 31.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nature genetics. 2000;26:268–70. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 32.Thukral SK, Eisen A, Young ET. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Molecular and cellular biology. 1991;11:1566–77. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes & development. 1991;5:670–82. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 37.Hubbard T, et al. The Ensembl genome database project. Nucleic acids research. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.v d La MJ, P K, D S. Multiple Testing Procedures: R multtest Package and Applications to Genomics. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2004 [Google Scholar]
- 41.Chen CL, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome research. 2010;20:447–57. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.