SUMMARY
We have solved two families of crystal structures of human Dicer ‘platform-PAZ-connector helix’ cassette in complex with siRNAs. The structures possess two adjacently positioned pockets: a 2-nucleotide 3′-overhang-binding pocket within the PAZ domain (3′-pocket) and a newly identified phosphate-binding pocket within the platform domain (phosphate pocket). One family of complexes contain a knob-like α-helical protrusion, designated ‘hDicer-specific helix’, that separates the two pockets and orients the bound siRNA away from the surface of Dicer, which could be indicative of a product release/transfer state. In the second complex, the helical protrusion is melted/disordered and the bound siRNA is aligned towards the surface of Dicer, suggestive of a cleavage-competent state. These structures allow us to propose that the transition from the cleavage-competent to the postulated product release/transfer state may involve release of the 5′-phosphate from the phosphate pocket, while retaining the 3′-overhang in the 3′-pocket.
The RNase III endoribonucleases (Carmell and Hannon, 2004; Robertson et al., 1968), Drosha in the nucleus (Lee et al., 2003) and Dicer in the cytoplasm (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001), play key roles in the miRNA biogenesis pathway (Kim, 2005; Kim et al., 2009) by functioning as molecular rulers, that cleave the substrate dsRNA within helical segments with site-selectivity, leading to products of defined length with a 5′-phosphate and a 2-nt overhang at the 3′-end (Elbashir et al., 2001; Han et al., 2006). Drosha in the presence of DiGeorge syndrome critical region 8 (DGCR8) processes primary miRNA (pri-miRNA) hairpins into their precursor miRNA (pre-miRNA) hairpin counterparts (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004; Lee et al., 2003), which are subsequently processed by Dicer and trans-activation response RNA-binding protein (TRBP) into mature miRNA duplexes (Chendrimada et al., 2005; Haase et al., 2005).
Human Dicer (hDicer) is a large multi-domain enzyme whose C-terminal half includes a PAZ domain, a pair of tandem RNase III domains and a double-stranded RNA-binding domain (dsRBD) (Figure 1A, upper panel). Dicer also contains a DEXD/H helicase domain towards its N-terminus that has been assigned an auto-inhibitory function (Ma et al., 2008). Dicer exhibits cleavage activity by generating siRNAs and miRNAs of defined length (21- to 28-nucleotides) as measured from the helical ends of dsRNA (Elbashir et al., 2001; Hutvagner et al., 2001), which in turn are loaded into the RNA induced silencing complex (RISC) for gene silencing by targeted mRNA degradation (Hammond et al., 2001), translational repression (Olsen and Ambros, 1999) and heterochromatin formation (Volpe et al., 2002).
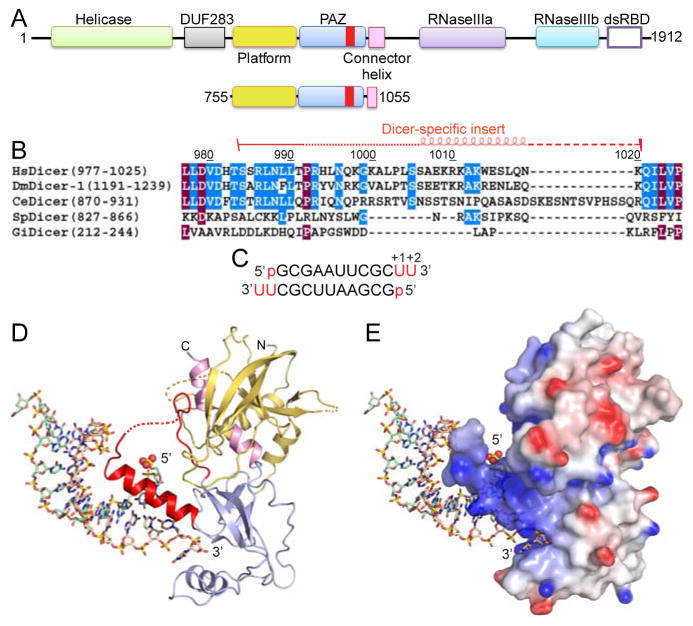
Figure 1. Dicer Domain Architecture, Species-Specific Sequences of ‘Dicer PAZ Insertion Element’ and Structure of hDicer PAZ Cassette Bound to a 12-mer siRNA.
(A) Domain architecture of hDicer (top) and hDicer ‘platform-PAZ-connector helix’ cassette (below), with the latter used for structural studies of complexes with bound siRNAs and dsRNA. The Dicer-specific insert is shown in red.
(B) Comparison of sequences (human, D. melanogaster-1, C. elegans, S. pombe and G. intestinalis) of the ‘Dicer PAZ insert’ element.
(C) Sequence of the self-complementary 12-mer siRNA containing 5′-p and UU-overhangs at 3′-ends.
(D) Structure of hDicer PAZ cassette bound to a 12-mer siRNA. The platform, PAZ and connector helix are colored in yellow, blue and pink, respectively. The ‘Dicer PAZ-insertion element’ is colored in red and composed of a disordered segment followed by the ‘hDicer-specific helix’. The 5′-phosphate is shown in a space-filling representation.
(E) A view of the complex in the same orientation as in panel D, with the protein in an electrostatic surface representation.
See also Figure S1, S2 and S6, and Table S2.
The first insights into the mechanism of catalytic cleavage by Dicer emerged from biochemical (Zhang et al., 2004), structural (Macrae et al., 2006) and modeling (Macrae et al., 2006) studies, supplemented by subsequent biochemical studies that included mutation, cleavage kinetics and domain swap experiments (MacRae et al., 2007), which proposed that the PAZ domain anchored one end of the double-stranded RNA (dsRNA), with the length of the cleavage product defined by the separation between the PAZ pocket and the pair of RNase III pockets of the single composite processing center (Macrae et al., 2006; Zhang et al., 2004). Each RNase III domain catalyzes the hydrolytic cleavage of one strand of the substrate dsRNA, a process facilitated by the participation of two Mg2+ cations in the catalysis at each active site (Macrae et al., 2006). Site-directed mutagenesis studies highlighted that the primary interaction between Dicer and its dsRNA substrates is mediated by the flat positively charged surface that connects the PAZ and the composite pair of RNase III domains (MacRae et al., 2007). In addition, a 10-residue ‘positioning loop’ located at the junction between the platform and pair of catalytic domains contributes to the correct placement of the substrate within the enzyme active sites (MacRae et al., 2007). These recognition and cleavage concepts are consistent with the intermolecular interactions elucidated from the crystal structure of homodimeric Aquifex aeolicus RNase III composed of a RNase III and a dsRBD bound to dsRNA (Gan et al., 2006).
To precisely locate the cleavage site, Dicer utilizes multiple features of RNA substrates. The most critical features are the 3′ and 5′ ends of RNA substrate. The 3′ end has been previously shown to be recognized by the PAZ domain (Lingel et al., 2004; Ma et al., 2004). Dicer also uses the 5′ end to locate the cleavage site (Park et al., 2011). The 5′ terminal phosphate binding activity has been mapped to basic residues near the 3′ end binding pocket but the structural basis of such recognition has not yet been described. Several additional factors modulate cleavage site choices, including the location of the loop (Gu et al., 2012; Tsutsumi et al., 2011) and the presence of dsRNA binding partners in some cases (Fukunaga et al., 2012).
There are no x-ray crystallographic structures to date on either higher eukaryotic Dicer or its components bound to siRNAs. This information is critically needed to address how Dicer interacts with its RNA targets and also to eventually definitively position the Dicer-RNA complex within available cryo-EM data in the context of the RNA induced silencing complex (RISC) (Lau et al., 2012; Taylor et al., 2013; Wang et al., 2009). Here we report on the crystal structure of the complex between the human Dicer ‘platform-PAZ-connector helix’ cassette (designated hDicer PAZ cassette) (Figure 1A, lower panel) bound to several siRNAs and dsRNAs containing 5′- and 3′-overhangs. The structures define two different alignments of the siRNA along the hDicer PAZ cassette and more importantly, identify a phosphate-binding pocket positioned within the platform domain.
RESULTS
Design of hDicer PAZ Construct and Crystallization of Complexes with Bound siRNAs
Our initial attempts to express the isolated PAZ domain of hDicer did not yield soluble protein. The structure of G. intestinalis Dicer in the free state (Macrae et al., 2006) established that linkers on either side of the PAZ module adopted folded domains, designated the ‘platform’ (for the linker on the N-terminal side) and the ‘connector helix’ (for the linker on the C-terminal side). Therefore, we generated hDicer constructs that included these linkers and identified one construct, extending from positions 755 to 1055 (Figure 1A, lower panel), that yielded soluble protein. This construct contains the entire ‘platform’, the complete PAZ domain, and more than half of the ‘connector helix’ (Figure S1A). We next attempted to crystallize this hDicer cassette with self-complementary siRNA duplexes of different length containing a 5′-phosphate and 2-nt overhangs at the 3′-end.
In addition, we noted the presence of an insert within the PAZ domain of Dicer proteins (Figure 1B and S1A) that was absent in the PAZ domain of Argonaute proteins (Figure S1B). This ‘Dicer-specific insert’ was found to be longer in higher eukaryotes (humans) when compared with their lower counterparts (G. intestinalis) (Figure 1B and S1A).
Initially we could successfully grow crystals of the complex of hDicer PAZ cassette (755-1055) with a 16-mer self-complementary siRNA duplex having a 5′-phosphate and UU 3′-overhangs that diffracted to 3.4 Å resolution, and built an initial model following collection of a single wavelength anomalous dispersion (SAD) data set on the seleno-methionine-labeled protein (x-ray statistics in Table S1). However, during refinement it became apparent that it was difficult to assign amino acids unambiguously for the platform domain, due to limited resolution and relatively poor electron density for this segment of the complex. To improve diffraction quality, we introduced three separate pairs of surface mutations of large flexible residues to Ala to stabilize crystal contacts by reduction in surface entropy.
One of the three double mutants (755-1055; K822A/K823A surface Lys mutants), gave crystals of the complex of hDicer PAZ construct bound to a self-complementary 12-mer duplex containing a 5′-phosphate and UU 3′-overhangs in a different crystal form that diffracted to improved 1.95 Å resolution. The structure of this complex was solved by molecular replacement using the partially refined low-resolution structure of hDicer PAZ cassette with the 16-mer self-complementary siRNA duplex presented above.
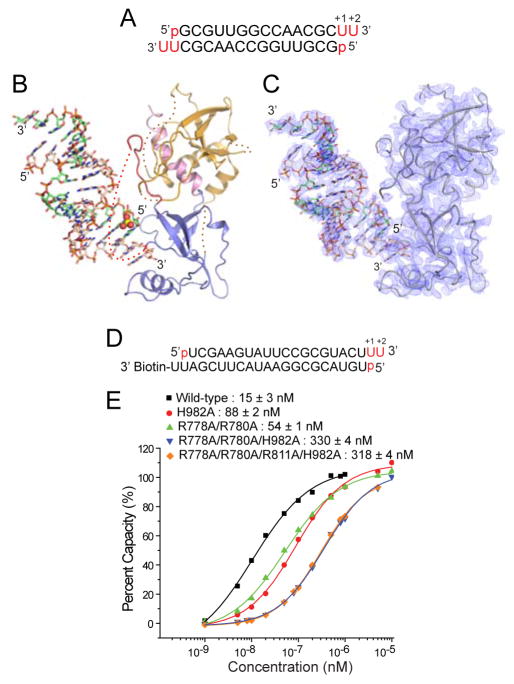
The sequence of the 12-mer siRNA was 5′-p-GCGAAUUCGCUU-3′ (Figure 1C and 2A) and hence four central A-U pairs are bracketed by three G-C pairs at either end. Thus, this siRNA duplex has stable base-paired ends, from which project a 5′-phosphate and UU 3′-overhangs. The structure of the hDicer PAZ cassette-siRNA 12-mer complex refined to 1.95 Å resolution (x-ray statistics in Table S2) is shown with the protein either in ribbon (Figure 1D) or electrostatic surface (Figure 1E) representations, and the bound RNA in a stick representation. Both the majority of the protein and the entire siRNA can be traced in the complex, with the omit map (1.2σ) for the bound siRNA shown in stereo in Figure S2A.
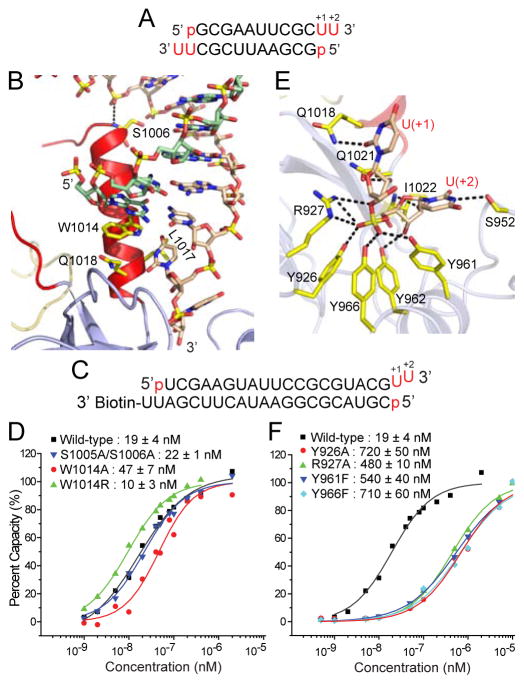
Figure 2. Structural Details of hDicer PAZ Cassette Bound to 12-mer siRNA and Impact of Mutations of Residues in the hDicer-Specific Helix and 3′-pocket.
(A) Sequence of the self-complementary 12-mer siRNA containing 5′-p and UU-overhangs at 3′-ends.
(B) Intermolecular hydrogen-bonding contacts involving either end of the ‘hDicer-specific helix’, with Trp1014 stacking over the terminal base pair.
(C) Sequence of immobilized siRNA used for SPR measurements containing 5′-p, UU 3′-overhang and stable terminal G-C pair at one end and biotin attached to the 3′-overhang at the other end.
(D) Measurement of SPR-based siRNA (sequence in panel C)-binding affinities for hDicer PAZ cassette mutants located within the ‘hDicer-specific helix’.
(E) Intermolecular hydrogen bonds involving the backbone phosphate and 2′-OH groups of the 2-nt overhang at the 3′-end of the 12-mer siRNA and residues lining the 3′-pocket in the complex.
(F) Measurement of SPR-based siRNA (sequence in panel C)-binding affinities for hDicer PAZ cassette mutants lining the 3′-pocket.
See also Figures S3, S4, S5 and S6 and Table S2.
Structure of hDicer PAZ Cassette-siRNA 12-mer Complex Containing an α-Helical ‘hDicer-Specific’ Segment
One of the unexpected features of the structure of the hDicer PAZ cassette-siRNA 12-mer complex is the identification of a disordered segment (993-1003), followed by an α-helical segment (1006-1020), with the latter designated the ‘hDicer-specific helix’ (in red, Figure 1D and 2B), that spans the large insert segment (982-1032) observed in higher eukaryotic Dicers (Figure 1B). This helix extends out from the surface of the protein in a knob-like manner (Figure 1D, E), and in turn orients the bound siRNA duplex at an angle (approximately 60°) away from the surface of the protein (Figure 1D, E).
There are a limited number of intermolecular contacts between this ‘hDicer-specific helix’ and the bound siRNA. First, the terminal base pair of the duplex is capped through stacking of G1 over the indole ring of Trp1014 at one end of the ‘hDicer-specific helix’ (Figure 2B). Ser1005 and Ser1006 at the other end of the ‘hDicer-specific helix’ form hydrogen bonds to adjacent phosphates of the bound duplex (Figure 2B).
We have superposed our high resolution structure of ‘platform-PAZ-connector helix’ cassette of human Dicer in the RNA-bound state with that of the same cassette within the structure of G. intestinalis Dicer in the free state reported previously (MacRae et al. 2006) (Figure S3). In the structure of the hDicer PAZ cassette-siRNA 12-mer complex, the mutated surface residues (K822A/K823A) are located far from the RNA binding site (Figure S3).
Impact on Binding of ‘hDicer-Specific Helix’ Mutants
To understand the significance of the observed contacts between siRNA and ‘hDicer-specific helix’, we introduced mutations to Dicer and examined its interaction with a biotin-labeled RNA duplex (Figure 2C) by surface plasmon resonance (SPR). One accessible end of the siRNA contained 5′-p, UU-3′ ends and a terminal G-C Watson-Crick pair, while the other end was biotin labeled to the 3′-overhang segment (Figure 2C). Compared to the wild-type protein (Kd = 19.5 nM), the W1014A mutant binds approx. 2.5-fold weaker (Kd = 47 nM), while the W1014R mutant, where the long hydrophobic side chain of Arg replaces the indole ring of Trp, binds approx. 2.5-fold stronger (Kd = 9.6 nM) (Figure 2D). There is essentially no change in binding affinity for the dual S1005A/S1006A mutant (Kd = 22.1 nM). Thus, the intermolecular contacts involving the ‘hDicer-specific helix’ contribute minimally to binding affinity of complex formation. These results are consistent with our previous observations that the same set of mutations (S1005A/S1006A, W1014A, and W1014R) did not affect pre-miRNA processing by full-length hDicer (Park et al., 2011; Park et al., unpublished data), suggesting that the ‘hDicer-specific helix’ is not necessary for Dicer processing per se.
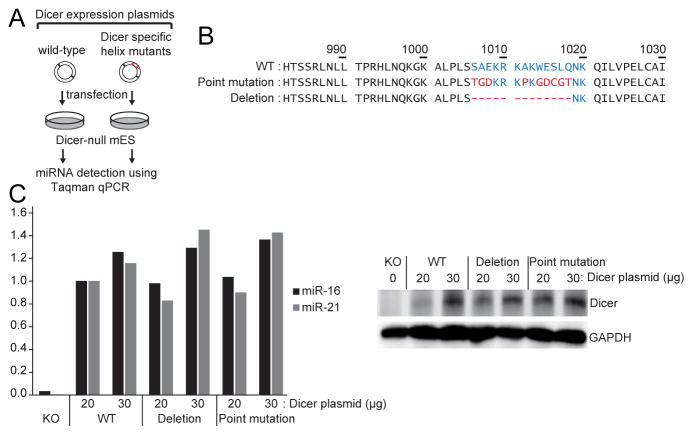
We also generated two mutants in which ‘hDicer-specific helix’ was either deleted or disrupted by point mutation. When these mutants were introduced into Dicer knockout cells, the mutants were as competent as the wild type Dicer protein in miRNA production (Figure 3). These results suggest that ‘hDicer-specific helix’ is unlikely to be actively involved in miRNA processing.
Figure 3. Dicer rescue assay in Dicer knockout embryonic stem cells using ‘hDicer-specific helix’ mutants.
(A) Experimental scheme of the Dicer rescue assay.
(B) Sequences of ‘hDicer-specific helix’ mutants. Residues comprising the ‘hDicer-specific helix’ are colored in blue. Mutant residues are in red.
(C) Results of Taqman miRNA qPCR using miR-16 and miR-21 probes (left panel). The plasmids were transfected in two different concentrations (20 μg and 30μg). Western blot showed comparable expression of Dicer (right panel).
3′ Overhang Recognition by 3′-Pocket and Impact on Binding of 3′-Pocket Mutants
The 2-nt UU overhang at the 3′-end [labeled U(+1)-U(+2) for clarity] inserts into its 3′-pocket within the hDicer PAZ domain and is anchored in place by a multitude of intermolecular hydrogen bonds to its sugar 2′-hydroxyl and internucleotide phosphate groups. Of specific note are the hydrogen bonds formed between four tyrosine hydroxyls (Tyr926, Tyr961, Tyr962 and Tyr966), together with the guanidinium of Arg927, primarily to the non-bridging phosphate oxygens of the internucleotide phosphate (Figure 2E).
Further, the first overhang base (designated U+1) forms a hydrogen bond with the side chain of Gln1018 (Figure 2E), while the second overhang base (designated U+2), forms a hydrogen bond with the side chain of Ser952 (Figure 2E). The overhang bases U(+1) and U(+2) do not stack on each other in the hDicer PAZ cassette-siRNA complex (Figure 2E), as they do in the hAgo PAZ-RNA complex reported previously (Lingel et al., 2004; Ma et al., 2004) (Figure S4).
We have investigated the impact of mutating residues lining the 3′-pocket on binding affinity using SPR measurements on the biotin labeled siRNA shown in Figure 2C. Compared to the wild-type protein (Kd = 19.5 nM), the single mutants Y926A (Kd = 720 nM), Y966F (Kd = 710 nM), Y961F (Kd = 540 nM) and R927A (Kd = 460 nM), result in an approx. 30-fold reduction in binding affinity (Figure 2F). These results imply that the intermolecular contacts involving the 3′-overhang binding pocket contribute significantly to binding affinity of complex formation. Notably, the mutations in the 3′-pocket (Y926F, R927A, Y961F, and Y966F) impaired Dicer processing in vitro, indicating that these residues are indeed critical for the interaction with dsRNA substrates (Park et al., 2011; Park et al., unpublished data).
We have also grown 2.1 Å diffracting crystals of the complex (x-ray statistics in Table S3) where the second 3′-overhang base at the +2 position was changed from a pyrimidine (U) to a bulkier purine (A). We note that the adenine at the +2 position can be accommodated by a slight expansion of the pocket through lateral movement of the Pro951-Ser952-Pro953 segment that forms one of the walls of the pocket (Figure S5). In line with this observation, variations in the 3′ terminal nucleotide (U, A, or C) did not influence Dicer processing of pre-let-7 (Park et al., 2011).
Identification of a Phosphate-Binding Pocket in the Platform Domain
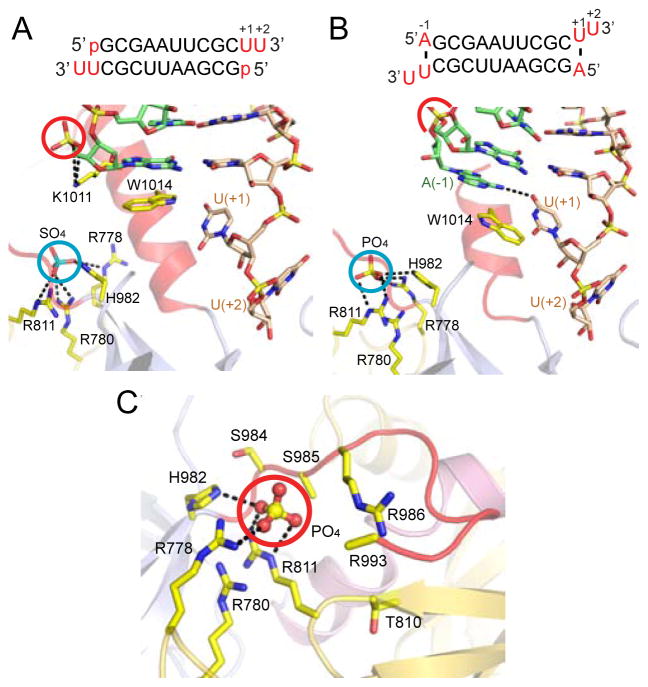
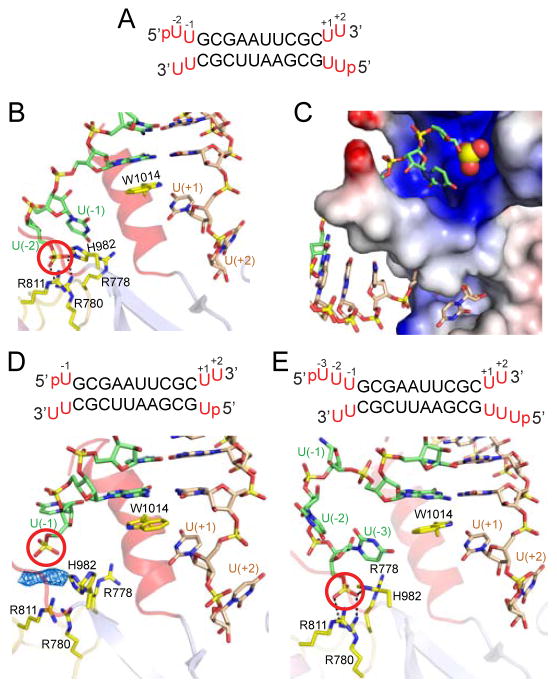
A striking feature of the hDicer PAZ cassette-siRNA 12-mer complex involved the identification of a bound sulfate (crystallization condition contained Li2SO4) coordinated to the side chains of Arg778, Arg780, Arg811 and His982 (cyan circle, Figure 4A; x-ray statistics in Table S2), which is suggestive of a basic phosphate-binding pocket. This phosphate/sulfate-pocket is positioned within the platform domain and separated by the ‘Dicer-specific helix’ from the 3′-pocket that is positioned within the PAZ domain. The two pockets are located adjacent to each other on the same face of the hDicer PAZ cassette. Notably, the 5′-phosphate of the bound 12-mer siRNA in the complex is hydrogen-bonded to Lys1011 (red circle, Figure 3A), which is far from the phosphate/sulfate-pocket (blue circle, Figure 3A) in the complex. Mutations to this region (K1009A/R1010A/K1011A/W1014A) did not affect Dicer processing in vitro (data not shown), indicating that Lys1011 is unlikely to interact with the 5′-phosphate during Dicer cleavage reaction.
Figure 4. Structural Details of hDicer PAZ Cassette Bound to 12-mer and 13-mer siRNAs that Identifed a Phosphate-binding Pocket Within the Platform Domain.
(A) Details of hDicer PAZ cassette bound to 12-mer siRNA (sequence in insert), containing a 5′-phosphate. Note the positioning and hydrogen-bonding of a bound sulfate (cyan circle) in a basic pocket, designated the ‘phosphate pocket’.
(B) Details of hDicer PAZ cassette bound to 13-mer siRNA (sequence in insert), containing an adenine, labeled A(−1), which base pairs with U(+1), with the purine ring of A partially inserting between the terminal base pair and Trp1014. Note the positioning and hydrogen-bonding of a bound phosphate (cyan circle) in the basic phosphate pocket.
(C) Basic patches on the surface of the platform domain in the structure of the hDicer PAZ cassette bound to 13-mer siRNA, with one basic patch bound to phosphate originating from the crystallization (Na/K phosphate) buffer. The phosphate is anchored in a pocket composed of Arg778, Arg780, Arg811 and His982. A second basic patch is composed of Arg986, Arg993 and His994 (disordered in the structure) and is positioned adjacent to the first patch. The side chain atoms of Ser985 and Arg993 are disordered in this structure.
See also Table S3.
We also observe an inorganic phosphate positioned inside the basic pocket in the 2.6 Å structure of the hDicer PAZ cassette bound to a 13-mer siRNA (Figure 4B, x-ray statistics in Table S3), where the bound phosphate (crystallization buffer contained Na/K-phosphate) is coordinated in a manner similar to the bound sulfate in Figure 3A. We also note that in addition to the basic patch (Arg778, Arg780, Arg811 and His982) that generates the phosphate/sulfate pocket, there is a second basic patch composed of Arg986, Arg993 and His994 (with the side chains of Arg993 and His994 disordered in the structure) in close proximity to it (Figure 4C).
An additional complex with a 17-mer siRNA was also successfully solved (3.1 Å resolution, x-ray statistics in Table S2). This 17-mer siRNA complex exhibits all the recognition features observed in the above discussed 12-mer siRNA complex (Figure S6A), with a pair of hDicer PAZ cassette modules bound at both ends of the 17-mer siRNA complex (Figure S6B) as they are in the 12-mer siRNA complex (Figure S6A).
Positioning the 5′-Phosphate of dsRNA-Containing a Pair of Disrupted U•U Pairs at Either End into the Phosphate Pocket
In an effort to mimic an RNA duplex containing thermodynamically unstable ends, we attempted to crystallize a complex of hDicer PAZ with a self-complementary 14-mer RNA duplex containing a pair of adjacent non-canonical U•U pairs at either end (Figure 5A). We retained the same self-complementary 10-mer duplex as the core and added pU(−2)-U(−1) at the 5′-end and U(+1)-U(+2) at the 3′-end, where the uracils are numbered for clarity. Crystals of this complex diffracted to 2.55 Å resolution and the structure was solved by molecular replacement (x-ray statistics in Table S2).
Figure 5. Structural Details of hDicer PAZ Cassette Bound to RNAs containing pUU (14-mer), pU (13-mer) and pUUU (15-mer) Overhangs at 5′-ends and UU Overhangs at 3′-ends.
(A) Sequence of the self-complementary 14-mer RNA containing pUU and UU-overhangs at 5′ and 3′-ends, respectively.
(B) A view of the positioning of the p-U(−1)-U(−2) at the 5′-end (green) and U(+1)-U(+2) at the 3′-end (wheat) on either side of the ‘hDicer-specific helix’ in the complex. Note that the 5′-phosphate (red circle) is positioned in the phosphate pocket and forms hydrogen bonds with basic residues. The base at position U(−2) is disordered.
(C) An expanded view of the phosphate and 3′-pockets in the 14-mer RNA complex, with the protein represented in an electrostatic surface representation. Note that the ‘hDicer-specific helix’ separates the basic phosphate pocket (above) from the hydrophobic 3′-overhang-binding pocket (below). The base at position U(−2) is disordered.
(D) Sequence of the 13-mer RNA duplex containing pU overhangs at 5′-ends and UU-overhangs at 3′-ends. A view of the positioning of the p-U(−1) at 5′-end (green) and U(+1)-U(+2) at 3′-end (wheat) on either side of the ‘hDicer-specific helix’ in the complex. The 5′-phosphate does not reach into the phosphate pocket that is represented by cluster of His and Arg residues. Unaccounted electron density is shown in a blue mesh representation, which could have originated from an additive used to facilitate crystallization.
(E) Sequence of the 15-mer RNA duplex containing pUUU overhangs at 5′-ends and UU-overhangs at 3′-ends. A view of the positioning of the p-U(−1)-U(−2)-U(−3) at 5′-end (green) and U(+1)-U(+2) at 3′-end (wheat) on either side of the ‘hDicer-specific helix’ in the complex. The base at position U(−1) is disordered. Intermolecular hydrogen-bonds are observed between the 5′-phosphate and the side chains of His982 and Arg780, with the side chains of Arg788 and Arg911 contributing to the basic nature of the phosphate-binding pocket in the complex.
See also Figure S7 and Table S2 and S3.
The structure of the hDicer PAZ cassette-dsRNA 14-mer complex is shown in Figure S7, with the protein in a ribbon representation and the RNA in a stick representation. An expanded view highlighting the 5′-phosphate and 3′-overhang ends anchored in their respective binding pockets separated by the ‘hDicer-specific helix’ is shown in Figure 5B, together with a related expanded view from a different perspective with the protein in an electrostatic surface representation shown in Figure 5C. We can identify two binding pockets, with one of them accommodating the 2-nt overhang at the 3′-end (lower pocket in Figure 5C) and the other accommodating the 5′-phosphate (whose phosphate group is highlighted in a space-filling representation) positioned in a basic patch (upper pocket in Figure 5C). The 5′-p-U(−2)-U(−1) and U(+1)-U(+2)-3′ segments bifurcate on either side of the ‘hDicer-specific helix’ (Figure 4B, C) and are not paired with each other in the complex (the base of U(−2) is disordered). The G1 base of the terminal G-C base pair stacks on the indole ring of Trp1014 (Figure 5B), similar to what was observed in the complexes presented above. The 5′-phosphate is anchored in the phosphate-binding pocket through intermolecular hydrogen bonds to the side chains of Arg780 and His982 (Figure 5B), while nearby Arg778 and Arg811 contribute to the basic patch.
Constraints Governing Positioning of 5′-Phosphate into the Phosphate Pocket
We next inquired into the minimal number of base pairs at the end of the duplex that need to be disrupted so as to facilitate positioning of the 5′-phosphate into the phosphate-binding pocket. To address this question, we studied complexes with self-complementary 13-mer and 15-mer RNAs containing p-U(−1) and p-U(−3)-U(−2)-U(−1) 5′-ends, while retaining the same U(+1)-U(+2) 3′-ends.
We solved the 2.60 Å structure of the complex containing 13-mer with 5′-p-U(−1) and U(+1)-U(+2)-3′ ends (Figure 5D; x-ray statistics listed in Table S2). It is readily apparent that the 5′-phosphate in the case of a 5′-overhang containing a single U is directed towards but unable to directly reach into the arginine-rich basic phosphate pocket (Figure 5D). We do observe unassigned electron density (blue mesh in Figure 5D; could be an additive used to facilitate crystallization) between the 5′-phosphate and the basic amino acids lining the phosphate pocket.
We have also solved the 2.55 Å structure of the complex containing 15-mer with 5′-p-U(−3)-U(−2)-U(−1) and U(+1)-U(+2)-3′ ends (Figure 4E; x-ray statistics listed in Table S2). Here, a portion of the 5′-overhang loops out prior to insertion of the 5′-phosphate into the phosphate pocket (Figure 5E). The U(−1) base is disordered in the complex, while the 5′-phosphate is anchored through hydrogen-bonding to the side chains of Arg780 and His982 (Figure 5E).
Conformational Features of hDicer PAZ Cassette-siRNA Complex Containing Melted/Disordered ‘hDicer-Specific’ Segment
Using the high-resolution structure of hDicer PAZ cassette-siRNA 12-mer complex (space group: C2; Table S2), the 3.4 Å structure of hDicer PAZ cassette-siRNA 16-mer complex (space group: I4122; Table S1) was refined to Rwork of 27.8 and Rfree of 31.1 (Table S1). While the bound 16-mer siRNA (Figure 6A), PAZ domain and connector helix can be traced with reasonable confidence, loop regions of the platform domain are poorly defined in this complex (Figure 6B). The composite omit map of the protein and siRNA in the complex are plotted at the 1σ level in Figure 6C, with those for the bound siRNA in the complex are plotted at the 1σ level in stereo in Figure S2B.
Figure 6. Structural Details of hDicer PAZ Cassette Bound to 16-mer siRNA and Impact of Phosphate-Pocket Mutants in the hDicer PAZ Cassette on Binding Affinity to a siRNA duplex.
(A) Sequence of the self-complementary 16-mer siRNA containing 5′-p and UU-overhangs at 3′-ends.
(B) Structure of hDicer PAZ cassette bound to a 16-mer siRNA. The platform, PAZ and connector helix are colored in yellow, blue and pink, respectively. The ‘Dicer PAZ-insertion element’ is disordered in this complex. The 5′-phosphate is highlighted in a space-filling representation.
(C) Omit map (1σ) of the protein and RNA in the hDicer PAZ cassette-16-mer siRNA complex.
(D) Sequence of immobilized RNA used for SPR measurements containing accessible 5′-p, UU 3′-overhang and terminal U•U mismatch at one end and biotin attached to the 3′-overhang at the other end.
(E) Measurement of SPR-based RNA-binding affinities for hDicer PAZ cassette mutants located within the phosphate pocket.
See also Table S1.
Surprisingly, the hDicer-specific-segment, that forms the ‘hDicer-specific helix’ in the structure of hDicer PAZ cassette-siRNA 12-mer complex (Figure 1D) is melted/disordered (spanning residues 1005-1019) in the hDicer PAZ cassette-siRNA 16-mer complex (Figure 6B, dotted red line). The bound siRNA is positioned towards the surface of the hDicer PAZ cassette (approximate angle of 30°) and would clash with the ‘Dicer-specific-helix’, were it to be present.
In the structure of hDicer PAZ cassette-siRNA 16-mer complex, the 2-nt 3′-overhang is positioned in the 3′-pocket, while the 5′-phosphate is closer to the phosphate pocket (side chains of Arg778 and Arg811 cannot be traced due to disorder) but does not directly coordinate to its basic residues in this complex, at least for this case where the siRNA contains a stable terminus with G-C pairs (Figure 6A). Our previous biochemical study showed that Dicer cannot recognize the 5′ terminal phosphate if the terminus is stable with a G-C pair (Park et al., 2011), presumably since disruption of base-pairing could allow the terminal phosphate to better reach into the 5′-pocket. Despite sustained systematic efforts, we were unable to grow diffraction quality crystals of hDicer PAZ cassette (including length changes at the N- and C-terminal ends) bound to self-complementary siRNAs of varying lengths (10- to 26-nt) containing unstable terminal base pairs (A•U pairs, and U•U and C•C mismatches).
Impact on Binding of Phosphate Pocket Mutants
In an earlier section we demonstrated that basic residues Arg778, Arg780 and Arg811 and polar His982 lined the walls of the phosphate pocket. To test whether the basic phosphate pocket is indeed required for RNA binding, we have undertaken SPR-based binding studies on the hDicer PAZ cassette-siRNA complex. One accessible end of the siRNA contained 5′-p, UU-3′ ends and a terminal U•U mismatch, while the other end was biotin labeled to the 3′-overhang segment (Figure 6D). SPR-based binding studies establish that single H982A and dual R778A/R780A mutants result in 5.9-fold and 3.6-fold loss in affinity, with much weaker binding for triple R778A/R780A/H982A (22-fold) and quadruple R778A/R780A/H982A/R811A mutants (21-fold) (Figure 6E). This implies that intermolecular contacts involving recognition of the 5′-phosphate by basic side chains lining the phosphate pocket contribute significantly to binding affinity. Notably, when we introduced the same mutations to full-length hDicer, the protein could no longer utilize the 5′ end of RNA to determine the cleavage site (Park et al., 2011). Taken together, our data indicate that the residues in the phosphate pocket are responsible for the recognition of the 5′ terminal phosphate of the siRNA.
DISCUSSION
Choice of hDicer Cassette for Complex Formation with siRNAs
Dicer serves in two key steps during small RNA biogenesis. It initially cleaves long dsRNAs (or pre-miRNAs) into small RNA duplexes and subsequently participates in RNA loading and strand selection within the RISC-loading complex. Biochemical studies support a model whereby hDicer uses distinct RNA-binding sites for dsRNA processing and for RISC loading (Noland et al., 2011). The model can be tested at the molecular level provided structural information becomes available. To date, crystallographic structural information has been restricted to G. intestinalis Dicer in the free state (Macrae et al., 2006), and as yet no crystal structures are available for Dicer or its domains in the RNA-bound state. Our structural efforts have focused on human Dicer, and given its large size, been restricted to RNA complexes with constructs containing the PAZ domain and its adjacent structured linker platform and connector helix elements. To that end, our research has focused on siRNA complexes with the hDicer domain composed of platform, PAZ and connector helix, culminating in the structure determination of the hDicer PAZ cassette bound to a range of siRNAs and dsRNAs.
Comparison of G. intestinalis and Human Dicer PAZ cassettes
The relative positioning of the platform, PAZ and connector helix domains are approximately the same in the structure of the hDicer PAZ cassette-siRNA 12-mer complex reported in this study and in the structure of G. intestinalis Dicer in the free state reported previously (Macrae et al., 2006) (see overlay in Figure S3). One notable difference is the formation of a knob-like α-helix (Figure 1D, E and Figure S3), associated with the higher eukaryotic ‘hDicer-specific insert’ (Figure 1B). The helix protrudes from the surface of the hDicer PAZ cassette and separates the 3′-pocket and the newly identified phosphate-binding pocket (Figure 3 and 4).
Phosphate Pocket Positioned within the Platform Domain
A phosphate-binding pocket had not been previously considered for Dicer, since the existing paradigm had been that the PAZ domain of Dicer solely recognized the 2-nt overhang at the 3′-ends of RNA duplex (Macrae et al., 2006; Zhang et al., 2004). The first insights into the potential contribution of 5′-phosphate recognition emerged following observation of unexpected cleavage patterns for processing of pre-miRNAs containing thermodynamically unstable duplex ends (Park et al., 2011).
In the current study, we have identified a common binding pocket for sulfate (Figure 4A) and phosphate (Figure 4B) in crystals grown from sulfate and phosphate-containing buffers, respectively. This basic pocket located within the platform domain, is lined by three Arg and one His residue and is positioned on the same face as the 3′-pocket (Figure 5B, C).
Plasticity of 3′-Pocket within the PAZ Domain in hDicer PAZ Cassette
The 2-nt overhang at the 3′-end of the siRNA is positioned in the PAZ domain and anchored in place by hydrogen bonds primarily to Tyr hydroxyl groups (Figure 2E), similar to what was reported previously for hAgo PAZ-siRNA (Figure S4) (Ma et al., 2004) and PAZ-ssRNA (Lingel et al., 2004) complexes. The hydrogen-bonding network is more extensive in the hDicer PAZ-siRNA complex (Figure 2E) compared to the hAgo PAZ complex (Figure S4). In addition, the overhang UU bases at the 3′-end are less well-stacked in the hDicer PAZ-siRNA complex (Figure 2E) compared with the stacked alignment observed for the hAgo PAZ-siRNA complex (Ma et al., 2004). The duplex-single strand junctions also adopt distinct conformations in the two complexes.
The 3′-pocket exhibits plasticity since it is able to accommodate both UU and UA 2-nt overhangs at the 3′-ends, with the bulkier purine at the 3′-terminus accommodated through a slight expansion in the dimensions of the pocket (Figure S5).
Recognition of the 2-nt 3′-end by the PAZ domain in the hDicer PAZ cassette-siRNA 12-mer complex contributes to complex formation since mutations of multiple Tyr residues lining the 3′-pocket result in dramatic loss in binding affinity (Figure 2F), similar to results of in vitro cleavage assays with the same mutants in intact Dicer (Park et al., 2011).
Conformation of hDicer PAZ Cassette-siRNA Complex Indicative of a Cleavage-Competent Step
In the absence of a structure of G. intestinalis Dicer bound to dsRNA, a model has been proposed where one end of the dsRNA is anchored in the PAZ domain, with the double helix aligned along a flat surface of a compact hatchet-like architecture, extending towards the pair of RNase III composite pockets, thereby accounting for formation of a fixed-length siRNA product following cleavage (Macrae et al., 2006). This model, where the bound dsRNA is aligned approximately in parallel to the long axis of the platform-PAZ-connector helix cassette, has been validated from biochemical experiments (MacRae et al., 2007).
Recently, the Narry Kim group proposed and biochemically validated the concept that mammalian Dicers utilize a combination of 5′- and 3′-end counting mechanisms for site-specific cleavage of pre-miRNAs, with the former contribution enhanced for thermodynamically less stable duplex ends (Park et al., 2011). Such a concept required the existence of a 5′-phosphate binding pocket. Here we have identified such a phosphate-binding pocket within the platform domain (Figure 4). Indeed, the earlier research from the Kim laboratory established that mutations in the phosphate pocket reduced processing efficiency and altered cleavage sites in vitro, while miRNA biogenesis was perturbed in vivo when Dicer-null embryonic stem cells are replenished with phosphate pocket mutants (Park et al., 2011).
Equally importantly, the phosphate and 3′-pockets are positioned adjacent to each other on the same face of the hDicer PAZ cassette in the structures of the siRNA complexes reported in this study. Thus, it is conceivable that the 5′-phosphate and 2-nt overhang of siRNA ends can be simultaneously accommodated in the phosphate and 3′ pockets, respectively. In fact, the two pockets are separated by approximately 20 Å, which corresponds to the distance between the 5′ and 3′ ends when the RNA contains a 2-nt 3′ overhang. This may provide the molecular basis for the strong preference of hDicer for dsRNAs and pre-miRNAs with a 5′ terminal phosphate and a 2-nt 3′ overhang (Park et al., 2011).
We propose that our structure of the hDicer PAZ cassette-siRNA 16-mer complex (Figure 6B), where the Dicer-specific segment is disordered/melted, likely reflects the cleavage-competent conformation. For this conformation of the complex, the 2-nt 3′-overhang is positioned in the 3′-pocket, while the 5′-phosphate is positioned close to the phosphate-pocket. The 5′ end is not inserted into the phosphate-pocket in this structure most likely because the siRNA 16-mer that yielded crystals of the complex (Figure 6A) contains stable terminal G-C base pairs, rather than terminal mismatch pairs known to be required for engagement of the 5′-counting rule (Park et al., 2011).
The cleavage-competent conformation where the Dicer-specific helix is disrupted and the siRNA is aligned parallel to the long axis of Dicer, may require additional intermolecular contacts with the pair of RNase III domains and perhaps the helicase domain present in intact Dicer, so as to stabilize the complex. To this end, it has been recently shown that constructs composed of hDicer RNase IIIA and IIIB domains in the absence and presence of the C-terminal RNA-binding domain (RBD) can be expressed and purified as homogeneous and stable entities (Ma et al., 2012), providing opportunities in the future for attempts at crystallization of complexes of hDicer PAZ cassette, hDicer RNase IIIA/IIIB and bound siRNA.
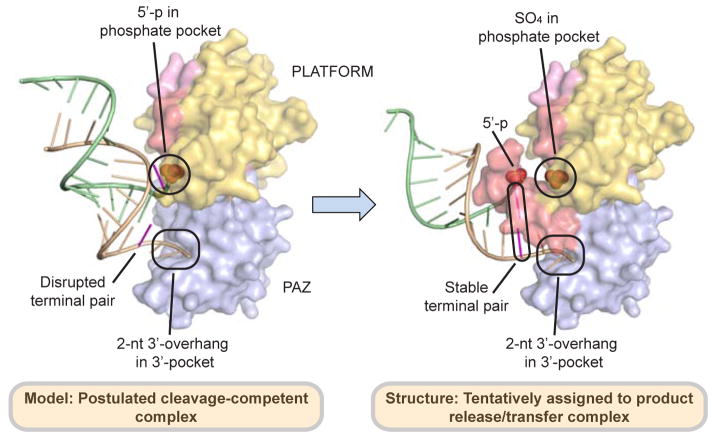
Proposed Transition from Cleavage-Competent Conformation to Postulated Product Release/Transfer Conformation
Biochemical studies indicated that siRNA undergoes a significant repositioning in Dicer complexes following the cleavage step, in the process biasing their orientation for guide strand selection based on the thermodynamic stability of the ends of the duplex (Noland et al., 2011). This model was further supported by a recent cryo-EM study, which showed that Dicer-RNA complexes can be in two different conformations, one representing cleavage-competent state and another indicative of loading intermediate or inactive state (Taylor et al., 2013). We postulate that the knob-like orientation of the hDicer-specific helix in hDicer PAZ cassette-siRNA 12-mer complex, wherein the bound siRNA is aligned at approximately 60° to the flat surface of the hDicer PAZ cassette, may represent the conformation associated with product release/transfer (Figure 7B), while the disruption of the hDicer-specific helix identified in the hDicer PAZ cassette-siRNA 16-mer complex, wherein the bound siRNA siRNA is aligned at an angle of approximately 30° with the flat surface of the hDicer PAZ cassette, most likely represents the conformation associated with the cleavage-competent step (Figure 7A). In such a concept, siRNA transfer following the cleavage step would be accompanied by formation of the Dicer-specific helix and release of the 5′-phosphate of the bound siRNA, while retaining the anchored 2-nt 3′-overhang in its pocket (Figure 7). Our in vivo data based on a Dicer rescue assay in Dicer knockout embryonic stem cells using ‘hDicer-specific helix’ mutants imply the ‘Dicer specific helix’ may not play active role for miRNA biogenesis. However, we cannot at this time rule out the possibility that our rescue assay is not sensitive enough to detect the defect in miRNA release, transfer or Ago loading step.
Figure 7. Proposed Alignments of hDicer PAZ Cassette and siRNA in Postulated Model of Cleavage-Competent and in the Structure Tentatively Assigned to the Product Release/Transfer Complexes.
(Left panel) The proposed features of the postulated model of the cleavage-competent complex are as follows: The Dicer-specific helix is disrupted, with the RNA duplex directed towards the protein. The 2-nt 3′-overhang is positioned in the 3′-pocket, with disruption of the terminal mismatch pair most likely facilitates positioning of the 5′-phosphate in the phosphate pocket. (Right panel) The unique features of the structure tentatively assigned to the product release/transfer complex are as follows: The Dicer-specific helix adopts a knob-like structure, thereby directing the RNA duplex away from the protein. The 2-nt 3′-overhang remains positioned in the 3′-pocket, while the 5′-phosphate is released from the phosphate pocket, which is now occupied by a sulfate/phosphate from the buffer.
A future challenge that will need to be addressed concerns how the structural characteristics observed in product release/transfer conformation contributes to the strand selection or effective loading of mature miRNA into the Ago complex.
EXPERIMENTAL PROCEEDURES
Please refer to the Supplemental Experimental Procedures for detailed methodology on protein expression and purification, RNA preparation, crystallization and data collection, structure determination, and surface plasmon resonance (SPR) binding assay.
Gene Cloning and Protein Preparation
The human Dicer “platform-PAZ-connector helix” cassette (residues 755-1,055) was cloned in pET-28b vector (EMD Biosciences) with a His6-SUMO tag at the N-terminus. All clones were overexpressed in E. coli strain BL21(DE3) cells. The proteins were first purified from the soluble fraction by a nickel-chelating affinity column, followed by SUMO protease Ulp1 treatment overnight at 4 °C to cleave the His6-SUMO tag, which was later removed via a second nickel-chelating column. Proteins were further purified by gel filtration chromatography.
Crystallization and Data Collection
The purified human Dicer ‘platform-PAZ-connector helix’ cassette (wild-type or surface lysine double mutant - K832A/K833A) was mixed with RNA in a molar ratio of 1:1.2 and incubated on ice for at least one hour. The complex crystals were grown using the hanging-drop vapor-diffusion method by mixing the protein-RNA complex with equal volume of reservoir solution. RNA sequences, which gave diffraction quality crystals, are listed in Table S5. The crystallization conditions for each protein-RNA complex are listed in Table S4. For data collection, crystals were flash frozen (100 °K) in the mother liquor supplemented with 30% (v/v) glycerol as cryo-protectant.
Structural Determination
The crystal structure of human Dicer PAZ cassette (residues 755-1055) in complex with self-complementary 16-mer siRNA containing a 5′-phosphate and a 2-nt overhang at 3′-end was solved by SAD method using data collected at Se-peak wavelength to a resolution of 3.4 Å in space group I4122 (see Supplementary Experimental Procedures for more details). In this structure, we observed relatively poor electron density for segments of the platform domain compared to the PAZ domain and bound RNA. Because of limited resolution and relatively poor electron density, it was difficult to assign amino acids unambiguously for the platform domain. Careful analysis of the data suggested no sign of twinning. Instead, poor crystal packing interactions for the platform domain and overall high B-factor of the data set, as well as anisotropic diffraction, appear to be responsible for the poor/noisy electron density for segments of the platform domain. Later, using the high resolution structure of hDicer PAZ cassette-siRNA 12-mer complex, amino acid sequence assignment was completed and the structure was further refined using Phenix.refine. The structure refinement was completed with cycles of minimization, group B-factor refinement along with TLS parameters, leading to a final structure with Rwork of 27.7% and Rfree of 31.7 %. Three loop regions and a few side-chain atoms for which no electron density was observed were omitted from the final model. The final map (2Fo–Fc) showed reasonably good quality electron density for the final model except for some segments in the platform domain, which has relatively poor density (see Table S1 for crystallographic statistics).
For improving diffraction quality of crystals, we made three different double mutants based on crystal contacts observed in the partially refined structure (see above), as well as predictions made by surface entropy reduction server (http://services.mbi.ucla.edu/SER/). One such mutant in which two surface lysines (Lys832 and Lys833) were mutated to Ala gave a crystal in complex with 12-mer siRNA having 5′-phosphate and 2-nt overhang at 3′-end which diffracted to a resolution of 2.25 Å. These two lysine residues are very far from the RNA binding site (Figure S3). This structure was solved by PHASER (McCoy et al., 2007) using protein atoms of partially refined low resolution structure of hDicer PAZ cassette-siRNA 16-mer complex as a search model. The initial model was further improved by manual and automated model building as well as refinement using the PHENIX program package (Adams et al., 2002; Afonine et al., 2005). This structure has clear and well interpretable electron density map for protein and RNA. All other crystal structures of hDicer PAZ cassette (wild-type or K832A/K833A) in complex with siRNAs having different overhangs at 5′- and 3′-ends (Table S5) discussed in this paper were solved by molecular replacement or by difference Fourier using protein atoms of the refined structure of human Dicer PAZ cassette (K832A/K833A) in complex with 12-mer siRNA having 5′-phosphate and 2-nt UU overhang at 3′-end (see Supplementary Experimental Procedures for more details). All crystallographic data and refinement statistics are summarized in Tables S1, S2 and S3.
Surface Plasmon Resonance (SPR) Binding Assay
RNA binding experiments were performed on BIAcore 2000 biosensor (BIAcore) at 25 °C (Katsamba et al., 2002). The RNA sequences used in this assay were biotinylated at the 3′-end on one strand of the duplex RNA (Figure S7). For detailed information, please see Supplementary Experimental Procedures.
Dicer rescue experiments
For transfection, Dicer knockout mouse ES cells were separated from feeder cells and 1,500,000 cells were seeded on gelatin-coated 6-well plates one day before transfection. 20~30 micrograms of plasmids were added to each well along with 10 μl of Lipofectamine 2000, according to the manufacturer’s protocol (Invitrogen). Protein and RNA was extracted at 48 h after transfection. To determine the protein levels, western blotting was performed using anti-Dicer and anti-GAPDH (Santa Cruz) antibodies. Expression of RNA was confirmed by Taqman MicroRNA Assays (Life Technologies).
PDB Accession Codes
The atomic coordinates and structure factors of the various complexes have been deposited in the Protein Data Bank with the following accession codes: Complexes containing hDicer PAZ cassette bound to 12-mer siRNA (Figure S5A) with UU-3′-ends (4NGB), 12-mer siRNA (Figure 1C) with 5′-p and UU-3′-ends (4NGD), 12-mer RNA (Figure S5A) with UA-3′ ends (4NGC), 13-mer RNA (Figure 4B) with 5′-A and UU-3′ ends (4NGG), 13-mer RNA (Figure 5D) with 5′-pU and UU-3′-ends (4NH3), 14-mer RNA (Figure 5A) with 5′-pUU and UU-3′-eds (4NH5), 15-mer RNA (Figure 5E) with 5′-p-UUU and UU-3′-ends (4NH6), 16-mer (Figure 6A) with 5′-p and UU-3′-ends (4NHA) and 17-mer siRNA (Figure S6B) with 5′-p and UU-3′-ends (4NGF).
Supplementary Material
HIGHLIGHTS.
Two families of crystal structures of hDicer PAZ cassette-siRNAs complexes
Dicer-specific loop region forms a knob-like α-helical protrusion
Identification of a phosphate-binding pocket in the platform domain of hDicer
Models of cleavage-competent and product release/transfer conformations
Acknowledgments
We are grateful to the staff of X-29 beamline at the National Synchrotron Light Source, Brookhaven National Laboratory and ID-24-C/E beamline at the Advanced Photon Source, Argonne National Laboratory for their help with data collection. D.J.P. was supported by NIH grant AI068776, while V.N.K. was supported by the Creative Research Initiatives Program of Korea (20090063603).
Footnotes
Author’s contributions
Y.T. with inputs from D.K.S., and J.B.M. designed and performed experiments leading to crystallization of hDicer ‘platform-PAZ-connector helix’ cassette bound to siRNAs, while D.K.S. was primarily responsible for structure solution and refinements of all complexes, with the above research carried out under the overall guidance and supervision of D.J.P. In addition, V.N.K. and members of her group (J.E.P. and I.H.) shared the results of their biochemical cleavage assays on wild-type and mutant intact human Dicer, during the structural experiments leading to the identification of the phosphate-pocket within the hDicer PAZ cassette. D.J.P. and V.N.K. participated in writing the paper with contributions from members of both their groups.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD. CCP4 Newsl 2005
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nature structural & molecular biology. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Gene Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan JH, Tropea JE, Austin BP, Court DL, Waugh DS, Ji XH. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO reports. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nature reviews Genetics. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JJ, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho YJ, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Katsamba PS, Park S, Laird-Offringa IA. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Current biology: CB. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nature structural & molecular biology. 2012;19:436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature structural & molecular biology. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. Journal of molecular biology. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA. Coordinated activities of human dicer domains in regulatory RNA processing. Journal of molecular biology. 2012;422:466–476. doi: 10.1016/j.jmb.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nature structural & molecular biology. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Molecular cell. 2011;43:110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Developmental biology. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HD, Webster RE, Zinder ND. Purification and properties of ribonuclease III from Escherichia coli. The Journal of biological chemistry. 1968;243:82–91. [PubMed] [Google Scholar]
- Taylor DW, Ma E, Shigematsu H, Cianfrocco MA, Noland CL, Nagayama K, Nogales E, Doudna JA, Wang HW. Substrate-specific structural rearrangements of human Dicer. Nature structural & molecular biology. 2013;20:662–670. doi: 10.1038/nsmb.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nature structural & molecular biology. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nature structural & molecular biology. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.