Abstract
The ydaO riboswitch, involved in sporulation, osmotic stress responses and cell wall metabolism, targets the second messenger c-di-AMP with subnanomolar affinity. We have solved the structure of c-di-AMP bound to the Thermoanaerobacter tengcongenesis ydaO riboswitch, thereby identifying a five-helical scaffold containing a zippered-up bubble, a pseudoknot and long-range tertiary base pairs. Highlights include the identification of two c-di-AMP binding pockets on the same face of the riboswitch, related by pseudo two-fold symmetry, with potential for cross-talk between sites mediated by adjacently-aligned base stacking alignments connecting pockets. The adenine rings of bound c-di-AMP molecules are wedged between bases and stabilized by stacking, base-sugar and sugar-sugar intermolecular hydrogen bonding interactions. The structural studies are complemented by ITC-based binding studies of mutants mediating key tertiary intermolecular contacts. The T. tengcongenesis ydaO riboswitch, like its B. subtilis counterpart, likely functions through a transcription termination mechanism, with the c-di-AMP bound state representing an ‘off’ switch.
A paradigm shift in our understanding of RNA-mediated gene regulation occurred with the independent discovery of metabolite-sensing RNAs that bound small ligands with high selectivity and discriminated against closely related analogs1,2. The fundamental elements of the riboswitch scaffold include a metabolite-sensing domain and a neighboring expression platform, with ligand-induced conformational changes in the sensing domain propagated to the expression platform, thereby regulating either transcription or translational control, and in some cases, ribozyme-like cleavage3-5. It remains quite remarkable that RNA containing only four nucleotides and bound Mg cations exhibits the capacity to generate exquisite pockets that recognize nucleotides, amino acids, and metabolites, as well as both cations and anions, and discriminates against closely related analogs. Structural biology has contributed significantly to our understanding of the higher order RNA architectures and intermolecular recognition elements within riboswitches that contribute to the selectivity of ligand recognition6-8.
A notable extension of riboswitch research to second messengers involved in signaling pathways emerged with the discovery that c-di-GMP, containing a pair of 3′,5′-internucleotide linkages, was targeted with picomolar affinity and 1:1 stoichiometry by the sensing domains of two distinct families of riboswitches9,10. c-di-GMP is an important second messenger molecule critical for the survival of many bacterial species with roles in biofilm formation and in control of virulence11,12. The c-di-GMP-I13,14 and c-di-GMP-II15 riboswitches adopt distinct junctional folds and ligand binding pockets. It was also shown that c-di-GMP containing 2′-OCH3 modifications could discriminate between type I and II riboswitches15, and that appropriate modifications could convert the c-di-GMP-binding riboswitch to its c-di-AMP-binding counterpart16.
Recently, attention has turned to the ydaO RNA motif initially categorized as an orphan riboswitch associated with genes that control sporulation, cell wall metabolism and osmotic stress17,18. Though initial efforts claimed to have identified ATP as the ligand19, more recent efforts definitively identified c-di-AMP containing a pair of 3′,5′ linkages as the ligand20 for the ydaO family of riboswitches. c-di-AMP is a recently identified second messenger21 impacting on the control of cell size and envelope stress22,23. More recent studies have evaluated the interplay between the c-di-GMP and c-di-AMP signaling pathways24. In-line probing experiments indicated that the c-di-AMP bound the ydaO riboswitch with 1:1 stoichiometry and with approx. 0.7 nM affinity20. Given that the proposed secondary fold of the sensing domain of the c-di-AMP riboswitch20 was completely distinct from both type I and II c-di-GMP riboswitches13-15, we set out to structurally define the tertiary architecture and ligand-binding pocket of the c-di-AMP riboswitch in the bound state, as well as identify the intermolecular contacts that could account for the recognition of the c-di-AMP ydaO riboswitch and discrimination against its c-di-GMP counterpart20 and related analogs. Structure-guided mutations could then identify tertiary interactions responsible for the structural integrity of the c-di-AMP bound ydaO riboswitch.
Our structural, binding and mutation studies on the sensing domain of the c-di-AMP riboswitch have identified novel RNA tertiary structures and binding pocket architectures, new ligand-RNA recognition principles, unanticipated ligand stoichiometry and the role of symmetry in defining ydaO riboswitch scaffolds and binding pocket architectures.
RESULTS
Riboswitch species selection and construct design
The sequence and secondary fold of the sensing domain of the B. subtilis ydaO riboswitch as deciphered previously20 is depicted schematically in Supplementary Results, Supplementary Fig. 1a. It is composed of seven helical segments, internal bubbles and one predicted pseudoknot segment. Our structural efforts have focused on the sensing domain of the ydaO riboswitch from Thermoanaerobacter tengcongenesis whose sequence and proposed secondary fold is shown in Supplementary Fig. 1b. We replaced non-GNRA hairpin loops (shown not to be involved in c-di-AMP recognition by in-line probing20) by their GAAA counterparts and also incorporated a 5′-GG step to facilitate in vitro transcription, with the sequence used for crystallization trials shown in Supplementary Figure 1c.
Crystal Structure of the c-di-AMP Riboswitch
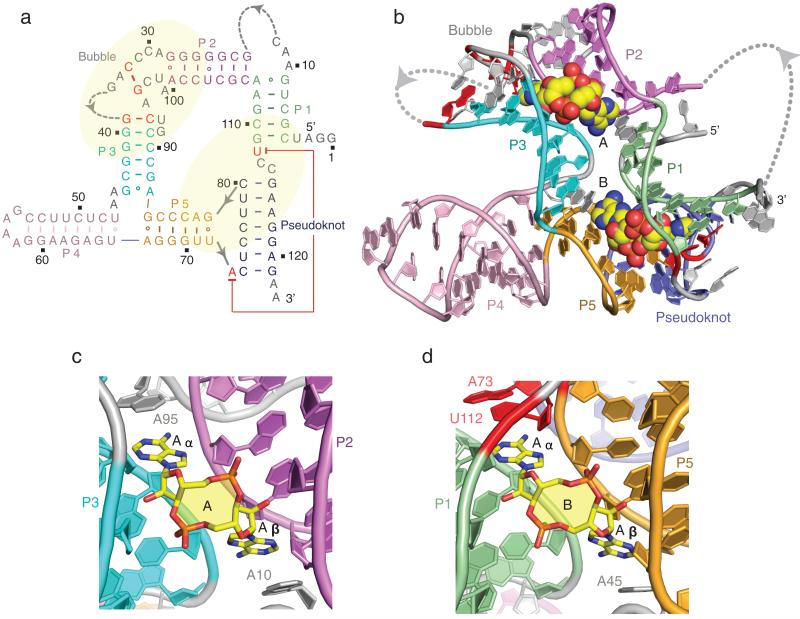
We grew 2.73 Å crystals of the sensing domain of the T. tengcongenesis ydaO riboswitch with bound c-di-AMP and determined the structure using iridium-hexamine to solve the phase problem (x-ray statistics in Supplementary Table 1). The observed secondary fold of the riboswitch deduced from the structure of the complex is shown in Supplementary Fig. 1d and redrawn as Fig. 1a. The complex of the sensing domain of the ydaO riboswitch with bound c-di-AMP contains five helices labeled P1 to P5, a partially zippered-up bubble, a pseudoknot, long-range tertiary pairs and two molecules of bound c-di-AMP (Fig. 1b and an alternate view rotated by 180 degrees in Supplementary Fig. 2a). All stems except P4 were elongated by a sheared G·A non-canonical base pair (Supplementary Fig. 1d and 2b-e). Segments that could not be traced in the electron density map include nucleotides (nts) 13-21 and 35-38 that are part of bubble or loop segments (Fig. 1a). The two molecules of bound c-di-AMP, labeled A and B (Fig. 1c, d), could be readily identified in omit maps shown in Supplementary Fig. 3a, b. The stoichiometry of two c-di-AMP molecules per ydaO riboswitch was unexpected, and even more with the identification that the binding pockets were related by pseudo-two fold symmetry (Fig. 1b), a feature neither apparent nor anticipated from the secondary fold (Fig. 1a). Thus, c-di-AMP molecule A is positioned in a binding pocket bracketed by helices P2 and P3 and capped by the partially zippered-up bubble segment (Supplementary Fig. 4a), while c-di-AMP molecule B is positioned in a binding pocket bracketed by helices P1 and P5 and capped by the helical pseudoknot segment (Supplementary Fig. 4b).
Figure 1. Secondary and tertiary structure of the T. tengcongensis ydaO riboswitch bound to c-di-AMP and alignment of c-di-AMP molecules A and B in their respective binding pockets.
a, The secondary fold of the sensing domain of the ydaO riboswitch. The stem segments are color-coded and labeled from P1 to P5, with additional labels marking segments containing a partially zippered-up bubble and a pseudoknot. The two c-di-AMP binding pockets identified from the crystal structure of the complex are marked in yellow background. b, The 2.73 Å crystal structure of the sensing domain of the ydaO riboswitch with two bound c-di-AMP molecules labeled A and B in space-filling representation. c, c-di-AMP molecule A (in yellow) bound within its pocket composed of stems P2 (in purple) and P3 (in cyan) and bubble (in grey) segments in the ydaO riboswitch complex. d, c-di-AMP molecule B (in yellow) bound within its pocket composed of stems P1 (in green) and P5 (in orange) and pseudoknot duplex (in blue) segments.
Both adenine rings of the bound c-di-AMP in an open, spread out conformation, insert into pockets within the riboswitch scaffold (Supplementary Fig. 5a, b), with both binding sites located on the same face and separated by a channel generated by side-by side arrangement of stacked bases connecting the sites (Supplementary Fig. 5c). It appears that the bound c-di-AMP molecules could stabilize the fold of both the partially zippered-up bubble segment and the pseudoknot, though in the absence of a structure of the free riboswitch, this conclusion requires validation.
Partially zippered-up bubble formation
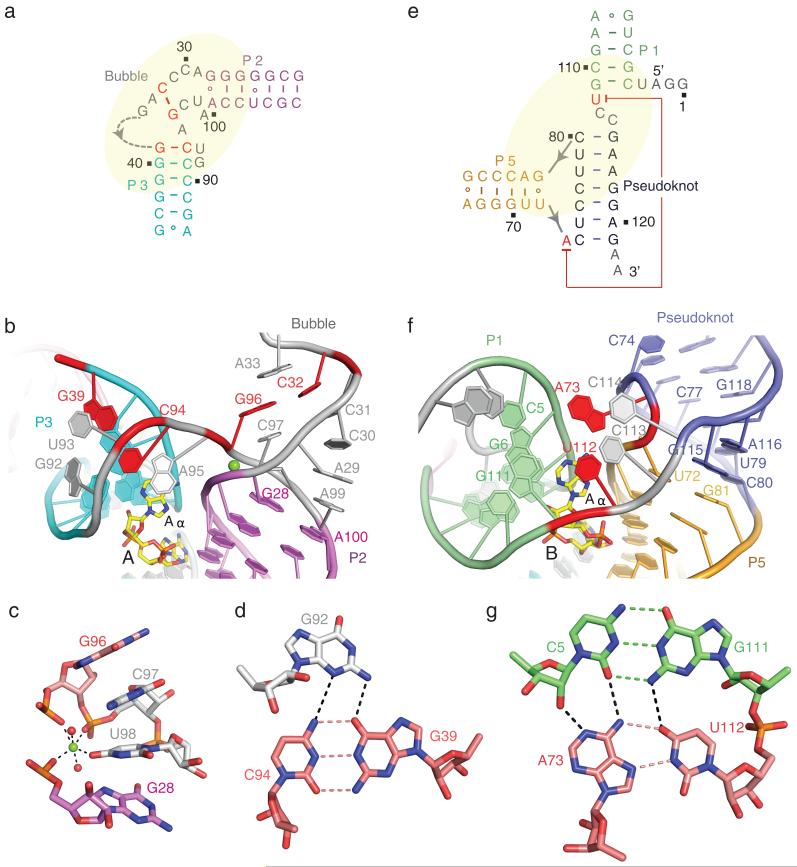
The partially zippered-up bubble and pseudoknot segments represent two novel features of the structure of the c-di-AMP bound riboswitch. The schematic and overall architecture of the bubble (in grey) and flanking helical stems P2 (in purple) and P3 (in cyan) is shown in Fig. 2a and 2b, respectively. This fold is stabilized in part by a bound Mg2+ cation (Fig. 2c; validated by monitoring the anomalous diffraction of crystals of the complex soaked in divalent Mn2+; Supplementary Fig. 6a), with the coordination geometry and bond lengths of the Mg2+ cation shown in Supplementary Fig. 6b. Replacement of U98, involved in direct coordination to the Mg2+, by guanine, disrupted c-di-AMP binding as monitored by in-line cleavage assays20. Isothermal titration calorimentry (ITC) binding curves for complex formation of the ydaO riboswitch with c-di-AMP as a function of Mg2+ concentration are plotted in Supplementary Fig. 7a), while the corresponding curves at 5 mM concentrations of Mg2+, Mn2+, Ba2+ and Ca2+ are compared in Supplementary Fig. 7b. The fold of the partially zippered-up bubble is further stapled together through formation of a G39(anti)-C94(syn) base pair (in red, Fig. 2a, b; positions 39 and 94 were protected in in-line probing experiments in the presence of c-di-AMP20) as part of a major groove triple with G92 (Fig. 2d; see bond distances in Supplementary Fig. 8a), which in turn stacks with the terminal base pair of stem P3 (Fig. 2a and 2b). The intervening base U93 forms a base-sugar interaction with G92 and stacks with base pair G39-C94 (Supplementary Fig. 9a). Base A95 participates in an A minor base triple interaction with a G-C base pair from stem P3 (Supplementary Fig. 9a, b). We also observed cross-strand stacking between unpaired bases A29 and A99 (Fig. 2b), while unpaired bases A33 and C97 stack with adjacent bases and sandwich the Watson-Crick C32(anti)-G96(anti) pair (in red, Fig. 2a, 2b; Supplementary Fig. 9c) further stapling the bubble segment.
Figure 2. Folding topologies adopted by the bubble and pseudoknot segments and their flanking stem segments in the structure of T. tengcongensis ydaO riboswitch bound to c-di-AMP.
a, b, Schematic (panel a) and folding topology (panel b) of the bubble segment (in grey) relative to flanking stems P2 (in purple) and P3 (in cyan) in the ydaO riboswitch complex. Key Watson-Crick pairs C32-G96 and G39-C94 are shown in red. The bound Mg2+ cation is shown as a green-colored ball. c, Octahedral coordination geometry of the Mg2+ cation (in green), with bound water molecules represented by red-colored balls. d, Pairing alignments of major groove-aligned G92•(G39-C94) base triple. Note that C94 adopts a syn alignment. e, f, Schematic (panel e) and folding topology (panel f) of the pseudoknot duplex segment (in blue) relative to flanking stems P1 (in green) and P5 (in orange) in the ydaO riboswitch complex. g, Pairing alignment within a tetrad formed by the long range Hoogsteen A73•U112 pair aligning with the minor groove edge of the Watson-Crick C5-G111 pair.
Pseudoknot formation
The schematic and overall architecture of the helical pseudoknot segment (in blue) and flanking helical stems P1 (in green) and P5 (in orange) is shown in Fig. 2e and 2f, respectively. A seven-base pair pseudoknot is formed by Watson-Crick pairing of nts 74-80 with nts 115-121 (Fig. 2e and 2f), as predicted previously from the secondary structure analysis of the ydaO riboswitch20 (Supplementary Fig. 1a, c). This architecture is stapled through formation of a critical long-range Hoogsteen A73•U112 base pair (Fig. 2e and 2g; see bond distances in Supplementary Fig. 8b; conserved positions 73 and 112 were protected in in-line probing experiments in the presence of c-di-AMP20), which participates in tetrad formation through alignment with the minor groove edge of a Watson-Crick G-C pair (Fig. 2g; the G at position 111 of this G-C base pair was protected in in-line probing experiments in the presence of c-di-AMP20). Further, U112 forms a continuous stack with unpaired bases C113 and C114 (Fig. 2f). Notably, the pseudoknot derived stem (C80-G115 pair) stacks continuously with one terminus (U72•G81 pair) of stem P5 at the junctional site (Fig. 2f). The other terminus (A67•G86 pair) of stem P5 in turn stacks with stem P4 (U47•U66 non-canonical pair) (Fig. 1a; Supplementary Fig. 9d).
c-di-AMP binding pockets and ligand recognition
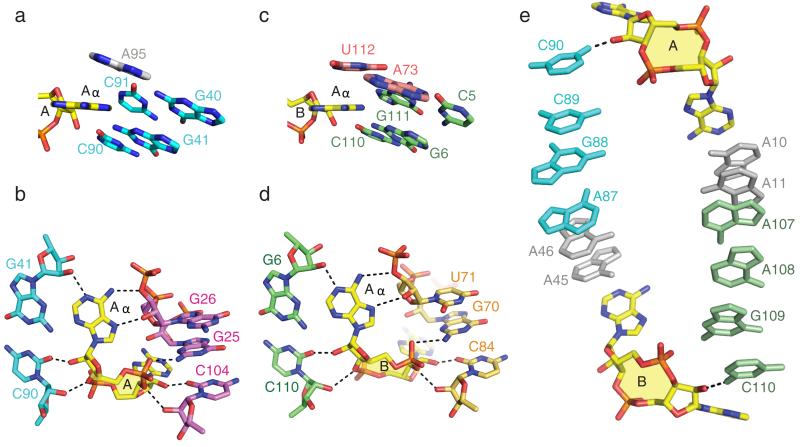
We next turn our attention to the binding pockets for c-di-AMP molecules A and B in the structure of the ydaO riboswitch complex. The overall architecture of the pocket for c-di-AMP molecule A generated by stems P2, P3 and partially zippered-up bubble segment (Supplementary Fig. 4a) is shown in Fig. 1c and Supplementary Fig. 10a. The bases Aα and Aβ of c-di-AMP are stacked on and bracketed by bases A95 and A10, respectively. Both Aα and Aβ of c-di-AMP molecule A are wedged between bases of the RNA (Fig. 3a for Aα and Supplementary Fig. 11a for Aβ) and their base and cyclic dinucleotide backbone form similar patterns of intermolecular hydrogen bonds with the sugar-phosphate backbone (Fig. 3b for Aα and Supplementary Fig. 11b for Aβ). There is also a potential for non-planar hydrogen bonds between the N3 nitrogen of A and the 2-amino groups of guanine residues (not drawn in Fig. 3b and Supplementary Fig. 11b). The lengths of the intermolecular hydrogen bonds involved in c-di-AMP molecular recognition are summarized in Supplementary Fig. 12. Thus, the intermolecular recognition features of Aα and Aβ of bound c-di-AMP molecule A, both of which adopt a C3′-endo sugar pucker, are related by pseudo two-fold symmetry.
Figure 3. Similarities in recognition features between Aα rings of c-di-AMP molecules A and B bound within their respective binding pockets and a pair of parallel base stacking alignments connecting binding pockets in the structure of T. tengcongensis ydaO riboswitch bound to c-di-AMP.
a, Wedge-shaped insertion of the Aα ring of c-di-AMP molecule A (in yellow) between bases of stem P3 (in cyan), with the Aα ring stacking on A95. b, Intermolecular hydrogen bonds between the Aα ring of c-di-AMP molecule A (in yellow) and the sugar rings of stems P2 (in purple) and P3 (in cyan). c, Wedge-shaped insertion of the Aα ring of c-di-AMP molecule B (in yellow) between bases of stem P1 (in green), with the Aα ring stacking on U112. d, Intermolecular hydrogen bonds between the Aα ring of c-di-AMP molecule B (in yellow) and the sugar rings of stems P1 (in green) and P5 (in orange). e, Pair of parallel base stacking alignments connecting c-di-AMP pockets A and B in the complex.
Strikingly, the same recognition patterns are observed in the overall architecture of the pocket for c-di-AMP molecule B generated by stems P1, P5 and the helical pseudoknot segment (Supplementary Fig. 4b) as shown in Fig. 1d and Supplementary Fig. 10b. The bases Aα and Aβ are stacked on and bracketed by bases U112 and A45, respectively. The wedging and intermolecular recognition features for Aα are shown in Fig. 3c, d and for Aβ in Supplementary Fig. 11c, d. Here again, the intermolecular recognition features of Aα and Aβ of bound c-di-AMP molecule B are related by pseudo two-fold symmetry.
We note that the two c-di-AMP pockets are connected by a side-by-side arrangement of stacked bases associated with strand (silver) and helical (color-coded) segments as depicted in Fig. 3e. An adenine ring of c-di-AMP stacks with a base at one end of the stacked array and the hydroxyl of a sugar ring of c-di-AMP forms a hydrogen bond with the base at the other end of the stacked array (Fig. 3e).
Pseudo two-fold symmetry
More importantly, despite the pocket for c-di-AMP molecule A being capped by a partially zippered-up bubble element (Supplementary Fig. 4a) and that for c-di-AMP molecule pocket B being capped by a pseudoknot (Supplementary Fig. 4b), the intermolecular contacts defining these pockets are also related by pseudo two-fold symmetry as outlined above (Fig. 1c, d). Identification of such elements of symmetry within a binding pocket and between binding pockets was completely unanticipated, as was formation of two c-di-AMP molecules bound per sensing domain of the riboswitch.
Local symmetry has also been identified in the central region between the two c-di-AMP binding sites of the ydaO riboswitch (Supplementary Fig. 13a). The base A10 participates in an A-minor motif base triple with a G-C base pair from stem P2 (Supplementary Fig. 13b), while A11 is involved in sugar-base pair edge recognition with G-C base pair also from stem P2 (Supplementary Fig. 13c). A10 and A11 form a continuous stack with a sheared G•A non-canonical terminal pair from stem P1(Supplementary Fig. 13a). Accordingly, higher order pairings involve nts A45 (A-minor motif base triple with a G-C pair from stem P5; Supplementary Fig. 13d) and A46 (sugar-sugar interactions with a sheared G•A non-canonical terminal pair of stem P5; Supplementary Fig. 13e), which in turn stack with a sheared G•A non-canonical terminal pair from stem P3 (Supplementary Fig. 13a). These symmetrical interactions together bridge the two c-di-AMP binding pockets (Supplementary Fig. 13a).
Structure and ITC parameters for c-di-AMP analogs
We have also solved the 2.65 Å structure of c-di-dAMP (DNA analog lacking 2′-OH groups) bound to the T. tengcongenesis ydaO riboswitch (x-ray statistics listed in Supplementary Table 1), with two molecules of the c-di-dAMP bound in the same pockets as c-di-AMP. Given that c-di-AMP has 2′-OH groups, there are additional intermolecular hydrogen bonds to its cyclic dinucleotide scaffold (Supplementary Fig. 14a; red arrows indicate hydrogen bonds to 2′-OH groups of c-di-AMP) than to the c-di-dAMP scaffold (Supplementary Fig. 14b).
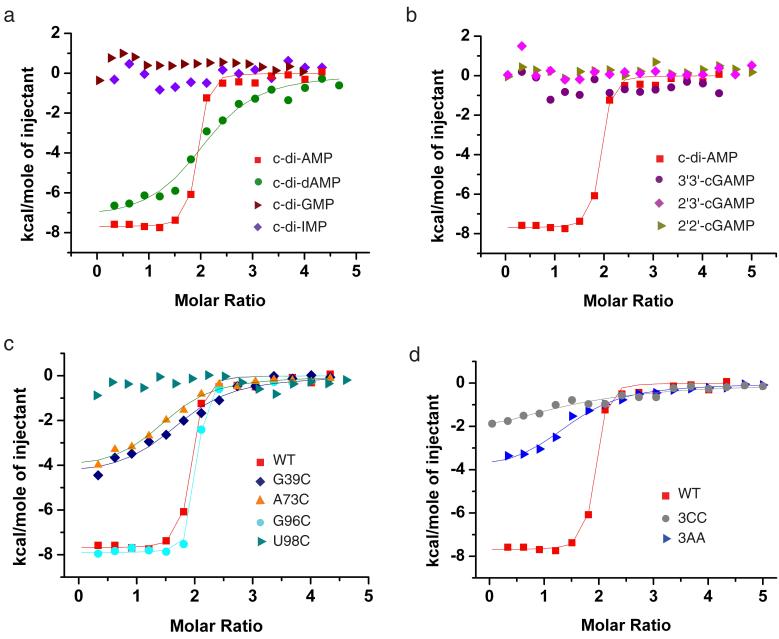
We have compared the binding of the T. tengcongenesis c-di-AMP ydaO riboswitch to c-di-AMP, c-di-dAMP, c-di-GMP and c-di-IMP by ITC (Fig. 4a). The ITC data were processed assuming that c-di-AMP binding to the two sites (displaying similar binding pocket architectures and intermolecular contacts) occurred with similar affinity and enthalpic change. Such a ‘one set of sites’ model yielded Kd = 0.066 M and ΔH = −7.7 Kcal/mol for c-di-AMP (reproducibility of the binding curves are depicted in Supplementary Fig. 15a), and a weaker Kd = 2.9 μM and ΔH = −7.4 Kcal/mol for c-di-dAMP. By contrast, the Kd values are too weak to monitor for c-di-GMP and c-di-IMP (Fig. 4a and Supplementary Table 2), a trend similar to that reported from in-line probing experiments20.
Figure 4. Comparison of ITC binding curves with other second messengers and analogs and mutants involved in tertiary pairing in the T. tengcongensis ydaO riboswitc.
a, ITC binding curves of ydaO riboswitch bound to c-di-AMP, c-di-dAMP, c-di-GMP and c-di-IMP. b, Comparative ITC binding curves for complex formation between the sensing domain of the ydaO riboswitch with c-di-AMP, and cGAMP linkage isomers containing both 2′,5′ linkages (designated 2′,2′-cGAMP), both 3′,5′ linkages (designated 3′,3′-cGAMP) and G(2′,5′)A/A(3′,5′)G linkages (designated 2′,3′-cGAMP). c, ITC binding curves for mutation G39C (disruption of G39-C94 pair), G96C (disruption of C32-G96 pair), A73C (disruption of A73•U112 pair) and U98C (disruption of Mg2+ coordination). d, A triple mutant where C39-G94, C32-G96 and A73•U112 were simultaneously replaced either by three C•C pairs (designated 3CC) or three A•A pairs (designated 3AA). In panels a, b, c and d, the cyclic dinucleotides were added gradually to the T. tengcongensis ydaO riboswitch and mutants.
We also investigated by ITC the binding of the T. tengcongenesis ydaO riboswitch to cGAMP linkage isomers containing both 3′,5′, both 2′,5′ and mixed GpA(2′,5′)/ApG(3′,5′) linkages25-28. The experimental data establish that the ydaO riboswtich does not bind any of the cGAMP linkage isomers (Fig. 4b).
Impact of mutating key tertiary pairs
There are two tertiary pairs, G39-C94 (Fig. 2a, b, d) and C32-G96 (Figure 2a, b and Supplementary Fig. 9c) associated with the partially zippered-up bubble segment and the conserved A73•U112 pair (Fig. 2e, f, g) associated with the pseudoknot segment, that appear to contribute to the stabilization of the global fold of the c-di-AMP bound riboswitch (Fig. 1a, pairs indicated in red). We have used ITC to monitor the impact of disrupting these tertiary contacts following single replacement of G39, G96 and A73 by C, as well as U98, mutated in the earlier in-line probing studies20, by C (Fig. 4c). The disruption of two of these tertiary interactions, resulted in a pronounced change from Kd = 0.066 μM and ΔH = − 7.7 kcal/mol for the wild-type ydaO riboswitch to Kd = approx. 3.0 μM and ΔH = approx. −4.4 Kcal/mol for both G39C and A73C mutant riboswitches (Fig. 4c; Supplementary Table 3). By contrast, replacing G96 by C in the C32-G96 pair associated with the partially zippered-up bubble segment had no impact on the thermodynamic parameters (Fig. 4c, Supplementary Table 3). Replacement of U98 resulted in complete loss in binding affinity (Fig. 4c).
We have also monitored by ITC, the impact of simultaneously replacing three tertiary pairs G39-C94, C32-G96 and A73•U112 by potential C•C (designated 3CC) or A•A (designated 3AA) pairs (Fig. 4d). We note that Kd is reduced to 4.5 μM for 3AA and further reduced to 19.8 μM for 3CC triple mutants (Supplementary Table 3).
Finally, we have monitored by ITC the impact of replacement of adenines A10 (Supplementary Fig. 13b), A45 (Supplementary Fig. 13d) and A95 (Supplementary Fig. 9b) involved in A-minor base triples by C (Supplementary Fig. 15c). The Kd values are reduced to 0.24 μM for A45C and 0.36 μM for A10C, and reduced by another order of magnitude to 3.9 μM for A95C (Supplementary Table 3).
DISCUSSION
The sensing domain of the ydaO riboswitch in the c-di-AMP bound state adopts a novel scaffold involving collinear alignment of stems P4, P5 and the pseudoknot (boxed segment, Supplementary Fig. 16a), of stem P2 and elements of the zippered up bubble (boxed segment, Supplementary Fig. 16b), and stems P1 and P3 with the adenine ring of c-di-AMP mediated by a pair unpaired bases (boxed segment, Supplementary Fig. 16c, d). An unanticipated observation was that all stems except P4 were elongated by a sheared G•A non-canonical base pair (Fig. 1a and Supplementary Fig. 2b-e). The C1′-C1′ separation is shorter in a sheared G•A pair (9.5 Å) relative to a Watson-Crick (10.5 Å) pair, thereby compressing the helical inter-strand separation, a structural feature presumably favored at junctional sites.
Notably, the residues lining the binding pockets and involved in c-di-AMP recognition are highly conserved (boxed red segments in Supplementary Fig. 17) for both pocket A (generated by stems P2, P3 and bubble) and pocket B (generated by stems P1, P5 and pseudoknot). Of special note was our unanticipated identification of separate c-di-AMP binding pockets that are related by elements of pseudo two-fold symmetry, both within individual binding pockets and between binding pockets, despite no evidence of such pseudo-symmetry in the secondary structure of the ydaO riboswitch. Aspects of pseudo two-fold symmetry within the RNA scaffold have also been reported in an earlier structural study of the six-helical junction flavin mononucletide (FMN) riboswitch, which contained a single binding pocket for ligand FMN29. Structural studies on the tetrahydrofolate (THF) riboswitch30, 31 have also identified two binding pockets occupied by the ligand within the sensing domain fold31; however they are not related by pseudo two-fold symmetry, as reported in the present study of the c-di-AMP riboswitch.
Our structural and binding studies strongly support the concept that the ydaO riboswitch targets c-di-AMP20 rather than ATP19. Our structural results also support formation of a pseudoknot as proposed previously from secondary fold and alignment studies20. We have identified a single strong Mg2+ site, that stabilizes the zippered-up bubble fold, with binding also facilitated by other divalent cations such as Mn2+, Ba2+ and Ca2+ (Supplementary Fig. 7). The structure of the ydaO riboswitch with bound c-di-AMP clarifies issues related to the secondary fold, whereby five helical segments were observed in the crystal structure (Fig. 1a and Supplementary Fig. 1d) in contrast to seven helical segments proposed previously (Supplementary Fig. 1b)20. Our ITC based binding studies yielded a Kd = 66 nM for the T. tengcongenesis c-di-AMP-ydaO riboswitch complex, compared to a Kd = 0.7 nM for the B. subtilis c-di-AMP-ydaO riboswitch complex estimated from in-line probing data20. The reason for this difference is not clear at this time, but it should be noted that the in-line probing studies were undertaken at lower concentrations than their ITC counterparts. The most striking difference is the 2:1 c-di-AMP:ydaO riboswitch stoichiometry observed both in the structure (Fig. 1b) and the ITC binding curves (Fig. 4a) for the T. tengcongenesis c-di-AMP ydaO riboswitch reported in this study, which contrasts with the 1:1 stoichiometry estimated from in-line probing data on the B. subtilis c-di-AMP ydaO riboswitch reported previously20.
The ydaO riboswitch binds c-di-dAMP with a 50-fold weaker Kd compared to c-di-AMP (Fig. 4a and Supplementary Table 2). The additional intermolecular hydrogen bonds involving the 2′-OH groups of c-di-AMP (Supplementary Fig. 14a) compared to c-di-dAMP (Supplementary Fig. 14b) on complex formation provides an explanation for the difference in binding affinities. By contrast, the ydaO riboswitch binds neither c-di-GMP, c-di-IMP (Fig. 4a) nor any linkage isomers of c-GAMP (Fig. 4b). Examination of the pockets for Aα and Aβ of c-di-AMP molecules A (Fig. 1c) and B (Fig. 1d) in the structure of the c-di-AMP riboswitch complex highlights steric clashes involving the amino group of guanine with a sugar ring, explaining why c-di-GMP or c-GAMP cannot replace c-di-AMP as a ligand for the ydaO riboswitch. The 6-amino group of bound c-di-AMP forms an intermolecular hydrogen bond with the bridging O3′ phosphate oxygen in the complex. Replacement of the 6-amino group of adenine of c-di-AMP by the 6-carbonyl oxygen of inosine of c-di-IMP (or guanine of c-di-GMP and c-GAMP) would disrupt this hydrogen bond, thereby explaining why c-di-IMP cannot replace c-di-AMP as a ligand for the ydaO riboswitch.
Our mutation studies have established that the key tertiary pairs G39-C94 associated with the zippered-up bubble segment (Fig. 1a, 2a, b, c) and conserved A73•U112 associated with the pseudoknot segment (Fig. 1a, 2e, f, g), are key contributors to the binding of the ydaO riboswitch by c-di-AMP (Fig. 4c). Equally importantly, mutation of conserved U98 as monitored by in-line probing20 and ITC studies (Fig. 4c), established this position to be also critically important for binding of the ydaO riboswitch by c-di-AMP. The latter result can now be explained from a structural perspective due to direct coordination between U98 and the bound Mg2+ cation (Fig. 2c) in the structure of the complex. We also show that replacement of A by C within three A-minor base triples results in a decrease in affinity of the ydaO riboswitch for c-di-AMP, the magnitude of which varies with the A-minor triple under consideration (Supplementary Fig. 15c and Supplementary Table 3).
Since two molecules of c-di-AMP bind the ydaO riboswitch, it raises the question as to whether the ligands bind in a cooperative manner. Previous studies have established that the tetrahydrofolate (THF) riboswitch binds two THF molecules at separated locations within the same riboswitch in a cooperartive manner under physiological conditions31. We cannot address the issue of cooperativity for the binding of two molecules of c-di-AMP to the ydaO riboswitch at this time, given that it would require independent monitoring of the individual binding pockets on addition of c-di-AMP (these could be probed by monitoring NMR of assigned resonances of the ydaO riboswitch on gradual addition of c-di-AMP, a project planned for the longer term). We do note that the two c-di-AMP binding sites are located on the same face of the riboswitch and connected by side-by-side arrangement of stacked bases (Fig. 3e), potentially allowing for cross-talk between sites.
The c-di-AMP riboswitch in the bound state (2:1 c-di-AMP:riboswitch) reported in this study uses completely different recognition elements from those used by the c-di-GMP-I riboswitch (1:1 c-di-GMP:riboswitch) reported previously13,14. Thus, Aα and Aβ adopt an open, spread out conformation in the c-di-AMP riboswitch complex, are wedged between bases and their intermolecular contacts are related by pseudo two-fold symmetry (Fig. 3). By contrast, Gα and Gβ are stacked with an intercalated adenine base between them in c-di-GMP-I riboswitch complex, form in-plane pairing alignments with bases of the RNA, with the intermolecular contacts involving Gα and Gβ being of an asymmetric nature13,14. Given the widespread occurrence of the ydaO riboswitch, the identification of new recognition principles and the unexpected pseudo-symmetry within and between binding pockets, highlights the versatility of RNA to adopt novel scaffolds and generate unanticipated binding pockets.
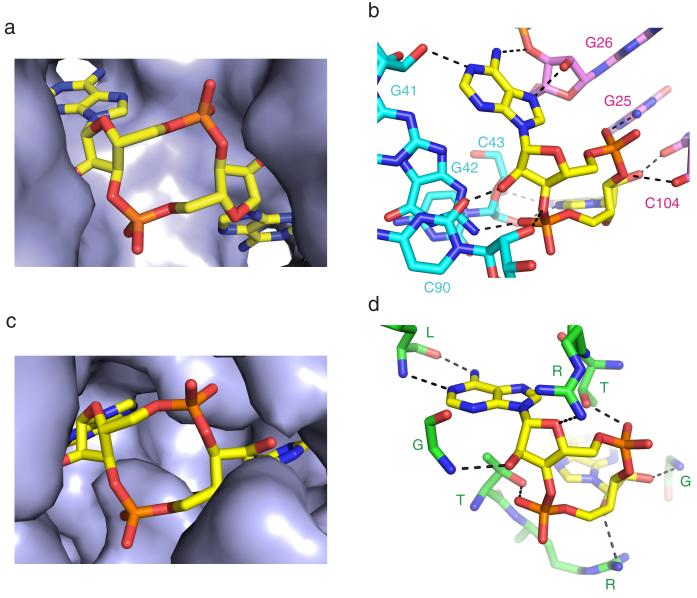
Recently, the protein DisA that controls B. subtilis sporulation checkpoint in response to DNA double-strand breaks, was shown to exhibit diadenylate cyclase activity regulated by DNA recombinant intermediates21. The crystal structure of DisA established formation of an octameric assembly with c-di-AMP molecules (3′,5′ linkages) bound at protein dimer interfaces21. We have compared the c-di-AMP binding pocket in the ydaO riboswitch (Fig. 5a, b) with its counterpart in DisA (Fig. 5c, d). We note that the sugar rings of c-di-AMP adopt 3′-endo sugar puckers in both complexes, with both adenine rings of c-di-AMP inserted into binding pockets in the riboswitch (Fig. 5a) and DisA (Fig. 5c) complexes. Further, intermolecular hydrogen bonds involving the adenine base edges, the 2′-sugar hydroxyls and phosphates of c-di-AMP are observed in both riboswitch (Fig. 5b) and DisA (Fig. 5d) complexes. Thus, the RNA and protein world have used common recognition principles to target the second messenger c-di-AMP.
Figure 5. Comparison of binding pockets and intermolecular hydrogen bonding contacts in complexes of c-di-AMP bound to the ydaO riboswitch and the protein DisA.
a, Insertion of both adenine rings of c-di-AMP within pockets in the complex with T. tengcongensis ydaO riboswitch. b, Intermolecular hydrogen bond interactions between c-di-AMP and the RNA in the ydaO riboswitch complex. c, Insertion of both adenine rings of c-di-AMP within pockets in the complex with DisA protein (PDB Code: 3C1Y). d, Intermolecular hydrogen bond interactions between c-di-AMP and the protein in the DisA complex (PDB Code: 3C1Y).
Functionally, it has been shown in vitro and in vivo that the B. subtilis ydaO riboswitch functions by transcription termination, with the c-di-AMP bound state representing a genetic ‘off’ switch and that riboswitch-mediated activation is associated with a decrease in c-di-AMP concentration20. The secondary structure of the sensing domain of the T. tengcongenesis c-di-AMP ydaO riboswitch, together with its 3′-flanking expression platform segment is drawn in Supplementary Fig. 18a. We note that when the P1 helix is formed, characteristic of the c-di-AMP bound state reported in this study, the 3′-flanking segment forms a transcription terminator (a stem-loop ending in a Un stretch), representing a transcription ‘off’ state. By contrast, disruption of stem P1, characteristic of the ligand free state, accompanied by disruption of the terminator stem loop, in turn can result in an alternate pairing alignment, termed antiterminator (Supplementary Fig. 18b), representative of a transcription ‘on’ state. Thus, like the B. subtilis ydaO riboswitch20, the T. tengcongenesis (this study) ydaO riboswitches could use a transcription termination mechanism, with the c-di-AMP bound state representing a potential genetic ‘off’ switch. As mentioned previously20, ydaO riboswitch-mediated detection of c-di-AMP using an ‘on’-‘off’ switch mechanism dependent on second messenger concentration, could control the expression of genes involved in germination, peptidoglycan biosynthesis and osmotic shock responses in diverse bacteria.
ONLINE METHODS
RNA preparation, purification and complex formation
The ydaO motif of the T. tengcongensis (Thermoanaerobacter tengcongensis MB4) c-di-AMP riboswitch followed by the hammerhead ribozyme was transcribed in vitro using T7 RNA polymerase32. To facilitate crystallization, loops of the wild type (wt) ydaO riboswitch (Supplementary Fig. 1b) were replaced by GAAA tetraloops and a GG step was added at the 5′-end (Supplementary Fig. 1c). The transcribed RNA was purified by denaturing polyacrylamide gel electrophoresis (PAGE) followed by anion-exchange chromatography and ethanol precipitation. The complex was generated by annealing the purified ydaO riboswitch at 70 °C with c-di-AMP or c-di-dAMP in a 1:2 molar ratio for 5 min in a buffer containing 100 mM K-acetate, pH 6.8, and 5 mM MgCl2 followed by incubation at 37 °C for 5 min and then cooling on ice for 1 h before setting up crystallization trials.
Crystallization
The crystals of the sensing domain of the ydaO riboswitch with bound c-di-AMP or c-di-dAMP in a 1:2 molar ratio were grown at 20 °C over a period of 2 weeks using the sitting-drop vapor diffusion approach after mixing the complex at an equimolar ratio with the reservoir solution containing 0.1 M Na-Bicine, pH 8.7-9.2 and 2.6-3.2 M (NH4)2SO4. For data collection, crystals were quickly transferred into a cryoprotectant solution containing 2.5 M Na-malonate pH 7.0 and flash-frozen in liquid nitrogen.
For Ir(NH3)63+ and Mn2+ soaking experiments, crystals were transferred into the crystallization solution containing 0.1 M Na-Bicine, pH 9.0 and 1.6 M (NH4)2SO4 and supplemented with 100 mM Ir(NH3)63+Cl3 or 100 mM MnCl2 at 4 °C for 24 h.
X-ray data collection and refinement
Iridium hexamine anomalous data of the ydaO riboswitch with bound c-di-AMP were collected at 100 K at the X29A beamline at the National Synchrotron Light Source (NSLS, Brookhaven) and processed with the program HKL2000 (HKL Research). X-ray diffraction native data and Mn2+ anomalous data were collected at NE-CAT beamlines at the Advanced Photon Source, Argonne National Laboratory and processed using the HKL2000 (HKL Research) and XDS programs. The structure (space group: C2221) was determined using SAD technique employing anomalous signal from two iridium atoms (see red arrows in anomalous map shown in Supplementary Fig. 19) using the HKL2MAP program33 and the PHENIX34 suite. RNA model was built in COOT35 and refined in PHENIX34 and REFMAC36 using 2.73 Å native data set of the ligand-bound c-di-AMP riboswitch. Metal ions and their coordinated waters were identified based on 2Fo-Fc and Fo-Fc maps guided by the coordination geometries. Mg2+ sites (Supplementary Fig. 6a) were identified by soaking crystals of the complex in Mn2+-containing solution and collecting an anomalous data set. c-di-AMP molecules were added to the model at the last stage based on the experimental and refined maps, coupled with electrostatic analysis. The x-ray statistics of the native, iridium hexamine-containing and Mn2+-soaked crystals are listed in Supplementary Table 1. The Mn2+-soaked structures were refined using c-di-AMP-bound structure as a starting model. We note that phosphates G34 and C43, as well as U8 and U71, from opposing strands are in close proximity in the structure of the complex It is conceivable that these pairs of phosphates may be bridged by cations.
Isothermal titration calorimetry
ITC experiments were performed on a Microcal ITC200 calorimeter at 35 °C. Prior to titration, 0.02-0.03 mM RNA samples were dialyzed overnight at 4 °C against an experimental buffer containing 50 mM K-acetate, pH 6.8, 100 mM KCl and 0 to 20 mM MgCl2 or other cations. RNAs were refolded by heating at 70 °C for 5 min and followed by cooling on ice. c-di-AMP and analogs powder were dissolved in the dialysis buffer at 0.4-0.6 mM concentration and typically titrated into the RNA in the sample cell (V = 207 μL) by 17 serial injections of 2.35 μl each, with a 0.5 μL/s rate, 180 s intervals between injections, and a reference power of 6 μcal/s. The thermograms were integrated and analyzed using Origin 7.0 software (Microcal, Inc.). The heat of ligand dilution from a control titration was subtracted from the experimental titration data. The Origin 7.0 software provides three built-in curve-fitting modules, namely ‘one set of sites’, ‘two sets of sites’ and ‘sequential binding sites’. Given that both binding pockets exhibit similar architectures and are related by pseudo two-fold symmetry, and that the bound c-di-AMPs exhibit the same intermolecular contacts, we assume that c-di-AMP binding to both sites are likely to exhibit similar Kd and ΔH values. Therefore, we utilized the ‘one set of sites’ model to estimate the binding constants and thermodynamic values listed in Tables S2 and S3.
Supplementary Material
Acknowledgements
We acknowledge assistance by staff at the X-29 beamline at the Brookhaven National Laboratory and NE-CAT beamlines at the Advanced Photon Source. The research was supported by NIH grant 1 U19 CA179564 to D.J.P.
Footnotes
Author Contributions
A.R. undertook all the crystallographic and ITC experiments under the supervision of D.J.P.
Competing financial interests
The authors declare no competing financial interests.
Accession codes: PDB: The atomic coordinates and structure factors have been deposited under accession codes as follows: T. tengcongensis ydaO riboswitch bound to c-di-AMP (4QLM) and c-di-dAMP (4QLN) .
Additional Information.
Supplementary Information is available in the on line version of the paper. Reprints and permissions information is available online at http://www.nature.com/reproints/index.html.
REFERENCES
- 1.Mironov AS, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 3.Nudler E, Miranov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Soc. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Ann. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Ann. Rev. Biophys. 2006;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serganov A, Patel DJ. Metabolic recognition principles and molecular mechanisms underlying riboswitch function. Ann. Rev. Biophys. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger c-di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee ER, et al. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Ann. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Smith KD, et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KD, et al. Structural basis of differential ligand recognition by two classes of bis-(3′,5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Scis. USA. 2011;108:7757–7762. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith KD, Strobel SA. Interactions of the c-di-GMP riboswitch with its second messenger ligand. Biochem. Soc. Trans. 2011;39:647–651. doi: 10.1042/BST0390647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrack JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Scis. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Block KE, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J. Bacteriol. 2010;192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson PY, Fedor MJ. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat. Chem. Biol. 2012;8:963–965. doi: 10.1038/nchembio.1095. [DOI] [PubMed] [Google Scholar]
- 20.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Science. 2010;329:845–848. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Scis. USA. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corrigan RM, et al. c-di-AMP is a new second messenger in S. aureus with a role in controlling cell size and envelop stress. PLOS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong JC, et al. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholera biofilm formation. J. Bacteriol. 2008;190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonicaL cyclic dinucleotide that activates human STING. Cell Reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ablasser A, et al. cGAS produces a 2′,5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkage is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a FMN riboswitch. Nature. 2011;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Ishibe-Murakami S, Patel DJ, Serganov A. Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch. Proc. Natl. Acad. Scis. USA. 2011;108:14801–14806. doi: 10.1073/pnas.1111701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand-binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pikovskaya O, et al. Preparation and crystallization of riboswitch-ligand complexes. Methods Mol. Biol. 2009;540:115–128. doi: 10.1007/978-1-59745-558-9_9. [DOI] [PubMed] [Google Scholar]
- 33.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- 34.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:212–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.