Abstract
Many areas of the central nervous system are organized into clusters of cell groups, with component cell groups exhibiting diverse but related functions. One such cluster, the superior olivary complex (SOC), is located in the ventral auditory brainstem in mammals. The SOC is an obligatory contact point for most projection neurons of the ventral cochlear nucleus and plays central roles in many aspects of monaural and binaural information processing. Despite their important interrelated functions, little is known about the embryonic origins of SOC nuclei, due in part to a paucity of developmental markers to distinguish individual cell groups. In this report, we present a collection of novel markers for the developing SOC nuclei in mice, including the transcription factors FoxP1, MafB, and Sox2, and the lineage-marking transgenic line En1-Cre. We use these definitive markers to examine the rhombic lip and rhombomeric origins of SOC nuclei and demonstrate that they can serve to uniquely identify SOC nuclei and subnuclei in newborn pups. The markers are also useful in identifying distinct nuclear domains within the presumptive SOC as early as embryonic day (E) 14.5, well before morphological distinction of individual nuclei is evident. These findings indicate that the mediolateral and dorsoventral position of SOC nuclei characteristic of the adult brainstem is established during early neurogenesis.
Keywords: auditory brainstem, fate mapping, lineage, rhombomere, superior olivary complex
INTRODUCTION
The medulla (myelencephalon) originates from the posterior subdivision of the hindbrain (rhombencephalon). Rhombomeres are defined by discrete, but transient, segmental boundaries (generally six rhombomeres (r)-r1-r6 and five pseudorhombomeres- r7-r11) along the anterior-posterior axis of the hindbrain and give rise to distinct cell lineages for neurons within the central nervous system and neural crest cells that migrate out of the neural tube. Taking advantage of the experimental accessibility of the dorsal surface of an embryo, fate maps of the brainstem have been produced using chick-quail chimeric transplantation (Tan and LeDouarin 1991; Marín and Puelles, 1995; Cambronero and Puelles, 2000) and by neuronal labeling in chick with lipophilic dyes (Cramer et al., 2000). These investigators have been able to determine that the early rostral-caudal arrangement of rhombomeres is mostly preserved in the adult brainstem, that neurons within individual nuclei can arise from multiple rhombomeres, and that the medial and lateral regions of a single rhombomere can give rise to distinct brainstem nuclei. This latter finding suggests that cell fates are already specified at or soon after the birth of neurons rather than during or after their migration into the final adult position (Cramer et al., 2000).
Due to the inherent difficulty of accessing mammalian embryos in utero, classical fate mapping studies are not common. More recent studies take advantage of transgenic lines that express Cre-recombinase in a rhombomere-specific fashion for r1 through r5 (Branda and Dymecki, 2004). Arising from these rostral rhombomeres are the nuclei of the auditory brainstem, including the cochlear nuclei and the nuclei that make up the superior olivary complex (SOC). The cochlear nuclei are integrative stations and relay centers for all auditory inputs from the cochlea (Young and Oertel, 2003), and include the dorsal, anteroventral, and posteroventral cochlear nuclei. The SOC is the earliest site for massive convergence of binaural information, and functions in both monaural and binaural information processing. The SOC of mammals consists of multiple, well-circumscribed nuclei, ranging in number from 6-11 cell groups depending upon species (Warr, 1972). The nuclei that are consistently found in species ranging in size from rodents to non-human primates include the lateral (LSO) and medial (MSO) superior olives and the medial (MNTB), ventral (VNTB), and lateral (LNTB) nuclei of the trapezoid body. The superior paraolivary nucleus (SPON) is typically found in rodents but may have a homologue in other species called the dorsomedial periolivary nucleus. These cell groups in mouse are born between embryonic days (E) 9-14, with peak birthdates earliest in the SPON and latest in the LSO (Taber-Pierce, 1973).
Fate maps of the mammalian cochlear nuclei have defined r2-r5 as their rhombomeric origins (Farago et al., 2006). Similar fate mapping of the SOC has determined that nuclei within the complex derive from r3-r5 (Maricich et al., 2009). Clues to SOC formation also can be gleaned from studies focused upon other neural systems. These additional studies revealed that distinct nuclei within the SOC originate within the rhombic lip, which is the lateral-most region of the developing hindbrain and is identifiable by its expression of the Wnt1 and Atoh1 genes (Nichols and Bruce, 2006; Fu et al., 2011; Maricich et al., 2009). SOC nuclei also reportedly derive from regions of the brainstem that express the Wnt3a and En1 genes (Louvi et al., 2007; Machold and Fishell, 2005). En1 has classically been used to label midbrain and r1 lineages (Zervas et al., 2004; Sgaier et al., 2005), but is also expressed within a specific dorsoventral domain of the spinal cord within the V1 interneurons, where it helps to define the axon trajectory of Renshaw and 1a inhibitory interneurons (Saueressig et al., 1999). We determined that a systematic study of SOC origins with appropriate developmental markers was necessary to piece together these many studies.
SOC cells have a postulated birth location at the rhombic lip, followed by ventral migration into their final position (Altman and Bayer, 1980). The finding that the LSO and MSO derive from the Atoh1 lineage (Maricich et al., 2009) supports this notion, but this situation does not necessarily apply to all SOC nuclei. MNTB cells are postulated to migrate from the ventral edge of the brainstem into their adult location, but these studies were based upon a time series of Golgi-stained tissue (Morest, 1969). In essence, very little has been firmly established with regard to the spatial origins and potential migratory patterns of the mammalian SOC. The absence of nuclei-specific and cell-specific markers for the developing central auditory brainstem has severely hampered efforts directed towards understanding the detailed mechanisms that govern its development and formation. In this report, we describe four early markers, FoxP1, MafB, Sox2, and En1, to facilitate observation of the presumptive SOC during embryonic development. Using these markers, we present the early embryonic organization of SOC nuclei prior to their identification as spatially distinct cell groups.
METHODS
Animals
Mice were maintained at West Virginia University in compliance with local IACUC and federal OLAW regulations on a 12-hour light/dark cycle. En1-Cre (courtesy of Dr. Mark Lewandoski), HoxB1-ires-Cre (courtesy of Dr. Mario Cappechi), Egr-2-Cre (courtesy of Dr. Patrick Charnay via Dr. Susan Dymecki), Wnt1-Cre (Jackson Laboratory), Rosa26R (Jackson Laboratory), RosaEGFP (Jackson Laboratory), Rosa-tdTomato (Ai9; Jackson Laboratory), FVB/NJ (Jackson Laboratory), and CD1 (Charles River) animals were used in this study.
Pregnant mice were sacrificed on the appropriate gestational day with midnight of timed mating considered E0. Embryos were collected, staged by size and morphology, and decapitated. Brains were either left in the head with their skulls split (for cryostat sectioning) or completely removed from the skull (for microtome sectioning). Samples were immersion fixed in a 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) solution (pH 7.4) overnight at 4°C. Postnatal mice were sacrificed by CO2 gas and decapitated, and brains were immersion fixed as described above.
Nissl Staining
Brains were cryoprotected in 30% sucrose/PBS solution, sectioned at 16 μm from caudal to rostral medulla, and dry-mounted onto slides. Sections were defatted by placing them into 1:1 alcohol/chloroform overnight and then rehydrated. Sections were then stained in 1% cresyl violet solution for 5-10 minutes, dehydrated in 95% ethanol and coverslipped.
In situ hybridization
The Engrailed-1 (En1) plasmid for preparing RNA probes was kindly provided by Dr. Mark Lewandoski. Antisense and sense RNA probes were prepared and labeled with digoxigenin according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). Digoxigenin-labeled riboprobes were purified by nucleotide removal kit (Qiagen, Valencia, CA) and diluted in hybridization buffer. At least four immersion-fixed CD1 brains from each of the following ages E11.5, E12.5, E13.5 and E14.5 were used in at least 3 hybridization studies (n≥12 at each age). Whole-mount in situ hybridization was performed as reported previously (Cygan et al., 1997). Briefly, brains were incubated with proteinase K, washed with glycine and refixed with a solution of 0.2% glutaraldehyde and 4% paraformaldehyde in PBS. Tissue was then placed into hybridization buffer at 63°C for 2 hours before incubation of brains with RNA probes at 65°C overnight. Brains were washed with a sodium citrate solution at 70°C, followed by incubation with RNase A (Sigma-Aldrich, St. Louis, MO) and RNase T1 (Sigma-Aldrich, St. Louis, MO), then further washed with sodium citrate at 65°C. Samples were blocked with lamb serum for 2 hrs and then incubated with an anti-digoxigenin antibody conjugated with alkaline phosphatase overnight at 4°C (1:4,000, Roche Applied Science). Incubation with BM purple (Roche Applied Science) was used to visualize the marker RNA. Digoxigenin-labeled sense RNA was used as a negative control. Samples were photographed using a stereomicroscope (MZFLIII, Leica) and a MicroFire digital camera (Optronics) and PictureFrame image capture software (Optronics).
Immunochemistry
Immersion fixed heads (n≥4 at each age) were cryoprotected in 30% sucrose in PBS and oriented in TBS tissue freezing medium, and stored at −80°C until sectioned in the coronal plane at 16 μm on a cryostat or on a freezing microtome at 40 μm. Antigen retrieval was performed by incubating cryostat sections on slides in 0.1M Tris HCL, pH 9.5 in a microwave for 2 minutes or by incubating microtome sections in 10mM citric acid, pH 6.0 for 30 minutes at 95°C. In order to block nonspecific labeling, some sections were incubated in PBS containing 0.6% H2O2 and 5% methanol for 1 hour at RT. Samples were then washed in PBS and blocked in PBS containing at least 5% heat-inactivated normal serum and 0.1% Triton X-100 at RT for 2 hours. Primary antibodies were diluted in PBS containing 4% serum and 0.1% Triton X-100. Samples were incubated overnight in the primary antibody solution at 4°C. The antibodies used in this study were: GFP (1:1000, Molecular Probes), MafB (1:2,000, SC-10022, Santa Cruz Biotechnology), Sox2 (1:1,000, Chemicon AB5603), Calbindin-D-28k (CaBP) (1:2,000, Swant CB38A) and FoxP1 (1:2000, Abcam 16645). Slides or wells were rinsed with PBS and incubated with the appropriate biotinylated (Vector, Laboratories, Burlingame, CA) or fluorescence-conjugated (Invitrogen) secondary antibodies in the same vehicle as the primary antibody. Biotin-labeled tissue was then rinsed with PBS and incubated with elite ABC reagent (Vector Laboratories), as described by the manufacturer. Immunoreactivity was visualized with 0.05% 3,3-diaminobenzidine (DAB) and photographed on an upright microscope (AX70, Olympus) with a MicroFire digital camera and PictureFrame image capture software (Optronics). Fluorescently labeled tissue was imaged on a Zeiss 510 Meta confocal microscope with a 10X (NA0.75) at 2.5μm or less Z-step size. Primary antibody was omitted in control tissue; no immunoreactivity was detected in these samples.
X-gal Staining
For ROSA26R tissue staining, brains were removed from skulls prior to fixation and stained for β-galactosidase. Samples were fixed in 4% paraformaldehyde solution in PBS for 0.5-4 hrs at RT, rinsed with PBS and incubated with X-gal staining solution (5 mM Potassium Ferricyanide, 5 mM Potassium Ferrocyanide, 5 mM Magnesium chloride, 1 mg/mL X-gal in PBS) at 4-22°C.
Marker Quantification
Coronal slices were sectioned at 100 μm thickness from the brainstem of postnatal day (P) 0 pups of the genotype En1-Cre; Rosa-tdTomato. Sections were reacted with anti-FoxP1 (1:2000, Abcam 16645) and anti-MafB (1:2,000, SC-10022, Santa Cruz Biotechnology) and appropriate, fluorescently labeled, secondary antibodies. Fluorescently labeled SOC (n=3) from two animals were imaged on a Zeiss 510 Meta confocal microscope. Z-stack projections were converted to TIFF images using Image J software 1.46r (National Institutes of Health, Bethesda, MD) and fluorescently labeled cells in each nucleus were counted using the Cell Counter plugin module within Image J.
Image Analysis
Images were stored as TIFF or JPEG files and examined with Adobe Photoshop (Adobe Systems, San Jose, CA), where brightness, contrast, and color balance were optimized uniformly and backgrounds of MafB in situ hybridizations were made uniformly black. Composite figures were prepared and final images were saved from Adobe Illustrator.
RESULTS
Rhombomeric origins of the superior olivary complex (SOC)
In an effort to explore the rhombomeric origins of the SOC using developmental markers, we employed the Egr2-Cre and Hoxb1-ires-Cre lines, which had been used previously to perform fate mapping of the developing auditory brainstem (Farago et al., 2005; Maricich et al., 2009). Parasagittal sections through the brainstem of Egr2-Cre; Rosa26R lacZ-reporter (Voiculescu et al., 2001; Soriano, 1999) animals were stained for β-galactosidase expression and revealed two distinct regions of labeling, corresponding to r3 and r5 (Fig. 1A). Based on morphology and location, we were able to identify the ventrally located, darkly labeled cells within the r5 region as MNTB (arrow in Figure 1A and compare to 1C,D). Similar parasagittal sections in animals carrying the Hoxb1-ires-Cre; RosaEGFP reporter (Arenkiel et al., 2003; Mao et al., 2001) were reacted with an anti-GFP antibody and showed Cre expression that was most prevalent in r4 (Fig. 1B). A small number of cells in the region of the MNTB showed Cre-mediated GFP expression in the Hoxb1-ires-Cre line (arrow in Fig. 1B). The location of SOC labeling was confirmed on adjacent sections stained with anti-calbindin (CaBP) and anti-Sox2 antibodies (Fig. 1C,D). Both proteins are expressed in cell bodies within the MNTB (arrow in Fig. 1C,D). Here we show for the first time that Sox2 is expressed in the embryonic and postnatal MNTB (Fig. 1D), as well as the VNTB and LNTB (Supplemental Fig. S1). Examination of coronal sections from the Egr2-Cre and Hoxb1-ires-Cre animals confirmed that more SOC cells derive from the Egr2-Cre lineage than the Hoxb1-ires-Cre lineage (Supplemental Fig. S2A,B compared to S2D), consistent with the findings of Maricich et al. (2009). To confirm the identities of distinct SOC nuclei in these coronal sections, we took advantage of antibodies against calbindin and the transcription factor, MafB (Fig. S2C, E, and F). Calbindin immunoreactivity labels cell bodies within the MNTB, but is also found in MNTB processes that extend into the LSO, MSO, and SPON (Supplemental Fig. S2C, E and Friauf, 1993). In addition to labeling the ventral cochlear nucleus (Farago et al., 2005; Howell et al., 2007), MafB expression is observed in the ventral brainstem in the region of the presumptive SOC (Supplemental Fig. S3) and the developing and mature LSO and MSO, with sparse labeling in the LNTB and SPON (Supplemental Figs. S2F and S4).
Figure 1.
Lineage labeling of the auditory brainstem in parasagittal sections of mouse hindbrain by Egr2-Cre; ROSA26R (A) and Hoxb1-ires-Cre; RosaEGFP (B-D). A) The region of the auditory brainstem, including the medial nucleus of the trapezoid body (MNTB, arrow), is strongly labeled by Xgal in the Egr2-Cre; ROSA26R line. The MNTB appears to be primarily within the r5 (caudal) rhombomere-labeling region. B) Activation of the GFP reporter by Hoxb1-Cre occurs primarily in a wedge of cells descending from r4. Modest labeling occurs in r3 and r5 domains, including light labeling in the MNTB region (arrow). C) A section proximal to panel B reacted with anti-CaBP antibody in the Hoxb1-ires-Cre; RosaEGFP line to reveal the MNTB (arrow). D) A section proximal to those in B and C reacted with anti-Sox2 antibody in the Hoxb1-ires-Cre; RosaEGFP line shows the specificity for Sox2 in the MNTB (arrow). Scale bar in A is 200 μm and applies to panels A-D.
Rhombic lip and dorsoventral origins of the SOC
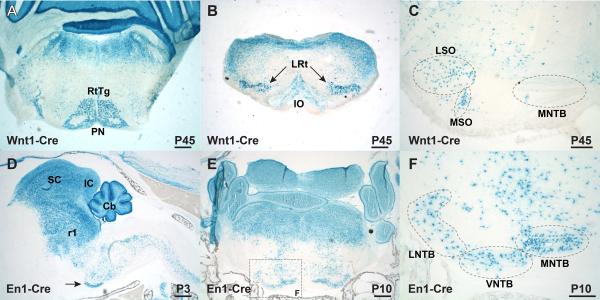
The rhombic lip in the mouse is defined by specific expression of growth and transcription factors, including Wnt1 and Atoh1 (Math1) (reviewed by Dun, 2012). Previous work on the origins of the mouse SOC suggested that the VNTB and LNTB derived from a Wnt1-positive lineage, along with a portion of the LSO (Fu et al., 2011) and that the MSO and LSO derive from an Atoh1-positive lineage (Maricich et al., 2009). Given that the expression domains of these two genes in the rhombic lip appear to overlap (Landsberg et al., 2005; see review by Dun, 2012), it would appear that these two findings are conflicting, especially if Wnt1 and Atoh1 expression occurs in the same cells. To clarify this discrepancy, we studied the cell lineages in Wnt1-Cre; Rosa26R adult animals (Fig. 2A-C). We analyzed sections in the coronal plane to discern SOC nuclei unambiguously. Comparable to previous findings (Landsberg et al., 2005; Fu et al., 2011), we observed strong labeling in the brainstem outside of the auditory region, including the pontine nuclei and inferior olivary complex, along with several other nuclei (Fig. 2A,B). Xgal-positive labeling within the SOC appeared in the LSO and MSO (Fig. 2C), with little, if any, labeling in the LNTB and VNTB, contrary to what was seen in Fu et al. (2011). Our Wnt1-Cre results are therefore in agreement with the labeling of the LSO and MSO in Atoh1-Cre animals (Maricich et al., 2009) and indicate that only a portion of the SOC derives from the rhombic lip.
Figure 2.
Lineage labeling with Wnt1-Cre and En1-Cre using Rosa26R reporter expression. A-F) Cre-mediated β-galactosidase expression in coronal (A-C, E, and F) or parasagittal (D) sections. Wnt1-Cre lineage (A-C) and En1-Cre lineage (D-F) marked by Xgal labeling. A) Wnt1-Cre marks P45 pontine and reticulotegmental nuclei, in addition to dorsal structures. B) Wnt1-Cre strongly labels the lateral reticular nuclei and weakly labels the inferior olive. C) Labeling within the SOC marks cells within the LSO and MSO (dotted ovals). The unlabeled MNTB is shown for orientation. D) En1-Cre lineage-labeled cells are found in the P3 superior and inferior colliculi, the cerebellum, r1-derived hindbrain, and spinal cord, as well as the ventral auditory brainstem, including the MNTB (arrow). E) Xgal-labeled coronal section through the P10 brainstem shows distinct labeling within the SOC. F) Boxed area from panel E shows Xgal labeling of the MNTB, VNTB, and LNTB. Scale bars in A, B, D, and E are 500 μm. Scale bars in C and F are 200 μm. Abbreviations: Cb- cerebellum; IC- inferior colliculus; LNTB- lateral nucleus of the trapezoid body; LSO- lateral superior olive; LRt- lateral reticular nucleus; MNTB- medial nucleus of the trapezoid body; MSO- medial superior olive; PN- pontine nucleus; r1- rhombomere 1 lineage; RtTg- reticulotegmental nucleus; SC- superior colliculus; VNTB- ventral nucleus of the trapezoid body.
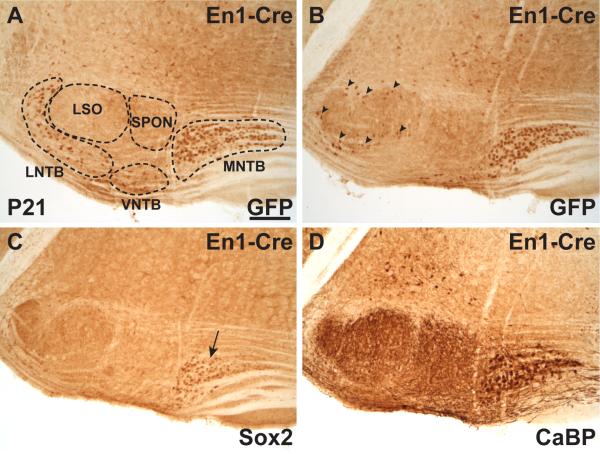
In addition to testing the Wnt1-Cre SOC lineage, we also analyzed the En1-Cre line for SOC lineage (Zervas et al., 2004; Sgaier et al., 2005), based on published images suggesting that one or more SOC nuclei might be lineage-labeled in this transgenic line (Fig. 2M in Machold and Fishell, 2005). In parasagittal sections from En1-Cre; Rosa26R samples at P3, the superior and inferior colliculi, the cerebellum, and the r1 hindbrain showed prominent Xgal labeling as expected (Fig. 2D). In addition to En1-Cre expression in these rostral regions, Xgal staining was observed diffusely in the reticular formation throughout the medulla and extending into the spinal cord, where En1 expression is found in V1 interneurons (Saueressig et al., 1999). Prominent En1-Cre expression is also observed in the ventral brainstem in a band consistent with the position of the MNTB (arrow in Fig. 2D). Coronal sections through this region at P10 showed prominent dorsal labeling, while only distinct nuclei along the ventral edge of the brainstem were stained with Xgal (Fig. 2E). Based on location and morphology, these ventral nuclei are likely the MNTB, VNTB, and LNTB (Fig. 2F). Marker analysis using the En1-Cre; RosaEGFP reporter strain (Mao et al., 2001) confirmed that the MNTB, VNTB, and LNTB are lineage-labeled by the En1-Cre line (Fig. 3A,B). The location of SOC labeling was confirmed on adjacent sections stained with anti-Sox2 (arrow in Fig. 3C) and anti-CaBP (Fig. 3D) antibodies. In addition, we observed En1-Cre-expressing cells surrounding the LSO (arrowheads in Fig. 3B), where the lateral olivocochlear shell neurons reside (reviewed by Brown, 2011).
Figure 3.
En1-Cre drives RosaEGFP reporter expression in the postnatal SOC in coronal sections at P21. A) Anti-GFP antibody shows En1-Cre lineage-labeling the MNTB, LNTB and VNTB. The unlabeled LSO and SPON are shown for orientation. B) A more caudal section from the same animal shows GFP labeling of the MNTB and cells peripheral to the LSO (arrowheads). C) Anti-Sox2 immunoreactivity is detected in the MNTB (arrow) on a proximal section to that in panel B, but does not label the cells surrounding the LSO. D) Anti-CaBP antibody in a section proximal to panels B and C labels the MNTB and its projections to the LSO, MSO, and SPON for positional reference. Scale in A is 200 μm and applies to panels A-D.
En1-Cre as a marker for embryonic SOC development
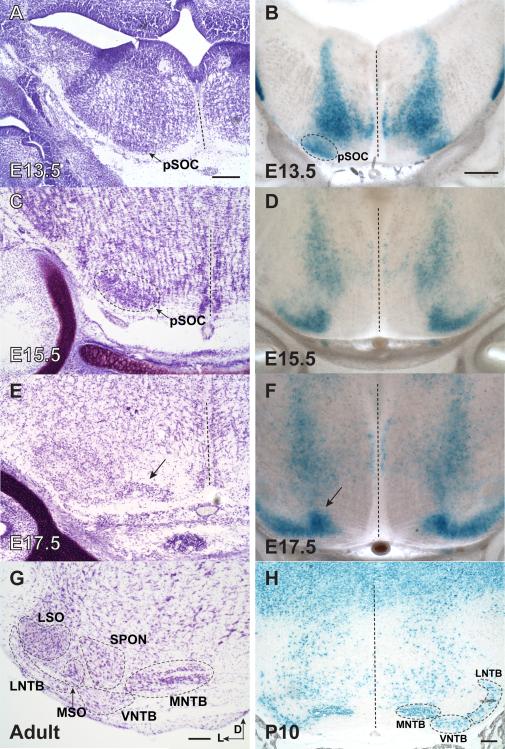
As a novel embryonic marker for a subpopulation of SOC nuclei, we postulated that the En1-Cre line could be useful in determining the origins of the SOC. Toward this goal, we analyzed the onset of En1 expression in the ventral brainstem. In situ hybridization with an En1 antisense riboprobe detected En1 expression beginning at E12.5 and continuing at 13.5 in a region that co-localizes with the presumptive SOC (Supplemental Fig. S5). Next, the Xgal-labeling pattern was examined on coronal sections from En1-Cre; Rosa26R embryos at E12.5-E18.5 and pups at P10 (Fig. 4 and data not shown). Cresyl violet-stained sections from E13.5-E17.5 and adult animals were analyzed for SOC morphology by comparison (Fig. 4). En1-lineage-labeled SOC cells are first distinguishable at E13.5 (Fig. 4B), as no Xgal-positive cells are present at the ventral edge of the auditory brainstem at E12.5 (data not shown). This area of staining colocalizes with the cluster of darkly Nissl-positive cells in the ventral brainstem that we identify as the presumptive SOC (pSOC) through lineage tracing (Fig. 4). These En1-Cre lineage-labeled cells at E13.5 appear soon after the completion of MNTB neurogenesis at E12 (Taber-Pierce, 1973). The one-day delay between En1 mRNA expression at E12.5 (Supplemental Fig. S5) and the Xgal labeling of the ventral brainstem at E13.5 likely represents an inherent delay in Cre recombination and subsequent accumulation of β-galactosidase to enable detection.
Figure 4.
En1-Cre lineage in Xgal-labeled coronal sections can be used to track formation of the SOC. A, C, E, G) Nissl-stained sections of auditory brainstem. B, D, F, H) En1-Cre; Rosa26R sections labeled with Xgal. A) At E13.5, darkly Nissl-stained cluster along the ventral edge of the brainstem marks the presumptive SOC (pSOC). B) Xgal-labeling first appears in the ventral brainstem at E13.5 in a region consistent with the presumptive SOC (pSOC). C) At E15.5, Nissl staining shows a uniform oblong cluster within the region of the pSOC (arrow). D) At E15.5, En1-Cre lineage cells in the pSOC exhibit a characteristic swoosh shape, which labels a subpopulation of the pSOC. E) By E17.5, a Nissl-stained cell group (arrow) appears medial to the main portion of the pSOC. F) By E17.5, the medially displaced cells are Xgal-labeled and in a region that is likely to become the future MNTB (arrow). G) The adult SOC with nuclei distinguished in Nissl-stained section. H) By P10, thin (16 μm), Xgal-labeled sections clearly identify MNTB, VNTB, and LNTB as the domains of En1-Cre expression. Note Xgal-labeled cells dorsally as in Fig. 2. Dashed lines indicate the midline. Scale bar in A is 200 μm and applies to A, C, and E. Scale in B is 200 μm and applies to B, D, and F. Scale bars in G and H are 200 μm. Abbreviations: D- dorsal; L- lateral; LNTB- lateral nucleus of the trapezoid body; LSO- lateral superior olive; MNTB- medial nucleus of the trapezoid body; MSO- medial superior olive; pSOC- presumptive superior olivary complex; SPON-superior paraolivary nucleus; VNTB- ventral nucleus of the trapezoid body.
The pSOC is uniform in its oblong appearance by Nissl-staining at E15.5 (Fig. 4A), but En1-expressing SOC cells appear in a “swoosh-shaped” cluster within the pSOC. This En1-Cre subdomain of the pSOC is present from E14.5 to E16.5 (Fig. 4D), suggesting that specification is already taking place within the pSOC with regard to orientation of SOC nuclei. By E17.5, the MNTB is distinguishable from the lateral cells within the pSOC (arrows in Fig. 4E,F), as we reported previously (Hoffpauir et al., 2010). These images suggest that cells migrate medially out of the pSOC region to establish the spatial domain of the adult MNTB. By P10, En1-Cre clearly distinguishes cells within the MNTB, VNTB, and LNTB (Fig. 4H).
Developmental markers for SOC nuclei
We next investigated whether the markers identified in this study could be used to successfully distinguish cells that will populate the individual SOC nuclei at birth, when the nuclear organization becomes adult-like. As shown above (Figs. 1-3 and S1-S4), we discovered markers for the LSO and MSO (MafB) and the MNTB, VNTB, and LNTB (En1-Cre and Sox2). The only major nucleus not identified by these markers is the SPON, a GABAergic nucleus located in the dorsal SOC between the MNTB and LSO (Kulesza and Berrebi, 2000). Based on RNA labeling patterns observed in the Allen Developing Mouse Brain Atlas (http://developingmouse.brain-map.org), we determined that the forkhead transcription factor, FoxP1 (Ferland et al., 2003), shows distinct labeling within the embryonic SOC. Antibodies targeted against FoxP1 mark early and localized expression in the presumptive SOC, starting as early as E14.5 (Supplemental Fig. S6). The FoxP1 expression pattern appears to resolve by P0 into labeling within the MNTB, SPON, and LNTB, along with modest labeling outside these nuclei (Fig. 5A). When combined with the labeling patterns at P0 for En1-Cre; Rosa-tdTomato (Fig. 5B), MafB (Fig. 5C) and Sox2 (Fig. 5D), a complete picture of labeling for SOC nuclei can be discerned. We quantified the labeling pattern at P0 for FoxP1-, En1-Cre-, and MafB-positive cells, along with double- and triple-labeled cells (Fig. 5F). Sox2 was omitted from this analysis given its overlap in expression with the En1-Cre line and the fact that the immunoreactivity of Sox2 in the SOC is unreliable prior to E16.5 (data not shown), rendering this marker ineffective for earlier embryonic studies.
Figure 5.
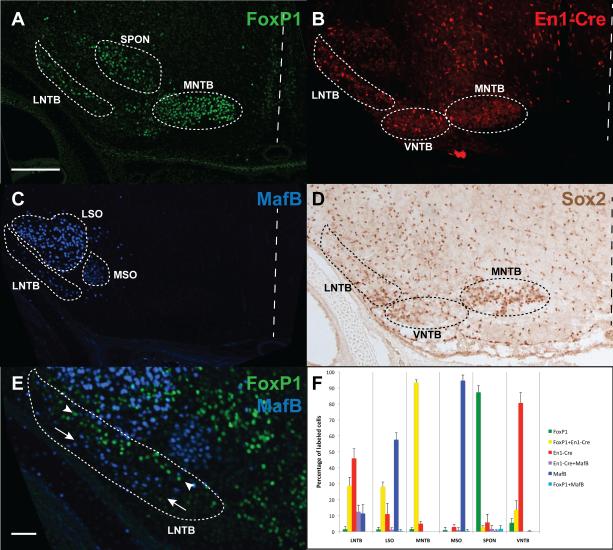
Developmental markers for SOC nuclei. A) Brainstem section from a P0 animal stained with anti-FoxP1 antibody strongly labels cells within the LNTB, SPON, and MNTB, along with sporadic labeling in the VNTB and LSO. B) Labeling in En1-Cre; Rosa-tdTomato P0 pups shows strong expression in the LNTB, VNTB, and MNTB. C) Labeling at P0 with anti-MafB antibody shows strong expression in the LSO, with weaker labeling in the MSO and LNTB. D) Labeling at P0 with anti-Sox2 antibody shows expression in LNTB, VNTB, and MNTB, along with weaker expression elsewhere in the SOC. E) Co-labeling of FoxP1 (green) and MafB (blue) in the LNTB shows two distinct longitudinal columns (arrowheads for FoxP1 vs. arrows for MafB). F) Quantification of labeled cells within each nucleus of the SOC at P0 for combinatorial En1-Cre, FoxP1, and MafB expression, along with double-labeled cells of each class. Triple-labeled cells represented fewer than 1% of labeled cells in any given nucleus. Dashed lines indicate the midline. The scale bar in A is 200 μm and applies to A-D. The scale bar in E is 50 μm.
In the colocalization experiments, we found that four nuclei were nearly homogenous in their labeling pattern, with the MNTB showing En1-Cre and FoxP1 double-labeling at 93.4% ±1.7%, the SPON being FoxP1-positive in 87.3% ±4.1% of labeled cells, MafB labeling the MSO in 94.6% ±3.5% of labeled cells (though at lower intensity levels than seen in the LSO), and the VNTB showing En1-Cre labeling in 80.6% ±6.5% of labeled cells (Fig. 5F). Conversely, the LSO and LNTB are comprised of several different labeled cell groups, with the LSO showing three different groups of cells each at greater than 10% of labeled cells. The LNTB is represented by four cell groups that meet the >10% labeling criterion. Interestingly, the LNTB labeling pattern for FoxP1 and MafB shows two longitudinally distinct groups running the length of the LNTB (Fig. 5E and see Fig. 6).
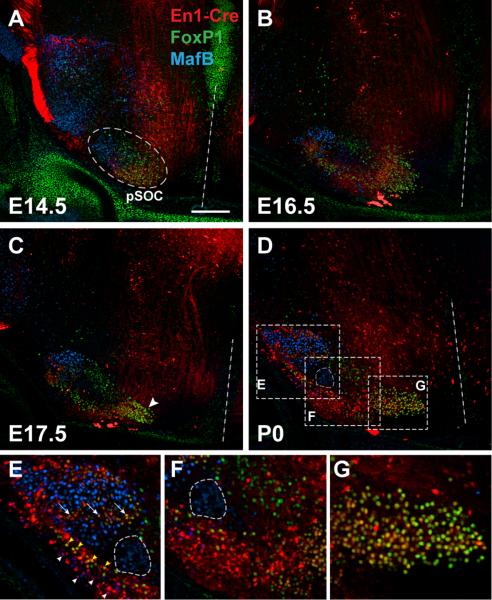
Figure 6.
Nuclear identities within the presumptive SOC (pSOC) are established at early embryonic ages. A-D) Coronal sections through the auditory brainstem were labeled with anti-FoxP1 (green) and anti-MafB (blue) antibodies and visualized using En1-Cre; Rosa-tdTomato reporter expression (red). A) Although the pSOC shows no visible distinction of nuclei at E14.5 (see Fig. 4), uniquely labeled cells are regionally distributed within the complex, with MafB-positive cells (future LSO and MSO) located laterally, FoxP1-positive cells (future SPON) located dorsally, and FoxP1+En1-Cre double-labeled cells (future MNTB) located medially and ventrally. B) By E16.5, the pSOC has elongated along the mediolateral axis and shows more distinct clusters of uniquely labeled cells in medial, central, and lateral regions. C) At E17.5, a cluster of cells positive for both FoxP1 and En1-Cre can be seen at the medial edge of the complex (arrowhead), where they are organizing into the mature MNTB. D) By P0, the adult-like structure of SOC nuclei is apparent in triple-labeled sections. The weakly labeled MSO is encircled in this panel and panels E and F as orientation to other cell groups; see Fig. 5 for individual nuclear labeling patterns at P0. Although there is some co-labeling within select nuclei, especially in the LSO (arrows in panel E) and LNTB, the general pattern for labeling is as follows- LSO is strongly MafB-positive with a central band of En1-Cre+FoxP1 double-labeled cells (arrows in panel E); LNTB is En1-Cre-positive with parts also being MafB-positive (white arrowheads in panel E) and FoxP1-positive (yellow arrowheads in panel E); MSO is weakly MafB-positive (encircled in panels D-F); SPON is FoxP1-positive (panel F); VNTB is En1-Cre-positive (panel F); and MNTB is FoxP1- and En1-Cre-positive (panel G). Scale bar in A is 200 μm and applies to A-D. Dashed white lines indicate the midline.
Embryonic organization of the presumptive SOC
The early labeling patterns of the En1-Cre lineage, along with anti-FoxP1 and anti-MafB immunoreactivity, provide a means to determine the spatial organization of the SOC during embryonic ages. We crossed En1-Cre-positive males to Rosa-tdTomato females and observed expression of the reporter allele. Cre-positive embryos and pups were selected for analysis of immunofluorescence at ages ranging from E14.5 to P0 (Figure 6). At P0, distinct nuclei within the SOC are observed both by Nissl-staining (data not shown) and fluorescence labeling (Fig. 5). As in Fig. 5, we observed strong co-localization between En1-Cre and FoxP1 labeled cells in the MNTB (nearly all cells in Fig. 6D,G exhibit co-localization). The majority of LSO cells are distinctly labeled by MafB expression (blue cells in Fig. 6E). However, within the LSO we found a cluster of cells that are co-labeled with En1-Cre and FoxP1, which could represent intrinsic lateral olivocochlear neurons (Brown, 2011; arrows in Fig. 6E). Similarly, the clusters of MafB-positive and FoxP1-positive cells in the LNTB (Fig. 5E) also appear to be labeled by En1-Cre (white vs. yellow arrowheads in Fig. 6E). Overall, the combined labeling pattern of these three markers can be utilized to identify individual SOC nuclei at birth (summarized in Fig. 5F).
Earlier embryonic ages presage the adult pattern by demonstrating populations of uniquely labeled cell groups within the presumptive SOC (Fig. 6A-C). The organization of labeled cell groups roughly approximates that of the adult SOC, even as early as E14.5 (Fig. 6A). At this early age, MafB-positive cells (presumptive LSO and MSO precursors) are located lateral to FoxP1-only labeled cells (presumptive SPON), which are themselves dorsolateral to FoxP1+En1-Cre double positive cells (presumptive MNTB). The only groups not regionally distinct at this age are the En1-Cre-positive cells (VNTB/LNTB), which are indistinguishable from presumptive MNTB cells. By E16.5, En1-Cre-positive cells elongate along the medial-lateral axis, and their pattern reveals a more defined separation of cell groups, as was seen in the swoosh pattern of Xgal labeling (Fig. 4). Between E16.5-E17.5, a medial cell group becomes spatially distinct (Fig. 6B,C), consistent with its emergence along the medial-lateral axis of Nissl-stained and En1-Cre-positive Xgal-stained cells during the same time frame (Fig. 4). Taken together, these findings indicate that the nuclear regionalization of the adult SOC is substantially complete by E14.5.
DISCUSSION
The coalescence of neurons and glia into a circumscribed and functionally specialized nuclear organization is one of the defining characteristics of the vertebrate CNS. Indeed, our nomenclature of brain anatomy relies on these structural boundaries, and yet little is known regarding the mechanisms that promote nuclear formation. In this report, we demonstrate that nucleogenesis occurs early for the developing auditory brainstem, and that the general orientation of SOC nuclei within the complex is patterned by marker expression well before morphogenetic boundaries are established. In the process of performing these studies, we have identified several novel markers for developing SOC nuclei that can aid future studies of cell specification and migration into the mature nuclear organization of the SOC. Through a combinatorial labeling strategy (Figs. 5 and 6 and Supplemental Table 1), we can identify cells that constitute the SOC nuclei at embryonic and perinatal ages, well before separation into clearly defined nuclear boundaries occurs. Furthermore, our studies offer an updated interpretation of data regarding the early origins for the SOC.
Lineage relationships of SOC nuclei
The embryonic origins of the SOC have been investigated for many years, but the findings have been less than definitive. Our investigations confirm many aspects of previous studies but refute certain findings regarding the origins of SOC nuclei. Although the SOC lies in the ventral brainstem, many investigators proposed that its origin arises via migration from the alar plate-derived rhombic lip, similar to migratory origins for the pontine and inferior olivary nuclei (reviewed by Kuhlenbeck, 1975). Along this line of reasoning, Altman and Bayer (1980) proposed the rhombic lip as the origin of SOC nuclei, followed by migration into the ventral brainstem. Our data indicate that the SOC nuclei may have multiple origins. As was previously observed (Fu et al., 2011; Nichols and Bruce, 2006), medial nuclei of the SOC do not arise from the Wnt1-positive rhombic lip lineage (Fig. 2C). We did observe labeling in Wnt1-Cre; Rosa26R animals in the region of the LSO and MSO (Fig. 2C). Fu et al. (2011) interpreted the SOC labeling that they observed in the parasagittal plane as marking the LSO, MVPO and LVPO (VNTB and LNTB, respectively). However, in coronal sections these latter two nuclei do not appear to be labeled (Fig. 2C), which is evident when compared with labeling of the VNTB and LNTB in the En1-Cre line (Fig. 2E,F). Our interpretation of these findings is that LSO and MSO derive from rhombic lip origins, while the MNTB, VNTB, LNTB, and SPON are most likely basal plate derivatives. We cannot, however, rule out the possibility that the Wnt1-Cre is activated outside of the lower rhombic lip, and that this secondary expression domain is the origin for LSO and MSO. The fact that the Atoh1-Cre lineage also labels LSO and MSO (Maricich et al., 2009) is suggestive that these two nuclei are likely rhombic lip derivatives.
It is intriguing that the LSO and MSO appear to be labeled consistently by Wnt1-Cre (Fig. 2), Atoh1-Cre (Maricich et al., 2009), and MafB (Fig. 5 and Supplemental Fig. 4). A similar overlap in expression of these three markers is seen in the ventral cochlear nucleus (Howell et al., 2007; Farago et al., 2005; Maricich et al. 2009; Fu et al. 2011; Landsberg et al. 2005; Wang et al., 2005). The overlap of Wnt1 and Atoh1 expression in the rhombic lip (Landsberg et al. 2005; reviewed by Dun, 2012) might suggest a hierarchical relationship, with high levels of Wnt1 inducing Atoh1 expression. We speculate that MafB might either respond directly to the Wnt1 signal or be induced secondarily by active expression of Atoh1 within the SOC and ventral cochlear nucleus. Such a regulatory hierarchy might help to explain the concurrent expression domains for these three genes.
The identification of labeling in the MNTB, VNTB, and LNTB by the En1-Cre lineage is novel, though careful observation of the results from Machold and Fishell (2005; see their Fig. 2M) indicates that they too labeled the MNTB, VNTB, and LNTB in their lineage preparations without mention of SOC structures. Expression of En1-Cre in the auditory brainstem appears by E13.5 and persists due to the lineage labeling in this assay. However, the En1-Cre labeling pattern at P10 and P21 very closely matches the En1 mRNA expression pattern seen at P4, P7, and P14 in the Allen Developing Mouse Brain Atlas, suggesting that the early expression pattern of En1 in the auditory brainstem is maintained into postnatal ages. The distinct pattern of dorsolateral labeling of the SOC with Wnt1-Cre and medioventral labeling of the SOC with En1-Cre is reminiscent of complementary labeling patterns for these two genes during Drosophila segmentation and vertebrate midbrain-hindbrain and limb patterning (Ingham et al., 1988; Ye et al., 2001; Kimmel et al., 2000).
En1-Cre appears to be activated in the auditory brainstem in a pattern that is similar to its dorsoventral expression domain in V1 interneurons of the spinal cord (Figs. 2D, 4B and Supplemental Fig. S5). Intriguingly, both the V1 interneurons and the trapezoid body nuclei exhibit glycine and GABA expression (see review by Goulding, 2009), suggesting that En1 activity may aid in defining inhibitory neuronal fate and/or synaptic connection patterns (Saueressig et al., 1999). In fact, the distinct domains of FoxP1-positive versus MafB-positive cells within the En1-Cre-expressing LNTB (see Figs. 5 and 6) fit nicely with the domains of expression for glycine and GABA within subdivisions of the adult cat LNTB (Spirou and Berrebi, 1996; 1997). Based on expression patterns within these subdivisions, we can speculate that MafB-positive cells in the pvLNTB would give rise to glycinergic LNTB neurons and FoxP1-positive cells in the mLNTB would give rise to LNTB neurons that are both glycinergic and GABAergic. Lineage-labeling studies would be needed with these markers to confirm this speculation.
Our work demonstrates a strong labeling of SOC nuclei from the Egr2-Cre lineage, and weaker cell contributions found in the Hoxb1-ires-Cre lineage (Fig. 1 and Supplemental Fig. S2). These results suggest that all SOC nuclei derive primarily from the r5 (and possibly r3) lineage, in agreement with the findings Maricich et al. (2009). However, the sparse labeling of the MNTB (Fig. 1) and more lateral SOC nuclei (Supplemental Fig. S2) in the Hoxb1-ires-Cre line suggests that the r4 lineage makes a minor contribution to SOC formation.
Embryonic organization of SOC nuclei
Previous studies on nucleogenesis have focused on groupings of neuronal cells into relatively uniform nuclei, including the mouse pontine nucleus and chick oculomotor complex (Nishida et al., 2011; Hasan et al., 2010). In contrast, organization of the functionally grouped, presumptive SOC requires positional alignment of at least six independent nuclei along the dorsoventral and mediolateral dimensions. In addition, these nuclei display distinct morphological changes along the rostrocaudal extent of the auditory brainstem (Kulesza et al., 2002). Therefore, the identification of cell-specific and region-specific SOC markers in this study is crucial for distinguishing among the multiple cell types within the pSOC at embryonic ages. A significant advantage of the markers selected for this study is that none appears to require terminal differentiation of the cells for labeling, which allows for early embryonic tracking of cells. As demonstrated in Fig. 6, FoxP1, MafB, and En1-Cre labeled cells are distinct as early as E14.5, and their relative position within the pSOC cluster at this age matches that seen in neonatal and postnatal samples.
The timing of neurogenesis for the SOC nuclei has been established in rodent embryos (Taber-Pierce 1973; Altman and Bayer, 1980; Kudo et al., 2000). Neurons of the mouse SOC are born between E9 and E14, with the progressive order of peak birthdates beginning with SPON, then MSO, MNTB and finally LSO (Taber-Pierce, 1973). Comparative neurogenesis in the rat SOC begins at E12 and ends around E16 (Altman and Bayer, 1980; Cant, 1998; Kudo et al., 2000). By E14.5 in the mouse, neurogenesis in the SOC is nearly complete (Taber-Pierce, 1973). These early birthdates of SOC neurons largely precede the onset for our markers. Therefore, it is difficult to determine where cells are born, because early-born cells would have ample time to migrate into their final position in the presumptive SOC along the ventral brainstem. The best indicator for the origins of the MNTB, VNTB, and LNTB comes from the expression of En1 mRNA at E12.5 in the ventral brainstem (Supplemental Fig. S5). The En1-Cre lineage distinctly labels these three nuclei, and the X-gal-positive cells are present along the ventral edge in the region of the presumptive SOC as early as E13.5 (Fig. 4). Given that neurons in these three nuclei are predominantly born between E11 and E12 (Taber-Pierce, 1973), it is possible that they are born in situ or migrate only a very short distance from their germinal zone. If true, then we would anticipate that neurons of the MNTB, VNTB, LNTB, and possibly SPON, would be derived from a region of the brainstem marked by a ventral domain of Ngn1 expression in the E11.5 rostral brainstem (see Landsberg et al., 2005). Further studies using an Ngn1-Cre line would be needed to confirm this speculation and to determine whether the lineage of these cell groups is distinct from that of the LSO and MSO.
At E14.5, the presumptive SOC shows no morphologically distinct features in Nissl-stained sections (Fig. 4), though the location of distinct markers clearly indicates that regional boundaries are established at this early age (Fig. 6). Nissl-stained sections, En1-Cre lineage labeling, and FoxP1 reactivity studies (Figs. 4, 5, and Supplemental Fig. 6) demonstrate that a transition in SOC organization begins to occur around E16.5, whereby distinct nuclei begin to coalesce into their adult locations. The most dramatic of these migrations appears to be the future MNTB, which emerges medially from the other SOC nuclei between E16.5 and E17.5 (Figs. 4 and 6). This observation is consistent with our previous finding that E17.5 marks the earliest age at which the MNTB is distinguishable for physiological studies (Hoffpauir et al., 2010). The migration of MNTB cells appears to take place along the mediolateral plane, with MNTB cells arriving in their medial adult location from the presumptive SOC domain that is present more laterally at earlier ages. An alternative interpretation of these data would be that the MNTB maintains its initial position and that the lateral nuclei in the SOC migrate laterally as the brain grows. Neither of these alternatives involves a dorsoventral movement of cells, and thus is in contrast to the proposed dorsoventral migration of MNTB cells occurring from the outer limiting membrane (the ventral brainstem surface) in the developing opossum (Morest, 1969). Using molecular markers (FoxP1+En1-Cre), we observe MNTB cells throughout late embryogenesis in the ventral brainstem, but clearly distinct from the ventral surface (see Figs. 4-6). Therefore, it seems that either the mouse MNTB cells behave differently from those of the opossum, or that the cells identified by Morest using Golgi stain represented non-MNTB neurons. Given his description of nuclear movement within the cell and end feet attachment to the outer limiting membrane, it is possible that the cells described as MNTB cells were instead neuronal stem cells. Taken together, the early organization of nuclei that we demonstrate here and the collection of cellular markers provide a basis to probe further into the earliest origins and migration patterns for the nuclei of the SOC.
Supplementary Material
Supplemental Figure 1. Sox2 protein expression as a cell-type specific marker in the developing SOC. A) Anti-Sox2 immunofluorescent labeling of the MNTB at P21. B) Anti- CaBP immunofluorescent labeling of the MNTB at P21. C) Co-immunofluorescence labeling with anti-CaBP and anti-Sox2 antibodies shows coincident expression in the P21 MNTB. D-F) Late embryonic (E17.5), early postnatal (P0) and adult (P50) expression of Sox2 in the SOC is restricted to the MNTB, VNTB, and LNTB.
Supplemental Figure 2. Rhombomeric origins of the SOC. A-F) Coronal sections through the SOC of Egr2-Cre (A-C) and Hoxb1-ires-Cre (D-F) animals. A) Egr2-Cre drives β-galactosidase expression from the Rosa26R reporter in all nuclei of the mouse SOC. Xgal staining at P22 shows labeled cells in the LNTB, LSO, MNTB, MSO, SPON, and VNTB. B) Immunoreactivity with anti-GFP antibody marks all nuclei in the SOC. C) Anti-CaBP reactivity on a section near to that shown in panel B labels the MNTB and terminals in the SPON, MSO, and LSO as reference for other panels. D) Hoxb1-ires-Cre drives RosaEGFP reporter expression in lateral regions of the SOC, including the LNTB, LSO, and MSO at P21. SPON, VNTB and MNTB show fewer cells that are labeled compared to lateral SOC nuclei. E) Anti-CaBP antibody marks the SOC in a section proximal to panel D as in panel C. F) Anti-MafB antibody labels cells in the LSO and MSO at P21, along with a few cells in the LNTB and SPON. Scale bars in A, B, and D are 200 μm. The scale in B is applies to B and C; the scale in D applies to D-F.
Supplemental Figure 3. MafB RNA expression in the ventral brainstem through embryogenesis. A) Schematic summary of MafB RNA expression showing labeling of the presumptive SOC (pSOC), trigeminal nucleus (TN), and the ventral cochlear nucleus (VCN). B-F) BM Purple-labeling of MafB RNA in the ventral brainstem of whole- mount embryos and neonatal pups. B) MafB is expressed in the VCN and TN, with relatively weak expression in the pSOC at E14.5. C) MafB expression is increased by E15.5 in the pSOC. D) Strong expression of MafB is seen in the VCN, TN, pSOC, along with the motor nucleus of VII (arrow). E) MafB RNA is prominent in the lateral pSOC at E18.5. F) At P0, MafB is still prominent and relatively specific in the ventral brainstem region corresponding to the lateral SOC.
Supplemental Figure 4. MafB protein expression as a cell-type specific marker in the SOC. A) MafB expression is restricted to the P10 lateral SOC, with labeling occurring in the LSO, LNTB, MSO, and sparse cells in the dorsal SPON. B) Immunohistochemical detection of CaBP in the P10 SOC. Axon labeling of MNTB principal cells highlights the LSO, MSO, and SPON. C-E) Co-immunofluorescence labeling with anti-MafB and anti- CaBP antibodies shows MafB expression in cell bodies of the lateral limb of the LSO. F) At P0, MafB expression is observed in regions corresponding to the future LSO, LNTB, and MSO. G) A similar pattern to that seen at P0 is seen at E17.5. H) Expression of MafB at E14.5 is seen in the VCN, TN, and pSOC. I) Expression of MafB at E13.5 is similar to that seen at E14.5, though the pSOC is more diffuse.
Supplemental Figure 5. En1 in situ hybridization in whole embryonic mouse brain. A) No labeling of the ventral brainstem is apparent at E11.5. B-C) Labeling is detected in the region of the presumptive SOC in two longitudinal bands at E12.5 and E13.5 (arrows).
Supplemental Figure 6. FoxP1 protein expression as a cell-type specific marker in the developing SOC. A-D) Immunofluorescence labeling with an anti-FoxP1 antibody detects expression in the presumptive SOC (pSOC) through multiple embryonic ages. A) FoxP1 expression is observed in a uniform pattern in the region of the pSOC along the ventral brainstem at E14.5. B) By E16.5, FoxP1 expression becomes asymmetric within the pSOC, with high level expression medially and two bands (dorsal and ventral; arrows) of expression in the lateral pSOC. C-D) Between E17.5 and E18.5, the medial expression domain of FoxP1 appears to resolve into the future MNTB. The dorsal band of FoxP1 expression appears to coincide with the SPON, with partial labeling of the LSO. The ventral band occurs within the future LNTB, with partial labeling of the VNTB.
Acknowledgements
We are grateful to Drs. Ariel Agmon, Mark Lewandoski, Susan Dymecki, and Mario Cappechi for sharing mice and/or reagents for the studies presented here. We would like to thank the reviewers of this manuscript for their helpful comments toward improving the presentation. The work was supported by NIH grants EY012152 (PHM), RR015574 subproject 5 (PHM), DC007695 (GAS), and DC010546 F32 (GM). The Center for Neuroscience at WVU supports the confocal facility under grant GM103503.
Footnotes
Conflict of interests- None
REFERENCES
- Altman J, Bayer SA. Development of the brain stem in the rat. III. Thymidine-radiographic study of the time of origin of neurons of the vestibular and auditory nuclei of the upper medulla. J Comp Neurol. 1980;194:877–904. doi: 10.1002/cne.901940410. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–386. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Brown MC. Anatomy of olivocochlear neurons. In: Ryugo DK, editor. Auditory and Vestibular Efferents. Vol. 38. Springer; 2011. pp. 17–37. [Google Scholar]
- Cambronero F, Puelles L. Rostrocaudal nuclear relationships in the avian medulla oblongata: a fate map with quail chick chimeras. J Comp Neurol. 2000;427:522–545. doi: 10.1002/1096-9861(20001127)427:4<522::aid-cne3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Cant NB. Structural development of the mammalian auditory pathways. In: Rubel EW, et al., editors. Development of the Auditory System. Vol. 9. Springer; New York: 1998. pp. 315–413. [Google Scholar]
- Cramer KS, Fraser SE, Rubel EW. Embryonic origins of auditory brain-stem nuclei in the chick hindbrain. Dev Biol. 2000;224:138–151. doi: 10.1006/dbio.2000.9779. [DOI] [PubMed] [Google Scholar]
- Cygan J, Neufeld-Kaiser W, Jara G, Daniel WL. Comparative biochemistry of a cytosolic artiodactyl glycosidase. Comp Biochem Physiol B Biochem Mol Biol. 1997;116:437–446. doi: 10.1016/s0305-0491(96)00290-8. [DOI] [PubMed] [Google Scholar]
- Dun XP. Origin of climbing fiber neurons and the definition of rhombic lip. Int J Dev Neurosci. 2012;30:391–395. doi: 10.1016/j.ijdevneu.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Friauf E. Transient appearance of calbindin-D28k-positive neurons in the superior olivary complex of developing rats. J Comp Neurol. 1993;334:59–74. doi: 10.1002/cne.903340105. [DOI] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Paxinos G, Watson C. Precerebellar cell groups in the hindbrain of the mouse defined by retrograde tracing and correlated with cumulative Wnt1-cre genetic labeling. Cerebellum. 2011;10:570–584. doi: 10.1007/s12311-011-0266-1. [DOI] [PubMed] [Google Scholar]
- Goulding M. Circuits controlling vertebrate locomotion: moving in the right direction. Nat. Rev. Neurosci. 2009;10:507–518. doi: 10.1038/nrn2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan KB, Agarwala S, Ragsdale CW. PHOX2A regulation of oculomotor complex nucleogenesis. Development. 2010;137:1205–1213. doi: 10.1242/dev.041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffpauir BK, Kolson DR, Mathers PH, Spirou GA. Maturation of synaptic partners: functional phenotype and synaptic organization tuned in synchrony. J Physiol. 2010;588:4365–4385. doi: 10.1113/jphysiol.2010.198564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell DM, Morgan WJ, Jarjour AA, Spirou GA, Berrebi AS, Kennedy TE, Mathers PH. Molecular guidance cues necessary for axon pathfinding from the ventral cochlear nucleus. J Comp Neurol. 2007;504:533–549. doi: 10.1002/cne.21443. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Baker NE, Martinez-Arias A. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Kudo M, Sakurai H, Kurokawa K, Yamada H. Neurogenesis in the superior olivary complex in the rat. Hear Res. 2000;139:144–152. doi: 10.1016/s0378-5955(99)00172-0. [DOI] [PubMed] [Google Scholar]
- Kuhlenbeck H. Spinal cord and deuterencephalon. Karger; Basel: 1975. [Google Scholar]
- Kulesza RJ, Jr, Berrebi AS. Superior paraolivary nucleus of the rat is a GABAergic nucleus. J Assoc Res Otolaryngol. 2000;1:255–269. doi: 10.1007/s101620010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesza RJ, Vinuela A, Saldana E, Berrebi AS. Unbiased stereological estimates of neuron number in subcortical auditory nuclei of the rat. Hear Res. 2002;168:12–24. doi: 10.1016/s0378-5955(02)00374-x. [DOI] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Louvi A, Yoshida M, Grove EA. The derivatives of the Wnt3a lineage in the central nervous system. J Comp Neurol. 2007;504:550–569. doi: 10.1002/cne.21461. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Xia A, Mathes EL, Wang VY, Oghalai JS, Fritzsch B, Zoghbi HY. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brainstem accessory auditory nuclei. J Neurosci. 2009;29:11123–11133. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Puelles L. Morphological fate of rhombomeres in quail/chick chimeras: a segmental analysis of hindbrain nuclei. Eur J Neurosci. 1995;7:1714–1738. doi: 10.1111/j.1460-9568.1995.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Morest DK. The differentiation of cerebral dendrites: A study of the post- migratory neuroblast in the medial nucleus of the trapezoid body. Z Anat Entwicklungsgesch. 1969;128:271–289. doi: 10.1007/BF00522528. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Bruce LL. Migratory routes and fates of cells transcribing the Wnt-1 gene in the murine hindbrain. Dev Dyn. 2006;235:285–300. doi: 10.1002/dvdy.20611. [DOI] [PubMed] [Google Scholar]
- Nishida K, Nakayama K, Yoshimura S, Murakami F. Role of Neph2 in pontine nuclei formation in the developing hindbrain. Mol Cell Neurosci. 2011;46:662–670. doi: 10.1016/j.mcn.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Saueressig H, Burrill J, Goulding M. Engrailed-1 and netrin-1 regulate axon pathfinding by association interneurons that project to motor neurons. Development. 1999;126:4201–4212. doi: 10.1242/dev.126.19.4201. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spirou GA, Berrebi AS. Organization of ventrolateral periolivary cells of the cat superior olive revealed by PEP-19 immunocytochemistry and Nissl stain. J. Comp. Neurol. 1996;368:100–120. doi: 10.1002/(SICI)1096-9861(19960422)368:1<100::AID-CNE7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Spirou GA, Berrebi AS. Glycine immunoreactivity in the lateral nucleus of the trapezoid body of the cat. J. Comp. Neurol. 1997;383:473–488. doi: 10.1002/(sici)1096-9861(19970714)383:4<473::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Taber-Pierce E. Time of origin of neurons in the brain stem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- Tan K, Le Douarin NM. Development of the nuclei and cell migration in the medulla oblongata. Application of the quail-chick chimera system. Anat Embryol (Berl) 1991;183:321–343. doi: 10.1007/BF00196834. [DOI] [PubMed] [Google Scholar]
- Voiculescu O, Taillebourg E, Pujades C, Kress C, Buart S, Charnay P, Schneider-Maunoury S. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development. 2001;128:4967–4978. doi: 10.1242/dev.128.24.4967. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Warr WB. Fiber degeneration following lesions in the multipolar and globular cell areas in the ventral cochlear nucleus of the cat. Brain Res. 1972;40:247–270. doi: 10.1016/0006-8993(72)90132-1. [DOI] [PubMed] [Google Scholar]
- Ye W, Bouchard M, Stone D, Liu X, Vella F, Lee J, Nakamura H, Ang SL, Busslinger M, Rosenthal A. Distinct regulators control the expression of the mid-hindbrain organizer signal FGF8. Nat Neurosci. 2001;4:1175–1181. doi: 10.1038/nn761. [DOI] [PubMed] [Google Scholar]
- Young ED, Oertel D. The cochlear nucleus. In: Shepherd GM, editor. Synaptic organization of the brain. Oxford University Press; New York: 2003. [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Sox2 protein expression as a cell-type specific marker in the developing SOC. A) Anti-Sox2 immunofluorescent labeling of the MNTB at P21. B) Anti- CaBP immunofluorescent labeling of the MNTB at P21. C) Co-immunofluorescence labeling with anti-CaBP and anti-Sox2 antibodies shows coincident expression in the P21 MNTB. D-F) Late embryonic (E17.5), early postnatal (P0) and adult (P50) expression of Sox2 in the SOC is restricted to the MNTB, VNTB, and LNTB.
Supplemental Figure 2. Rhombomeric origins of the SOC. A-F) Coronal sections through the SOC of Egr2-Cre (A-C) and Hoxb1-ires-Cre (D-F) animals. A) Egr2-Cre drives β-galactosidase expression from the Rosa26R reporter in all nuclei of the mouse SOC. Xgal staining at P22 shows labeled cells in the LNTB, LSO, MNTB, MSO, SPON, and VNTB. B) Immunoreactivity with anti-GFP antibody marks all nuclei in the SOC. C) Anti-CaBP reactivity on a section near to that shown in panel B labels the MNTB and terminals in the SPON, MSO, and LSO as reference for other panels. D) Hoxb1-ires-Cre drives RosaEGFP reporter expression in lateral regions of the SOC, including the LNTB, LSO, and MSO at P21. SPON, VNTB and MNTB show fewer cells that are labeled compared to lateral SOC nuclei. E) Anti-CaBP antibody marks the SOC in a section proximal to panel D as in panel C. F) Anti-MafB antibody labels cells in the LSO and MSO at P21, along with a few cells in the LNTB and SPON. Scale bars in A, B, and D are 200 μm. The scale in B is applies to B and C; the scale in D applies to D-F.
Supplemental Figure 3. MafB RNA expression in the ventral brainstem through embryogenesis. A) Schematic summary of MafB RNA expression showing labeling of the presumptive SOC (pSOC), trigeminal nucleus (TN), and the ventral cochlear nucleus (VCN). B-F) BM Purple-labeling of MafB RNA in the ventral brainstem of whole- mount embryos and neonatal pups. B) MafB is expressed in the VCN and TN, with relatively weak expression in the pSOC at E14.5. C) MafB expression is increased by E15.5 in the pSOC. D) Strong expression of MafB is seen in the VCN, TN, pSOC, along with the motor nucleus of VII (arrow). E) MafB RNA is prominent in the lateral pSOC at E18.5. F) At P0, MafB is still prominent and relatively specific in the ventral brainstem region corresponding to the lateral SOC.
Supplemental Figure 4. MafB protein expression as a cell-type specific marker in the SOC. A) MafB expression is restricted to the P10 lateral SOC, with labeling occurring in the LSO, LNTB, MSO, and sparse cells in the dorsal SPON. B) Immunohistochemical detection of CaBP in the P10 SOC. Axon labeling of MNTB principal cells highlights the LSO, MSO, and SPON. C-E) Co-immunofluorescence labeling with anti-MafB and anti- CaBP antibodies shows MafB expression in cell bodies of the lateral limb of the LSO. F) At P0, MafB expression is observed in regions corresponding to the future LSO, LNTB, and MSO. G) A similar pattern to that seen at P0 is seen at E17.5. H) Expression of MafB at E14.5 is seen in the VCN, TN, and pSOC. I) Expression of MafB at E13.5 is similar to that seen at E14.5, though the pSOC is more diffuse.
Supplemental Figure 5. En1 in situ hybridization in whole embryonic mouse brain. A) No labeling of the ventral brainstem is apparent at E11.5. B-C) Labeling is detected in the region of the presumptive SOC in two longitudinal bands at E12.5 and E13.5 (arrows).
Supplemental Figure 6. FoxP1 protein expression as a cell-type specific marker in the developing SOC. A-D) Immunofluorescence labeling with an anti-FoxP1 antibody detects expression in the presumptive SOC (pSOC) through multiple embryonic ages. A) FoxP1 expression is observed in a uniform pattern in the region of the pSOC along the ventral brainstem at E14.5. B) By E16.5, FoxP1 expression becomes asymmetric within the pSOC, with high level expression medially and two bands (dorsal and ventral; arrows) of expression in the lateral pSOC. C-D) Between E17.5 and E18.5, the medial expression domain of FoxP1 appears to resolve into the future MNTB. The dorsal band of FoxP1 expression appears to coincide with the SPON, with partial labeling of the LSO. The ventral band occurs within the future LNTB, with partial labeling of the VNTB.