Abstract
Dinoflagellates harboring diatom endosymbionts (termed “dinotoms”) have undergone a process often referred to as “tertiary endosymbiosis”—the uptake of algae containing secondary plastids and integration of those plastids into the new host. In contrast to other tertiary plastids, and most secondary plastids, the endosymbiont of dinotoms is distinctly less reduced, retaining a number of cellular features, such as their nucleus and mitochondria and others, in addition to their plastid. This has resulted in redundancy between host and endosymbiont, at least between some mitochondrial and cytosolic metabolism, where this has been investigated. The question of plastidial redundancy is particularly interesting as the fate of the host dinoflagellate plastid is unclear. The host cytosol possesses an eyespot that has been postulated to be a remnant of the ancestral peridinin plastid, but this has not been tested, nor has its possible retention of plastid functions. To investigate this possibility, we searched for plastid-associated pathways and functions in transcriptomic data sets from three dinotom species. We show that the dinoflagellate host has indeed retained genes for plastid-associated pathways and that these genes encode targeting peptides similar to those of other dinoflagellate plastid-targeted proteins. Moreover, we also identified one gene encoding an essential component of the dinoflagellate plastid protein import machinery, altogether suggesting the presence of a functioning plastid import system in the host, and by extension a relict plastid. The presence of the same plastid-associated pathways in the endosymbiont also extends the known functional redundancy in dinotoms, further confirming the unusual state of plastid integration in this group of dinoflagellates.
Keywords: tertiary endosymbiosis, dinotom, relict plastid, redundancy
Introduction
In the evolution of plastids, the dominant processes have been endosymbiotic events followed by integration and reduction (Keeling 2013). The initial endosymbiotic event that ultimately gave rise to plastids was the uptake of a cyanobacterium by a eukaryotic cell (Douglas and Turner 1991), whose integration and reduction resulted in the establishment of the primary plastid now found in glaucophytes, red algae, green algae, and land plants (Cavalier-Smith and Lee 1985; Palmer and Delwiche 1998; Keeling 2010). Subsequently, plastids spread by a process termed secondary endosymbiosis, where an alga already containing a primary plastid was taken up by another eukaryote and integrated within the host, accompanied by reduction of the endosymbiont to varying extents (McFadden 2001; Keeling 2004; Gould et al. 2008). Secondary endosymbiosis occurred more than once, as both red and green algae have been involved in secondary endosymbiotic events, but the exact number of those events is still contentious, particularly in the case of red secondary plastids (Keeling 2013). Red secondary plastids are found in cryptophytes, haptophytes, stramenopiles, apicomplexans, and dinoflagellates. Dinoflagellate plastids differ from other red secondary plastids in being bounded by three membranes; all other red algal plastids share a four-membrane structure (Dodge 1975; Keeling 2004). They are also unusual in containing the pigment peridinin, which constitutes the major carotenoid in dinoflagellates and appears to be specific to this group (Jeffrey et al. 1975).
Several dinoflagellate lineages have taken the endosymbiotic process one step further, and replaced their peridinin-containing plastid of red algal origin with secondary plastids from other algae, in a process termed “tertiary endosymbiosis.” Two groups of dinoflagellates are known to possess permanent tertiary plastids of red algal origin: The kareniaceae, harboring plastids of haptophyte origin (Tengs et al. 2000) and the “dinotoms,” containing diatom-derived endosymbionts (Tomas and Cox 1973; Kite and Dodge 1985; Chesnick et al. 1996, 1997). A third dinoflagellate lineage, Dinophysis, may have tertiary plastids of cryptophyte origin, but is still disputed whether this plastid is permanently integrated or a transiently retained organelle derived from prey (Hackett et al. 2003; Kim et al. 2012).
Although the plastid of the kareniaceae is reduced to a similar extent as most secondary plastids (Tengs et al. 2000), the dinotom endosymbiont is not: In addition to the plastid itself, it has retained a number of features usually lost in secondary or tertiary endosymbionts. Diatom-derived tertiary endosymbionts have preserved their cytosol, endoplasmatic reticulum, free and membrane-associated ribosomes, mitochondria, and a large, DNA-rich nucleus, all surrounded by a single membrane separating the endosymbiont from the host cytosol (Dodge 1971; Tomas and Cox 1973; Tomas et al. 1973; Horiguchi and Pienaar 1994b). Despite this lower degree of reduction, the endosymbiont is present in the host through all stages of the life-cycle and division of host and endosymbiont are synchronized (Tippit and Pickettheaps 1976; Figueroa et al. 2009), indicating that the relationship between host and endosymbiont is a permanent one.
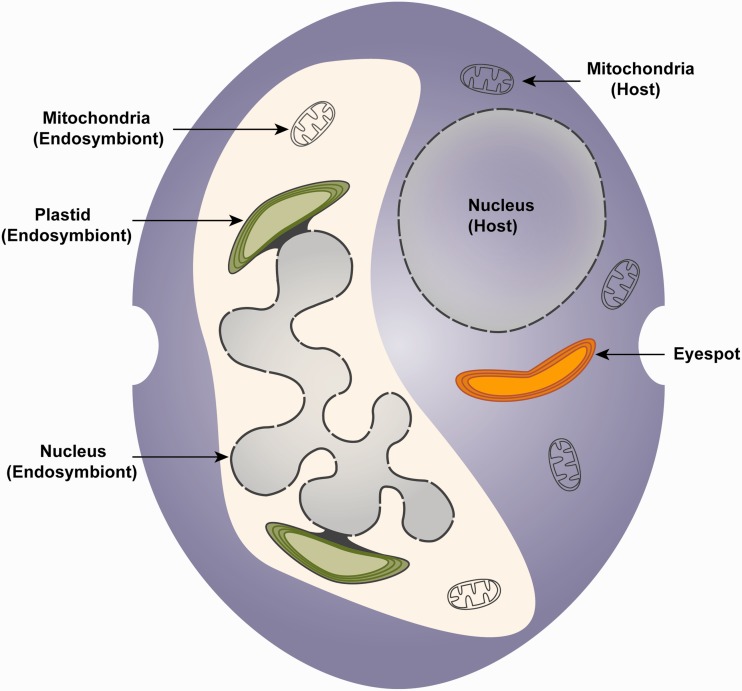
The low level of cellular reduction of the endosymbiont in dinotoms has resulted in a certain degree of redundancy (fig. 1). Genetic analysis of dinotom host and endosymbiont mitochondria not only revealed a functional overlap between these organelles but also showed that both the host and the endosymbiont mitochondria appear to be largely unaffected by the integration of the endosymbiont, retaining nearly all characteristics of free-living dinoflagellate and diatom mitochondria, respectively (Imanian and Keeling 2007; Imanian et al. 2012). Observed size variability of the endosymbiont nucleus containing varying amounts of DNA and reports of isolates lacking the endosymbiont nucleus prompted speculations that the endosymbiont nucleus of dinotoms may be no longer functional (Kempton et al. 2002; Figueroa et al. 2009). However, the existence of functional mitochondria in the endosymbiont suggests that an actively transcribing nucleus is present to maintain these organelles. A recent nuclear transcriptome analysis of dinotoms, despite using dinoflagellate-specific primers, identified several transcripts probably originating from the endosymbiont nucleus, implying a significant amount of DNA being transcribed in this nucleus (Burki et al. 2014). A specific analysis of tryptophan biosynthesis has similarly shown that both nuclei retain a complete and redundant set of biosynthetic enzymes for this pathway (Imanian and Keeling 2014).
Fig. 1.—
Schematic representation of a dinotom cell. The host cytosol contains a spherical nucleus, mitochondria and the triple membrane-bound eyespot. The cytosol of the endosymbiont contains a large, multilobed nucleus, mitochondria and four membrane-bound plastids, where the outermost chloroplast membrane is continuous with the outer membrane of the nucleus (Pienaar et al. 2007).
Dinotom plastid genomes have also retained numerous features present in free-living diatoms (Imanian et al. 2010), but unlike mitochondrial redundancy, there is no photosynthetic plastid present in the host cytosol, suggesting that in this case the endosymbiont plastid has functionally substituted that of the host, or at least has taken over photosynthesis. Whether this means the host plastid has been lost is, however, still an open question. Several dinoflagellate lineages harboring diatom-endosymbionts contain eyespots (fig. 1), a pigmented organelle that shows distinct structural variation even among different dinoflagellate species (Foster and Smyth 1980; Dodge 1984). The function of eyespots in dinoflagellates and other algal lineages has generally been presumed to be the reception of light stimuli and facilitation of phototactic responses, supported by the location of most eyespots close to the flagellar base (Dodge and Crawford 1969). Correspondingly, the eyespots described for several dinotom species are situated close to the flagella (Tomas and Cox 1973), but phototaxis in dinotoms is contentious (Withers and Haxo 1978; Horiguchi et al. 1999), suggesting other or additional functions for this organelle in dinotoms. Also, the structure of dinotom eyespots sets them apart from other dinoflagellate eyespots: They are triple-membrane bound (Tomas and Cox 1973; Horiguchi and Pienaar 1994a, 1994b; Pienaar et al. 2007), strongly resembling the membranes surrounding the peridinin-containing chloroplasts of dinoflagellates (Dodge 1968). It was therefore proposed that the eyespot in dinotoms is a relic of the host plastid that was present before acquisition of the endosymbiont, which in turn may have led to a reduction of the original plastid to the now existing eyespots (Dodge 1984; Horiguchi et al. 1999).
If the eyespot in dinotoms is a relict plastid, it may have retained plastid-associated functions, as is known for the nonphotosynthetic apicoplast in apicomplexan parasites, and the putative cryptic plastids in Perkinsus marinus and Oxyrrhis marina, two nonphotosynthetic lineages positioned at the base of dinoflagellates (Seeber and Soldati-Favre 2010; Fernández Robledo et al. 2011; Lee et al. 2014). Here, we investigated the transcriptomes of three dinotom species, Durinskia baltica, Glenodinium foliaceum, and Kryptoperidinium foliaceum, for the expression of metabolic pathways generally associated with plastids in the host nucleus. We show that the dinoflagellate host genome expresses genes that function in several plastid-associated pathways, and that homologous genes are also expressed in the endosymbiont. According to phylogenetic evidence and targeting peptide predictions, the products of these genes are targeted to a plastid-like organelle in the host. This is further supported by the identification of a key component of the plastid protein import machinery transcribed in the host nucleus, Tic110. Altogether this not only represents a new level of redundancy in dinotoms but also provides the first genomic evidence for the fate of the host plastid in a tertiary endosymbiont.
Materials and Methods
Cultures, Media, Growth, and Harvest Conditions
Cultures of D. baltica (Peridinium balticum) CSIRO CS-38, K. foliaceum CCMP 1326, and G. foliaceum CCAP 1116/3 were obtained from the Australian National Algae Culture Collection (CSIRO Marine and Atmospheric Research, Hobart, Australia), the Provasoli-Guillard National Center for Marine Algae and Microbiota (East Boothbay, ME), and Culture Collection of Algae and Protozoa (CCAP SAMS Research Services Ltd. Scottish Marine Institute, OBAN, Scotland, UK), respectively. Durinskia baltica culture was maintained in GSe medium at 22 °C in 12:12 light:dark cycles and harvested either during the light phase (light samples) or after 48 h in the dark (dark samples), whereas K. foliaceum and G. foliaceum cultures were maintained in and harvested from f/2-Si medium under the same conditions.
Nucleic Acid Extraction, Purification and Poly-A Library Construction, Sequencing, and Assembly
Exponentially growing cells were collected and ground as described elsewhere (Imanian and Keeling 2007). Cell lysis, nucleic acid extraction, precipitation, and purification were performed as described earlier (Imanian et al. 2010). The total RNA was cleaned up after DNase treatment (RNeasy MinElute Cleanup kit; Qiagen, Mississauga, ON), and poly-A RNA was purified from 25 µg of cleaned-up total RNA (Oligotex mRNA Mini Kit; Qiagen). Library preparation, sequencing, assembling, and annotation of the poly-A transcriptome of the three dinotoms were performed at the National Centre for Genome Resources (NCGR; see Imanian and Keeling 2014, supplementary material).
Taxonomic Analysis and Phylogenetic Pipeline
The translated transcriptome data sets were filtered for peptides longer than 100 amino acids and used as queries to perform a BLASTP search (Altschul et al. 1990; E value threshold ≤ 1e-5) against the nr protein database. The resulting hits were parsed for their GenBank taxonomy ID, leading to 28905/17859 (D. baltica light/dark), 26797/23125 (G. foliaceum light/dark), and 36839/22853 (K. foliaceum light/dark) transcripts that were sorted according to their putative taxonomy.
The NCBI protein database was searched for homologs of the MEP/DOXP and heme pathway in diatom and dinoflagellates (or other alveolates if no dinoflagellate sequence was available) to be used as bait to identify the respective homologs in our data sets (list of accession numbers available in supplementary table S1, Supplementary Material online). All seven steps of the MEP/DOXP pathway were investigated, from 1-deoxy-d-xylulose-5-phosphate synthase (dxs) to 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (ispH). The heme pathway was analyzed from glutamyl-tRNA reductase (hemA/GTR = first step unique for this pathway) to protoporphyrinogen oxidase (hemY/PPOX = catalyzes synthesis of protoporphyrin IX, the common precursor for chlorophyll and heme/bilin). The bait sequences were used as queries in a stringent BLASTP search (E value threshold ≤ 1e-25) against one dinotom data set only, G. foliaceum light, to reduce redundancy.
All hits were used in a reciprocal BLASTP search against the nr protein database to confirm the identity of the hit. The confirmed hits were used in the initial phylogenetic analysis: The recovered sequences were used to query a custom protein database (see supplementary table S2, Supplementary Material online, for a complete list of organisms) with BLASTP (E value threshold ≤ 1e-5). The database was subjected to cd-hit (Li and Godzik 2006) with a similarity threshold of 85% to reduce redundant sequences and paralogs, except for the six dinotom data sets themselves. The search results of the BLASTP step were parsed for hits with an E value threshold ≤ 1e-25 and a query coverage of ≥ 50% to reduce the possibility of paralogs and short sequence matches (e.g., protein domains only). Additionally, to limit the number of prokaryotic taxa, only a maximum of four taxa in each prokaryotic group was retained for alignment, except for the cyanobacteria and alphaproteobacteria, from each of which eight taxa were retained. The filtered fasta files were aligned using MAFFT with the L-INS-i option (Katoh et al. 2005; Katoh and Standley 2013) and poorly aligned regions were eliminated using trimAl (Capella-Gutierrez et al. 2009) with a gap threshold set at 80%. Initial maximum likelihood (ML) tree reconstructions were performed with FastTree using the default options (Price et al. 2010) on trimmed alignments with a query coverage of ≥ 50% and at least five sequences in the alignment. Diatom-derived and dinoflagellate-derived queries and multiple queries for the same pathway gene resulted in very similar trees and the most comprehensive tree was chosen for each gene for final tree building. The tree for the heme pathway gene glutamate-1-semialdehyde 2,1-aminomutase (hemL/GSA-AT) was manually trimmed to remove a clade consisting of homologs for the related protein ornithine-delta-aminotransferase that was also recovered with the parameters used in this pipeline. Additionally, the trees for heme biosynthetic enzymes uroporphyrinogen decarboxylase (hemE/UROD) and coproporphyrinogen III oxidase (hemF/CPOX) were manually modified by removing selected taxa from strongly represented phyla to reduce the complexity of the tree (see supplementary table S2, Supplementary Material online, for trimmed taxa). Retained taxa were realigned and trimmed as before and final ML trees for every gene were reconstructed using RAxML with the LG amino acid substitution model + Γ distribution with four rate categories and 100 bootstrap replicates (Stamatakis 2006; Le and Gascuel 2008). To search for the putative protein-channel of the inner membrane plastid import machinery, Tic110, the homolog of the diatom Phaeodactylum tricornutum (accession number: XP_002185478) was used as a query in a BLASTP search against all dinotom transcriptome data sets (E value threshold ≤ 1e-5). The best hit was used for initial phylogenetic analysis as described above. The longest dinoflagellate-derived Tic110 homolog, as identified by FastTree analysis, was used for final tree building as described above.
Localization Prediction
Prediction of signal peptides as part of N-terminal bipartite leader sequences in diatom- as well as in dinoflagellate-derived transcripts was performed with the Hidden Markov Model of SignalP3.0 (Nielsen and Krogh 1998; Bendtsen et al. 2004) using the default truncation setting of 70 residues. TMHMM v.2.0 transmembrane helix prediction (Sonnhammer et al. 1998) was used to predict transmembrane domains in dinoflagellate-derived presequences. Hydrophobicity scores for the dinoflagellate presequences were calculated with the Kyte–Doolittle amino acid scale from ProtScale (http://web.expasy.org/protscale/, last accessed September 5, 2014), using the default parameters. Dinoflagellate as well as diatom-derived transcripts were manually aligned using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html, last accessed September 5, 2014).
Results and Discussion
A Large Fraction of Transcripts Is Diatom-Associated
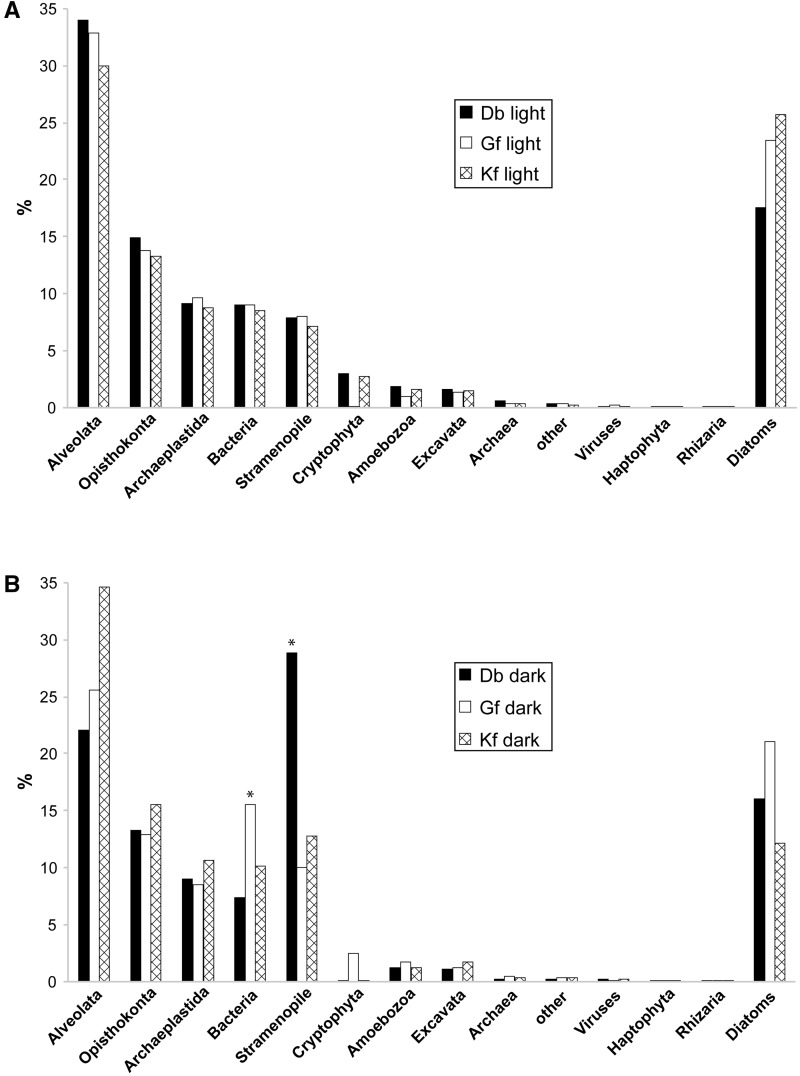
We investigated the transcriptomes of three different dinotom species, D. baltica, G. foliaceum, and K. foliaceum, under two different conditions: Cells harvested during the light phase or after an extended dark phase, resulting in three “light” and three “dark” data sets (see Materials and Methods). Due to the presence of the large, DNA-rich diatom endosymbiont nucleus in dinotoms, we expected the transcriptome data to contain not only dinoflagellate but also a considerable number of diatom transcripts. To estimate the fraction of transcripts encoded by the diatom nucleus, we first grouped sequences into putative dinoflagellate and diatom nucleus-encoded transcripts according to sequence similarity (E value < 1e-5) with dinoflagellate and diatom genes. Based on this, between 18% and 26% of transcripts are associated with diatoms in the light data sets, whereas the fraction of diatom-associated sequences in the dark data sets ranges from 12% to 21% (fig. 2A and B). Taking into account the relatively low stringency of the BLAST search, the distribution of E values was investigated: 78–83% of the transcripts in the data sets have a diatom best hit with an E value <1e-25, supporting the conclusion that those sequences are indeed of diatom-origin (supplementary fig. S1, Supplementary Material online). These results imply significant transcriptional activity of the endosymbiont nucleus in dinotoms, although to reliably pinpoint the origin of these transcripts large-scale phylogenetic analysis will be necessary.
Fig. 2.—
Taxonomic affinities of dinotom transcripts based on BLAST analyses. A bar chart shows the proportions of transcripts with top BLAST hits to various taxa for the transcriptome data sets of Durinskia baltica (Db), Glenodinium foliaceum (Gf), and Kryptoperidinium foliaceum (Kf) after filtering for peptides greater than 100 amino acids. Bars marked with an asterisk indicate subgroups deviating from the general pattern: (A) “light” data sets and (B) “dark” data sets.
Dinoflagellates have some of the largest eukaryotic genomes known (Wisecaver and Hackett 2011), a feature that has prevented the genomic exploration of these organisms: To date, only one draft genome of a core dinoflagellate has been assembled, that of Symbiodinium (Shoguchi et al. 2013). It is therefore not surprising that the majority of nondiatom-associated transcripts does not yield a dinoflagellate best hit. To distinguish between sampling bias and a possible contamination, the nondiatom-associated transcripts were grouped into several taxonomic categories according to their best-hit ID (fig. 2A and B). In all cases, except the “D. baltica dark” subset, the most abundant top hits corresponded to alveolates, as expected, and the overall distribution is similar between different dinotoms, altogether indicating that the majority of the transcripts probably originates from the host nucleus. Two subgroups deviate from the general pattern: The fraction of bacterial hits is distinctly higher in the “G. foliaceum dark” data and in the D. baltica dark subset the proportion of stramenopile hits is strongly increased (fig. 2A and B).
Plastid-Associated Pathways Are Present in Both Host and Endosymbiont
We searched the dinotom transcriptomes for pathways normally associated with plastids, and in particular those pathways that have been retained in other nonphotosynthetic plastids. The methylerythritol phosphate/1-deoxy-d-xylulose-5-phosphate (MEP/DOXP) pathway for the synthesis of isoprenoids is plastid-specific in all eukaryotes and has also been reported in the nonphotosynthetic plastids of apicomplexans and P. marinus (Clastre et al. 2007; Matsuzaki et al. 2008). Similarly, the heme biosynthesis pathway is located in the plastid of photosynthetic eukaryotes and also partially in the nonphotosynthetic apicoplast of apicomplexans, although here parts of the pathway are also found in the mitochondrion (Obornik and Green 2005; Nagaraj et al. 2010; Koreny et al. 2011). Therefore, those two pathways were deemed to be good candidates to have been retained in a relic, nonphotosynthetic plastid in the dinotom host.
Using homologs for genes in these two pathways from diatoms and dinoflagellates (or other alveolates if no dinoflagellate sequence was available), we retrieved sequences for the complete MEP/DOXP pathway and the first eight unique steps of the heme pathway from dinotoms (see Materials and Methods). To distinguish diatom- and dinoflagellate-derived transcripts, we carried out phylogenetic analyses for all genes in both pathways. For each step in both MEP/DOXP and heme pathways, at least one diatom-related gene was identified, suggesting that complete functional pathways of diatom-origin are present in dinotoms (table 1; figs. 3 and 4; supplementary figs. S2 and S3, Supplementary Material online). All sequences inferred to be diatom-derived by phylogenetic analyses were confirmed to have been identified as diatom-derived by our original BLAST-based filter.
Table 1.
Overview of the Phylogenetic Analysis of MEP/DOXP and Heme Pathway Transcripts
| Name | PA | Support PA (%) | Support Dino Clade (%) | Support Diatom Clade(s) (%) |
|---|---|---|---|---|
| dxs | α-proteo | >95 | >95 | >95 |
| dxr | cyano | >95 | >95 | >95 |
| ispD | PBE | >95 | 69 | >95 |
| ispE | PBE | >95 | >95 | >95 |
| ispF | PBE | >95 | 95 | >95 |
| ispG | PBE | >95 | >95 | >95 |
| ispH | cyano | >95 | >95 | >95 |
| hemA | cyano | >95 | >95 | >95 |
| hemL | cyano | >95 | <50 | >95 |
| hemB | cyano | 84 | >95 | >95 |
| hemC | α-proteo | 76 | >95 | >95 |
| hemD | PBE | <50 | >95* | 82 |
| hemE | cyano | >95 | >95 | >95/>95 |
| nuclear, prim. | >95 | 94* | >95 | |
| nuclear, dino | <50 | >95 | — | |
| hemF | nuclear, prim. | 92 | >95 | >95/<50 |
| unknown | >95 | — | 58 | |
| hemY | cyano | >95 | >95 | 51 |
Note.—The column “Phylogenetic Affinity (PA)” describes the putative origin of the respective transcript (α-proteo: α-proteobacterial; cyano: cyanobacterial; nuclear, prim.: primary algal nucleus; nuclear, dino: dinoflagellate nucleus) or the general phylogenetic affinity (PBE, Plastid Bearing Eukaryotes). Support values are bootstrap values as described in Materials and Methods. A dash indicates the complete absence of the respective clade. An asterisk next to the support value indicates the absence of dinotom sequences in the respective dinoflagellate clade.
Fig. 3.—
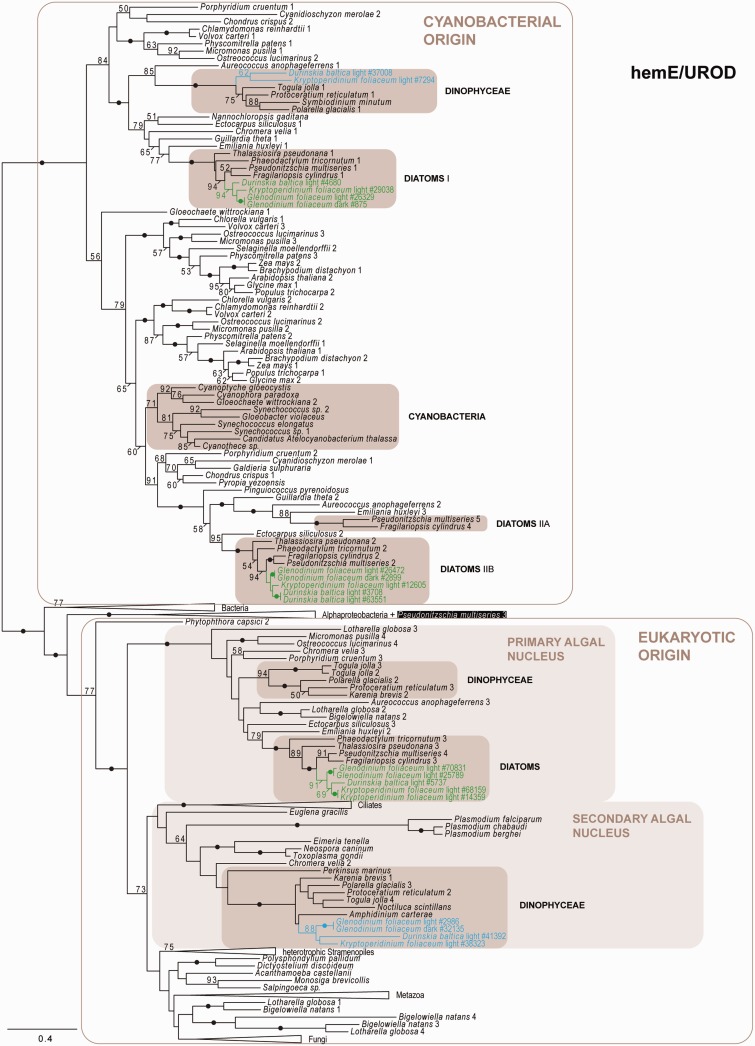
Phylogeny of hemE/UROD as inferred by ML (LG + Γ model), depicting several dinoflagellate- and diatom-derived hemE/UROD paralogs with different evolutionary origins: Cyanobacterial, from the primary algal nucleus (nucleus of a primary alga that may have been taken up as secondary endosymbiont), and/or the secondary algal nucleus (nucleus of a heterotrophic eukaryote that has taken up an alga). Black dots correspond to greater than 95% ML bootstrap support. Numbers at nodes represent bootstrap supports of greater than 50%. Shaded boxes indicate clades of interest. Blue and green texts indicate dinoflagellate- and diatom-derived transcripts, respectively; the numbers of the transcripts correspond to the original contig numbering of NCGR. The black box surrounding the paralog of “Pseudonitzschia multiseries 3” indicates the possibility of an alphaproteobacterial-derived form of hemE/UROD in this organism. The numbers after species names indicate different paralogs and are numbered according to node order. Similarly, the diatom clades of cyanobacterial origin are numbered according to node order. The scale bar represents the estimated number of amino acid substitutions per site.
Fig. 4.—
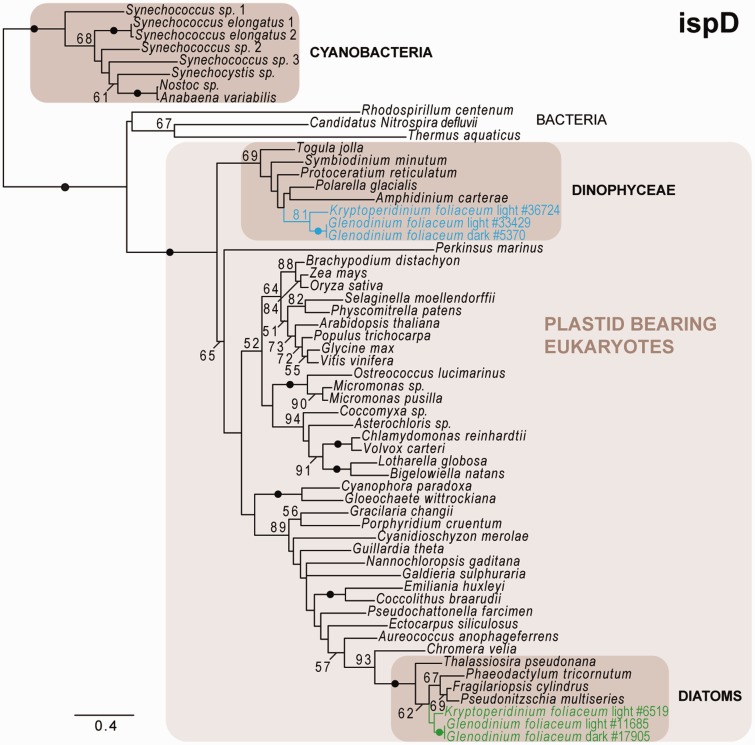
Phylogeny of ispD as inferred by ML (LG + Γ model), depicting dinoflagellate- and diatom-derived ispD homologs, with the Durinskia baltica homolog missing in both, the dinoflagellate- as well as the diatom-derived, clade. Dinoflagellate- and diatom-derived genes cluster with plastid-bearing eukaryotes but not with cyanobacteria. Black dots correspond to greater than 95% ML bootstrap support. Numbers at nodes represent bootstrap supports of greater than 50%. Shaded boxes indicate clades of interest. Blue and green texts indicate dinoflagellate- and diatom-derived transcripts, respectively; the numbers of the transcripts correspond to the original contig numbering of NCGR. The scale bar represents the estimated number of amino acid substitutions per site.
The plastid-associated hemE/UROD and hemF/CPOX have previously been shown to be encoded by several genes with different evolutionary origins (Koreny et al. 2011). Here, three versions of hemE/UROD were identified: Two group with cyanobacteria and photosynthetic eukaryotes, suggesting plastid-origin, whereas a third clusters with cytosolic homologs from primary and secondary algae, implying a nuclear origin, perhaps from the primary endosymbiont (fig. 3). hemF/CPOX is also represented by three different genes: One groups with photosynthetic eukaryotes but not cyanobacteria, and the other two hemF/CPOX fall into a large clade containing representatives from primary and secondary algae, suggesting a duplicated gene originating from the primary algal nucleus (supplementary fig. S3G, Supplementary Material online). Overall, diatom-derived homologs for every step in both pathways were identified in all three dinotom lineages; with the single exception that 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (ispD, the third step in the MEP/DOXP pathway) was not identified in D. baltica (fig. 4).
In addition to these diatom-related genes, phylogenetic analysis also revealed a distinct set of genes specifically related to dinoflagellate homologs (table 1). In the case of the MEP/DOXP pathway, a complete set of dinoflagellate-related genes was identified (fig. 4 and supplementary fig. S2, Supplementary Material online). The majority of these genes was found in all three dinotom lineages: The exceptions being 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (ispF; supplementary fig. S2D, Supplementary Material online), which was not identified in G. foliaceum, and ispD, which was not found in D. baltica (fig. 4). Whether these genes are truly absent or simply not sampled in these transcriptomes is unknown: The particular absence of both diatom and dinoflagellate ispD in D. baltica is noticeable, because the same gene appears to be absent in both expression studies and the genome of P. marinus (Matsuzaki et al. 2008). All dinoflagellate-related MEP/DOXP pathway genes cluster with plastid-bearing eukaryotes in the respective trees (supplementary fig. S2A–F, Supplementary Material online). The dinoflagellate-homologs of 1-deoxy-d-xylulose-5-phosphate reductoisomerase (dxr) and ispH also group with cyanobacteria, confirming the cyanobacterial origin for those genes also in dinotoms whereas the dxs tree supports the alphaproteobacterial origin for this gene. The dinoflagellate-homologs of (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ispG) cluster with plastid-bearing eukaryotes and cyanobacteria, but the latter form a separate clade with primary algae, as observed in the MEP/DOXP pathway study of P. marinus (Matsuzaki et al. 2008).
As with MEP/DOXP, the heme pathway in dinotoms is also almost completely represented by transcripts that group with high support with peridinin-containing dinoflagellates (fig. 3 and supplementary fig. S3, Supplementary Material online). One gene, encoding uroporphyrinogen-III synthase (hemD/UROS), could not be identified in any of the transcriptomes, even when applying less stringent BLAST search criteria before tree-reconstruction. Overall, however, relatively few hemD/UROS were identified (supplementary fig. S3E, Supplementary Material online), the gene shows a relatively low level of sequence conservation (Tan et al. 2008), and a high proportion of homologs were derived from complete genomes, so it is possible that these genes are simply difficult to identify and/or not expressed at a sufficiently high level to enable identification from transcriptomic data.
The remaining genes of the heme pathway were found in all three lineages, except hemF/CPOX, which was not identified in G. foliaceum (supplementary fig. S3G, Supplementary Material online), and hemY/PPOX, which was not identified in D. baltica (supplementary fig. S3F, Supplementary Material online). As with the diatom-derived genes, we found more than one dinoflagellate-derived gene encoding for hemE/UROD (fig. 3). One dinoflagellate hemE/UROD clusters with one of the diatom-derived plastid hemE/UROD genes, whereas a second is probably derived from the dinoflagellate nucleus. A third clade contains sequences from other dinoflagellates and branches with strong support with the diatom-associated hemE/UROD probably originating from the primary algal nucleus; however, this clade contains no dinotom sequences. Only a single dinoflagellate-derived hemF/CPOX was found (supplementary fig. S3G, Supplementary Material online). All other heme pathway genes (hemA/GTR, hemL/GSA-AT, porphobilinogen synthase [hemB/ALAD], and hemY/PPOX) cluster with plastid-bearing eukaryotes and cyanobacteria, corresponding to their cyanobacterial origin, with the exception of hydroxymethylbilane synthase (hemC/PBGD), forming a clade with plastid-bearing eukaryotes and alphaproteobacteria (supplementary fig. S3A–F, Supplementary Material online).
No Evidence for Endosymbiotic Gene Transfer in Dinotom Plastid Genes
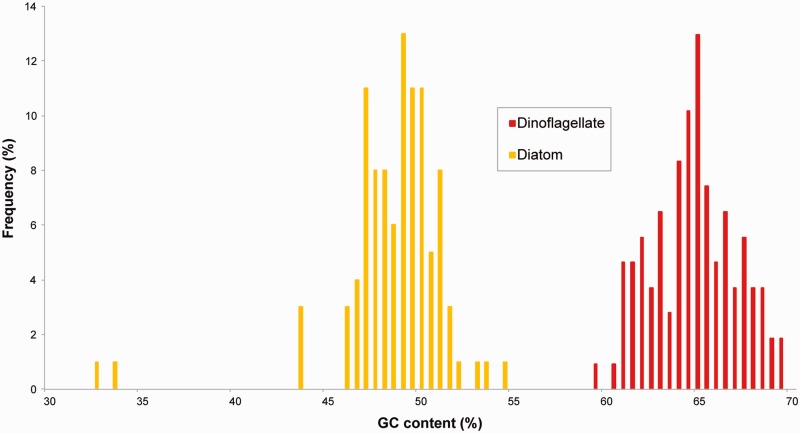
To date, there has been no strong evidence for endosymbiotic gene transfer in dinotoms, so the null hypothesis for diatom-related transcripts is that they are encoded in the endosymbiont nucleus. To investigate the possibility that any of these plastid-associated genes have been transferred to the host nucleus, we analyzed the GC content of diatom-derived as well as of dinoflagellate-derived transcripts from both pathways. It has recently been shown that the diatom and dinoflagellate genomes differ significantly in GC content, with diatom transcripts having a GC content less than 55%, whereas most dinoflagellate transcripts are above 55% (Burki et al. 2014; Imanian and Keeling 2014). The GC content of all genes involved in MEP/DOXP and heme pathways was consistent with this, falling into two distinct groups that correlate with phylogeny: The GC content of transcripts clustering with diatoms is constantly below 55%, ranging from 32% to 54 %, whereas the transcripts branching with dinoflagellates range from 58% to 69% GC (fig. 5). This distribution is not consistent with any endosymbiotic gene transfer, but rather suggests that the diatom-related genes are still encoded in the endosymbiont genome and dinoflagellate-related genes remain in the host genome. None of the transcripts investigated here was found to encode the dinoflagellate-specific spliced leader at the N-terminus (Zhang et al. 2007).
Fig. 5.—
GC contents of dinoflagellate-derived and diatom-derived transcripts do not support endosymbiotic gene transfer for the MEP/DOXP and heme pathway in dinotoms. A histogram shows the frequency distribution of the GC contents of all dinoflagellate-derived and diatom-derived transcripts present in the phylogenetic analyses.
Plastid-Pathway Proteins Carry N-Terminal Plastid Targeting Peptides
The presence of transcripts for plastid-derived genes in the dinotom host genome raises the question whether the protein products are potentially targeted to a plastid-derived organelle, such as the eyespot may represent.
The MEP/DOXP and the heme pathway transcripts with diatom-origin are expected to be targeted to the endosymbiont plastids, as that is the case in free-living diatoms (Obornik and Green 2005). To confirm this, we sought to identify the characteristic N-terminal bipartite leaders similar to those used by diatoms to target nucleus-encoded proteins to the plastid (Apt et al. 2002; Kilian and Kroth 2004b). These sequences consist of a signal peptide followed by a transit peptide-like sequence, with a highly conserved phenylalanine in an “ASAFAP” motif between them that is essential for plastid import in diatoms (Kilian and Kroth 2004a; Gruber et al. 2007).
All diatom-derived transcripts for every step in the heme pathway are intact at the 5′-end in all three dinotom lineages, and in every case they were predicted to encode a signal peptide. The conserved phenylalanine is present at every cleavage site, except in one CPOX paralog in G. foliaceum, where it is substituted by a tryptophan residue. The “ASAFAP”-like motif is conserved in all three lineages, with the second position being the least conserved, as observed in free-living diatoms (Gruber et al. 2007) (supplementary fig. S4, Supplementary Material online). All three forms of UROD encode bipartite leaders in all three dinotoms, as do all three forms of CPOX if present, however the 5′-ends of some CPOX genes were missing in some lineages. Similarly, all diatom-derived transcripts for genes in the MEP/DOXP pathway also encode a signal peptide and contain the cleavage site motif, with the exception of the D. baltica dxr and ispE, which are truncated (supplementary fig. S5, Supplementary Material online). Altogether, these data show, not surprisingly, that the diatom endosymbiont retains a functional plastid targeting system and targets proteins for heme and isoprenoid biosynthesis to the plastid.
To assess the presence of possible plastid-targeting sequences in the dinoflagellate host, we used a slightly different strategy, because targeting to peridinin-containing plastids differs from targeting to other secondary plastids of red algal origin and is not as well understood. First, we examined all MEP/DOXP and heme biosynthetic genes for the presence of N-terminal extensions in comparison to cyanobacterial homologs. Next, we analyzed the transcripts with extensions for features typically associated with dinoflagellate peridinin plastid-targeted sequences. Protein-targeting to peridinin plastids also uses a signal peptide and a transit peptide-like domain (Nassoury et al. 2003); however, identifying presequences is more complex because there are two different types of transit peptide (Patron et al. 2005). Class I transit peptides contain a hydrophobic transmembrane domain in the transit peptide before the predicted start of the mature protein, whereas class II peptides lack such a domain in the transit peptide. We found N-terminal extensions in six of seven MEP/DOXP pathway proteins and four of eight heme pathway proteins. Of those, all but one also encoded a transmembrane domain before the start of the mature protein. Moreover, the predicted transmembrane helices were immediately followed by a region with a strong negative hydrophobicity score, dominantly featuring arginine residues, which is also consistent with the observations made in dinoflagellates with peridinin-containing plastids (Patron et al. 2005) (fig. 6A). Overall, these features are consistent with these proteins being targeted to a plastid-derived organelle, mediated by a dinoflagellate class I transit peptide.
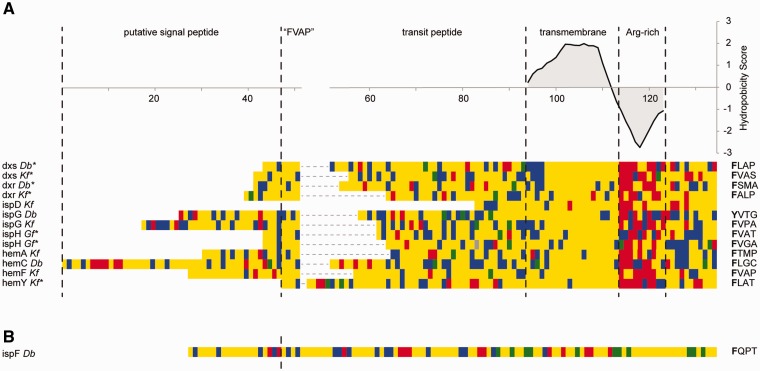
Fig. 6.—
Dinoflagellate-derived transcripts contain characteristic dinoflagellate-plastid targeting sequences. (A) Class I transit peptides, containing a transmembrane domain: Manual alignment of N-terminal regions of MEP/DOXP and heme pathway transcripts at the “FVAP” motif and their transmembrane regions, respectively. The average hydrophobicity score for each column in the transmembrane and arginine-rich domain is plotted above the alignment. The “FVAP” motif for every sequence is displayed next to the alignment. Note: The phenylalanine in the Durinskia baltica ispG was substituted by tyrosine, another hydrophobic residue. An asterisk next to the transcript name indicates a truncated signal peptide. Amino acid color code: Yellow, hydrophobic; blue, polar; green, negatively charged; red, positively charged. (B) Class II transit peptide: The dinoflagellate-derived presequence lacks a transmembrane domain in the transit peptide but contains the characteristic “FVAP” motif at the signal peptide cleavage site.
Distinguishing signal peptides and transmembrane domains can be difficult as both have similar characteristics, so we manually evaluated each to identify cases where a transmembrane region was misidentified as a possible signal peptide (fig. 6A). In several sequences, the region between the N-terminus and the transmembrane region is comparatively short, suggesting a truncated signal peptide caused false signal peptide prediction in a transmembrane domain. To estimate the true length and position of the signal peptide, we therefore searched the sequence upstream of the transmembrane helix for the most conspicuous characteristic of dinoflagellate transit peptides: A conserved phenylalanine residue at or close to their N-terminus with none of the three following residues being basic or acidic (Patron et al. 2005; Patron and Waller 2007). This “FVAP” motif was found in the N-terminal region of all transcripts with transmembrane domains, except for K. foliaceum ispD, which is truncated four amino acids upstream of the predicted transmembrane region (fig. 6A). Assuming that this motif is at or close to the cleavage site of the signal peptide, this suggests that the transcripts for dxs, dxr, and ispH as well as hemY are truncated in the signal peptide.
The transcript for ispF of the MEP/DOXP pathway completely lacked any evidence for a transmembrane domain, but it does encode a complete 5′-end, including a predicted signal peptide followed by the phenylalanine-containing motif typical for dinoflagellate transit peptides (fig. 6B). This suggests that ispF encodes a class II transit peptide.
In contrast to the dinoflagellate-derived transcripts, no transmembrane helices were predicted in the transit peptides in any of the diatom-derived transcripts (supplementary fig. S5, Supplementary Material online), further supporting the presence of two different targeting systems and in extension, the possible presence and maintenance of two distinct plastids.
When comparing the dinoflagellate-derived sequences in dinotoms with their corresponding homologs in photosynthetic dinoflagellates harboring a canonical peridinin-plastid (see supplementary table S2, Supplementary Material online, for a list of dinoflagellate data sets available), we observed the same distribution of class I and class II transit peptides. All homologs that contain a transmembrane domain in their transit peptide in the investigated dinotom species also contain a transmembrane region in other peridinin-plastid harboring dinoflagellates (supplementary fig. S6A, Supplementary Material online). Similarly, the transcript for ispF, which lacks a transmembrane domain in dinotoms, does not contain such a domain in other dinoflagellates either (supplementary fig. S6B, Supplementary Material online). Altogether this conservation of class I and class II transit peptides in dinotoms supports that the plastid targeting system found in dinotoms is derived from the original peridinin-plastid.
The Host Expresses an Essential Component of the Plastid Import Machinery
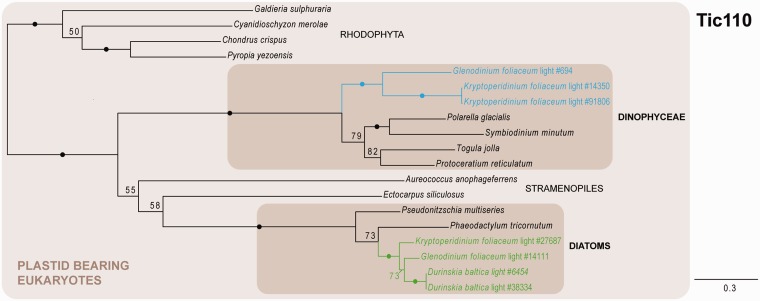
The import of nuclear-encoded proteins into the plastid is facilitated by specialized protein complexes, the translocons of the inner and outer chloroplast envelope membrane (Tic and Toc, respectively). Although only one of the components of the Toc machinery (Toc75/Omp85) could be identified in organisms with secondary plastids so far (Bullmann et al. 2010; Hirakawa et al. 2012), several components of the translocon at the inner membrane were found in secondary plastid-harboring algae, including the nonphotosynthetic apicomplexans (McFadden and van Dooren 2004; van Dooren et al. 2008; Glaser et al. 2012; Petersen et al. 2014). Among the Tic proteins, the putative protein-conducting pore Tic110 represents one of the central components of the inner membrane apparatus, directly interacting with the Toc complex and all other Tic components (Gross and Bhattacharya 2009). We were able to identify dinoflagellate-derived homologs of Tic110 in G. foliaceum and K. foliaceum based on similarity, however not in D. baltica (fig. 7). Overall we recovered only a relatively small number of homologs from our custom protein database, even when applying less stringent parsing criteria on the BLAST search results before tree building, identifying only organisms with a secondary plastid from red algal origin and several red algal species. This is probably reflecting the low degree of conservation of Tic110 (Kalanon and McFadden 2008), which seems to be particularly divergent between red and green algae. Altogether, the presence of a protein with such a central function in the translocation process into the plastid adds further support to the idea that the dinotom host retained a plastid-like organelle.
Fig. 7.—
Phylogeny of Tic110 as inferred by ML (LG + Γ model), depicting dinoflagellate- and diatom-derived Tic110 homologs. Black dots correspond to greater than 95% ML bootstrap support. Numbers at nodes represent bootstrap supports of greater than 50%. Shaded boxes indicate clades of interest. Blue and green texts indicate dinoflagellate- and diatom-derived transcripts, respectively; the numbers of the transcripts correspond to the original contig numbering of NCGR. The scale bar represents the estimated number of amino acid substitutions per site.
Concluding Remarks
The fate of the peridinin-containing plastid in the dinoflagellate host of dinotoms has been the subject of some conjecture as the morphological similarities between the eyespot and peridinin plastids were first observed (Dodge and Crawford 1969). Here, we provide evidence that the dinoflagellate host genome has retained genes for at least two metabolic pathways derived from this plastid, and show that their protein products are likely targeted to a relict plastid structure. The eyespot is indeed the most obvious candidate for a relict plastid, and in the future this could be tested by localizing proteins in the MEP/DOXP and heme pathways using antibodies specific to the dinoflagellate-derived proteins. The fact that the same pathways are also present in the endosymbiont extends the functional redundancy that was already shown for the dinotom mitochondria to certain plastidial functions. As to why specifically those two pathways were retained in both the endosymbiont and the host we can for only speculate. Both pathways produce precursors for numerous diverse metabolites that need to be transported into different subcellular compartments and are highly regulated (Mochizuki et al. 2010; Zhao et al. 2013). Loss of those pathways in either host or endosymbiont may therefore require several transporters to provide the host and/or endosymbiotic partners with the necessary intermediates in a well-regulated manner. It is possible that dinotoms have not (yet) evolved a sufficiently well-integrated set of transporters and the retention of redundant pathways is the results. Whether this is an “indermediate stage” in an ongoing process of integration or not is an interesting question, as dinotoms in several ways blur the distinction between endosymbiont and organelle.
Supplementary Material
Supplementary figures S1–S6 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (227301 to P.J.K.), and a grant to the Centre for Microbial Diversity and Evolution from the Tula Foundation. This work was also supported by a prospective researcher postdoctoral fellowship from the Swiss National Science Foundation to E.H. and a doctoral scholarship from NSERC to B.I. P.J.K. is a Fellow of the Canadian Institute for Advanced Research. The authors thank Martin Kolisko for valuable discussion about and comments on the phylogenetic analyses.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apt KE, et al. In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci. 2002;115:4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bullmann L, et al. Filling the gap, evolutionarily conserved omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, et al. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryot Cell. 2014;13:246–255. doi: 10.1128/EC.00299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Lee JJ. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J Protozool. 1985;32:376–379. [Google Scholar]
- Chesnick JM, Kooistra WHCF, Wellbrock U, Medlin LK. Ribosomal RNA analysis indicates a benthic pennate diatom ancestry for the endosymbionts of the dinoflagellates Peridinium foliaceum and Peridinium balticum (Pyrrhophyta) J Eukaryot Microbiol. 1997;44:314–320. doi: 10.1111/j.1550-7408.1997.tb05672.x. [DOI] [PubMed] [Google Scholar]
- Chesnick JM, Morden CW, Schmieg AM. Identity of the endosymbiont of Peridinium foliaceum (Pyrrophyta): analysis of the rbcLS operon. J Phycol. 1996;32:850–857. [Google Scholar]
- Clastre M, et al. The methylerythritol phosphate pathway for isoprenoid biosynthesis in coccidia: presence and sensitivity to fosmidomycin. Exp Parasitol. 2007;116:375–384. doi: 10.1016/j.exppara.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Dodge JD. The fine structure of chloroplasts and pyrenoids in some marine dinoflagellates. J Cell Sci. 1968;3:41–48. doi: 10.1242/jcs.3.1.41. [DOI] [PubMed] [Google Scholar]
- Dodge JD. Dinoflagellate with both a mesocaryotic and a eucaryotic nucleus. 1. Fine structure of nuclei . Protoplasma. 1971;73:145–157. doi: 10.1007/BF01275591. [DOI] [PubMed] [Google Scholar]
- Dodge JD. A survey of chloroplast ultrastructure in the dinophyceae. Phycologia. 1975;14:253–263. [Google Scholar]
- Dodge JD. The functional and phylogenetic significance of dinoflagellate eyespots. Biosystems. 1984;16:259–267. doi: 10.1016/0303-2647(83)90009-6. [DOI] [PubMed] [Google Scholar]
- Dodge JD, Crawford RM. Observations on the fine structure of the eyespot and associated organelles in the dinoflagellate Glenodinium foliaceum. J Cell Sci. 1969;5:479–493. doi: 10.1242/jcs.5.2.479. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Turner S. Molecular evidence for the origin of plastids from a cyanobacterium-like ancestor. J Mol Evol. 1991;33:267–273. doi: 10.1007/BF02100678. [DOI] [PubMed] [Google Scholar]
- Fernández Robledo JA, et al. The search for the missing link: a relic plastid in Perkinsus? Int J Parasitol. 2011;41:1217–1229. doi: 10.1016/j.ijpara.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa RI, Bravo I, Fraga S, Garcés E, Llaveria G. The life history and cell cycle of Kryptoperidinium foliaceum, a dinoflagellate with two eukaryotic nuclei. Protist. 2009;160:285–300. doi: 10.1016/j.protis.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Foster KW, Smyth RD. Light antennas in phototactic algae. Microbiol Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, et al. Tic22 is an essential chaperone required for protein import into the apicoplast. J Biol Chem. 2012;287:39505–39512. doi: 10.1074/jbc.M112.405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D. Revaluating the evolution of the Toc and Tic protein translocons. Trends Plant Sci. 2009;14:13–20. doi: 10.1016/j.tplants.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Gruber A, et al. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- Hackett JD, Maranda L, Yoon HS, Bhattacharya D. Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae) J Phycol. 2003;39:440–448. [Google Scholar]
- Hirakawa Y, Burki F, Keeling PJ. Genome-based reconstruction of the protein import machinery in the secondary plastid of a chlorarachniophyte alga. Eukaryot Cell. 2012;11:324–333. doi: 10.1128/EC.05264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi T, Kawai H, Kubota M, Takahashi T, Watanabe M. Phototactic responses of four marine dinoflagellates with different types of eyespot and chloroplast. Phycol Res. 1999;47:101–107. [Google Scholar]
- Horiguchi T, Pienaar RN. Ultrastructure and ontogeny of a new type of eyespot in dinoflagellates. Protoplasma. 1994a;179:142–150. [Google Scholar]
- Horiguchi T, Pienaar RN. Ultrastructure of a new marine sand-dwelling dinoflagellate, Gymnodinium quadrilobatum sp. nov. (Dinophyceae) with special reference to its endosymbiotic alga. Eur J Phycol. 1994b;29:237–245. [Google Scholar]
- Imanian B, Keeling PJ. The dinoflagellates Durinskia baltica andKryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evol Biol. 2007;7:172. doi: 10.1186/1471-2148-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Keeling PJ. Horizontal gene transfer and redundancy of tryptophan biosynthetic enzymes in dinotoms. Genome Biol Evol. 2014;6:333–343. doi: 10.1093/gbe/evu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Pombert J-F, Dorrell RG, Burki F, Keeling PJ. Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One. 2012;7:e43763. doi: 10.1371/journal.pone.0043763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Pombert J-F, Keeling PJ. The complete plastid genomes of the two “dinotoms” Durinskia baltica and Kryptoperidinium foliaceum. PLoS One. 2010;5:e10711. doi: 10.1371/journal.pone.0010711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey SW, Sielicki M, Haxo FT. Chloroplast pigment patterns in dinoflagellates. J Phycol. 1975;11:374–384. [Google Scholar]
- Kalanon M, McFadden GI. The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics. 2008;179:95–112. doi: 10.1534/genetics.107.085704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91:1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- Kempton JW, et al. Kryptoperidinium foliaceum blooms in South Carolina: a multi-analytical approach to identification. Harmful Algae. 2002;1:383–392. [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2004a;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- Kilian O, Kroth PG. Presequence acquisition during secondary endocytobiosis and the possible role of introns. J Mol Evol. 2004b;58:712–721. doi: 10.1007/s00239-004-2593-z. [DOI] [PubMed] [Google Scholar]
- Kim M, Nam SW, Shin W, Coats DW, Park MG. Dinophysis caudata (Dinophyceae) sequesters and retains plastids from the mixotrophic ciliate prey Mesodinium rubrum. J Phycol. 2012;48:569–579. doi: 10.1111/j.1529-8817.2012.01150.x. [DOI] [PubMed] [Google Scholar]
- Kite GC, Dodge JD. Structural organization of plastid DNA in two anomalously pigmented dinoflagellates. J Phycol. 1985;21:50–56. [Google Scholar]
- Koreny L, Sobotka R, Janouskovec J, Keeling PJ, Obornik M. Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell. 2011;23:3454–3462. doi: 10.1105/tpc.111.089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Lee R, et al. Analysis of EST data of the marine protist Oxyrrhis marina, an emerging model for alveolate biology and evolution. BMC Genomics. 2014;15:122. doi: 10.1186/1471-2164-15-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: a plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol Biol Evol. 2008;25:1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J Phycol. 2001;37:951–959. [Google Scholar]
- McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, et al. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 2010;15:488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Nagaraj VA, Arumugam R, Prasad D, Rangarajan PN, Padmanaban G. Protoporphyrinogen IX oxidase from Plasmodium falciparum is anaerobic and is localized to the mitochondrion. Mol Biochem Parasitol. 2010;174:44–52. doi: 10.1016/j.molbiopara.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- Obornik M, Green BR. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol. 2005;22:2343–2353. doi: 10.1093/molbev/msi230. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Delwiche CF. The origin and evolution of plastids and their genomes. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II. Boston (MA): Springer US; 1998. pp. 375–409. [Google Scholar]
- Patron NJ, Waller RF. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. BioEssays. 2007;29:1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348:1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Petersen J, et al. Chromera velia, endosymbioses and the rhodoplex hypothesis—plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages) Genome Biol Evol. 2014;6:666–684. doi: 10.1093/gbe/evu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienaar RN, Sakai H, Horiguchi T. Description of a new dinoflagellate with a diatom endosymbiont, Durinskia capensis sp. nov. (Peridiniales, Dinophyceae) from South Africa. J Plant Res. 2007;120:247–258. doi: 10.1007/s10265-006-0047-y. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010;281:161–228. doi: 10.1016/S1937-6448(10)81005-6. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tan F-C, et al. Identification and characterization of the Arabidopsis gene encoding the tetrapyrrole biosynthesis enzyme uroporphyrinogen III synthase. Biochem J. 2008;410:291. doi: 10.1042/BJ20070770. [DOI] [PubMed] [Google Scholar]
- Tengs T, et al. Phylogenetic analyses indicate that the 19'Hexanoyloxy-fucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin. Mol Biol Evol. 2000;17:718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- Tippit DH, Pickettheaps JD. Apparent amitosis in binucleate dinoflagellate Peridinium balticum. J Cell Sci. 1976;21:273–289. doi: 10.1242/jcs.21.2.273. [DOI] [PubMed] [Google Scholar]
- Tomas RN, Cox ER. Observations on the symbiosis of Peridinium balticum and its intracellular alga. I. Ultrastructure. J Phycol. 1973;9:304–323. [Google Scholar]
- Tomas RN, Cox ER, Steidinger KA. Peridinium balticum (Levander) Lemmermann, an unusual dinoflagellate with a mesocaryotic and an eucaryotic nucleus. J Phycol. 1973;9:91–98. [Google Scholar]
- van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci U S A. 2008;105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. Dinoflagellate genome evolution. Annu Rev Microbiol. 2011;65:369–387. doi: 10.1146/annurev-micro-090110-102841. [DOI] [PubMed] [Google Scholar]
- Withers NW, Haxo FT. Isolation and characterization of carotenoid-rich lipid globules from Peridinium foliaceum. Plant Physiol. 1978;62:36–39. doi: 10.1104/pp.62.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci U S A. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chang WC, Xiao Y, Liu HW, Liu P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem. 2013;82:497–530. doi: 10.1146/annurev-biochem-052010-100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.