Abstract
Integrative analysis of multi-level molecular profiles can distinguish interactions that cannot be revealed based on one kind of data in the analysis of cancer susceptibility and metastasis. DNA copy number variations (CNVs) are common in cancer cells, and their role in cell behaviors and relationship to gene expression (GE) is poorly understood. An integrative analysis of CNV and genome-wide mRNA expression can discover copy number alterations and their possible regulatory effects on GE. This study presents a novel framework to identify important genes and construct potential regulatory networks based on these genes. Using this approach, DNA copy number aberrations and their effects on GE in lung cancer progression were revealed. Specifically, this approach contains the following steps: (1) select a pool of candidate driver genes, which have significant CNV in lung cancer patient tumors or have a significant association with the clinical outcome at the transcriptional level; (2) rank important driver genes in lung cancer patients with good prognosis and poor prognosis, respectively, and use top-ranked driver genes to construct regulatory networks with the COpy Number and EXpression In Cancer (CONEXIC) method; (3) identify experimentally confirmed molecular interactions in the constructed regulatory networks using Ingenuity Pathway Analysis (IPA); and (4) visualize the refined regulatory networks with the software package Genatomy. The constructed CNV/mRNA regulatory networks provide important insights into potential CNV-regulated transcriptional mechanisms in lung cancer metastasis.
Keywords: lung cancer, DNA copy number variation, mRNA gene expression, regulatory networks
Introduction
Lung cancer has the highest mortality rate for both men and women in the world. Lung cancer includes non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC constitutes 85% of all lung cancer cases, and the other 15% lung cancers are SCLC. NSCLC has two major subtypes of histology: squamous cell lung carcinoma and lung adenocarcinoma (AC).1 Surgery is the main treatment option for stage I NSCLC.2 Unfortunately, 35–50% of stage I NSCLC patients will die from relapse or metastasis within 5 years after surgery and adjuvant chemotherapy for patient stages II and III have not shown good results.3 It is a challenging issue for physicians to identify patients for chemotherapy. In spite of a few studies describing transcriptional profiling for lung cancer prognosis,3,4 currently, there is no fully validated clinical model for predicting lung cancer prognosis or chemoresponse.5 Therefore, it remains an important research issue to identify prognostic and predictive genes for improving lung cancer treatment.
In the search of clinically important biomarkers, many studies have ranked genes based on clinical outcome and used top-ranked genes in a classifier.6 However, rank-based feature selection methods cannot model the complex interactions among genes in disease.7 It has been shown that the genes that have a statistically significant association with outcome results are not necessarily good classifiers.8–10 Discovering networks of molecular interactions is much more important than finding a list of genes in personalized medicine.11 Molecular network analysis had been shown to be useful in disease classification12 and identification of novel therapeutic targets.13
DNA copy number variations (CNVs) refer to the copy number changes in a chromosomal segment, often observed in tumor tissues.14,15 Several studies have shown that CNV may be linked to the change of expression levels in some genes.14,16,17 Recent studies estimate that CNVs are responsible for >15% of heritable variation in gene expression (GE).18 The relation between GE and DNA copy number is complex. For cancer cells in general, the impact of CNV on GE is found to be important. Many studies have investigated the relation between CNV and GE. Many of them consider correlation between GE and CNV, in a manner of gene by gene across all samples.19,20 Nevertheless, the correlation between CNV and GE was not consistent across all genes.21 Moreover, each cancer is unique and has its own driver genes. Driver genes are involved in cancer pathogenesis, in contrast to “passenger genes,” which mutate during pathogenesis without having an effect on cancer.22 It is an important issue to identify driver genes and distinguish them from “passenger genes” that have no discerning advantage.23 Integrative analysis of CNV and GE could recognize potential cancer driver genes.24
A challenge in finding the candidate drivers is that the number of regulators (potential candidate drivers) is large and is related to the amount of available data. As CNV regions contain a large number of genes, it is difficult to identify a driver gene that is a correct regulator.25 Therefore, there is a need to use some information in addition to DNA copy number, such as GE, in the search for the functionally important driver genes. CONEXIC23 is a method that combines both CNV and GE to detect driver genes located in a deleted or amplified region and constructs regulatory networks based on the identified driver genes. Each potential driver gene changes in some tumors and is considered to have a major role in regulating expression of a group of genes.23 In this study, we present a novel framework based on CONEXIC to integrate CNV and GE and apply it to model lung cancer progression. First, candidate driver genes were selected from a pool of genes that were either amplified or deleted in NSCLC tumors, or were associated with metastasis at DNA copy number level or mRNA expression level. Second, CONEXIC was used to construct regulatory networks based on these candidate drivers and rank the importance of the driver genes. Third, because the predicted regulatory networks were large, Ingenuity Pathway Analysis (IPA) was used to reduce them to the networks of experimentally validated interactions. Finally, these refined regulatory networks were visualized with Genatomy.26
Materials and Methods
DNA copy number and mRNA profiles in patient cohorts
Two datasets were used in this study. The first dataset contains 271 NSCLC tumor samples with DNA copy number profiles. These 271 samples are histologically divided into lung AC (n = 179) and squamous cell carcinoma (SQCC; n = 92). This dataset also contains GE profiles for 49 samples (n = 29 for AC; n = 20 for SQCC). This dataset is available in NCBI Gene Expression Omnibus (GEO) with accession number GSE31800. DNA copy number profiles of the first dataset were quantified for each sample with whole-genome tiling path array comparative genomic hybridization (aCGH). Details of the genomic array, DNA extraction, labeling, and hybridization were described previously.27 aCGH is a technique for measuring the changes in chromosomal segments.28 The main difference between CGH and mRNA expression is that DNA is hybridized rather than mRNA transcript.29 CGH consists of log-ratio normalized intensities from disease versus normal samples. With resolution enhancements, aCGH is becoming more powerful. Consequently, this method has more advantages comparing to cytogenetic techniques such as fluorescence in situ hybridization (FISH).29 The GE for this dataset was generated by Affymetrix GeneChips30 and is available at GEO with accession number GSE31800.
The second dataset includes SQCC tumors with DNA copy number profiles (n = 201) and mRNA expression profiles (n = 132). DNA copy number values were generated with Agilent 415 microarrays.31 Copy number estimations for each tumor were refined using tangent normalization. Tangent normalization divides tumor signals by signal intensities from the linear combination of all normal samples.31 GE of this dataset was quantified using Agilent 244k microarrays. Preparation, hybridization, and processing to produce GE was previously described,32 and the dataset is available in The Cancer Genome Atlas (TCGA) data portal1 Lung SQCC section.31 Clinical information of these two patient cohorts is shown in Table 1.
Table 1.
Clinical information of patient cohorts analyzed in this study.
| VARIABLES | TCGA DATASET (31) (n=201) | GEO DATASET (GSE31800; n=271) |
|---|---|---|
| Gender | ||
| Male | 28% | NA |
| Female | 72% | NA |
| Race | ||
| White | 65% | NA |
| Black or African American | 5% | NA |
| Asian | 2% | NA |
| NA | 28% | NA |
| Histological type | ||
| Lung adenocarcinoma | 0 | 179 |
| Lung squamous cell carcinoma | 201 | 92 |
| Age | ||
| Mean ± std | 67.5 ± 8.5 | NA |
| [Min, Max] (Median) | [39, 85] (68) | NA |
| Vital status | ||
| Alive | 58% | NA |
| Dead | 42% | NA |
| Pathological tumor stage | ||
| Stage I | 55% | NA |
| Stage II | 24% | NA |
| Stage III | 21% | NA |
| Ethnicity | ||
| Not Hispanic or Latino | 59% | NA |
| Hispanic or Latino | 2% | NA |
| NA | 39% | NA |
| Tobacco smoking history | ||
| Current Smoker | 16% | NA |
| Reformed smoker for < or = 15 years | 56% | NA |
| Reformed smoker for > 15 years | 21% | NA |
| Non-Smoker | 4% | NA |
| NA | 3% | NA |
Matching DNA probes to genes
DNA copy number values were assigned to the corresponding genes with a mapping scheme. DNA copy number datasets contain information about the chromosomal location of each probe and its copy number value. There is no chromosomal information of genes in GE datasets, requiring the use of some public repositories such as UCSC Genome Browser2 and MatchMiner3. To find the chromosomal location for each gene, gene names should be entered to the software in a batch mode, and the output includes chromosomal location. The DNA copy number probes must then be matched to the corresponding genes. Among different methods to match genes to copy number probes, distance matching33 was used. This method matches each gene to the closest probe that is on the same chromosome.
Detecting DNA copy number aberrations
There is a need to convert raw log-ratio values in the original aCGH data to absolute measures of DNA copy numbers. These absolute measures usually have three states: loss (less than two copies), normal (two copies), and gain (three to four copies). This process is named “calling.”34 CGHcall was used to detect these different DNA copy number states.34 CGHcall classifies different regions of CNV based on mixture models. First, it uses circular binary segmentation (CBS) to segment DNA copy number values of aCGH. The CBS method has been shown as one of the strongest methods for segmentation.35 Second, CGHcall considers the fact that loss, normal, and gain levels are not uniform across all the samples. It allows fluctuation by using random effects. Finally, it uses a combination of segmentation results and mixture model to figure out the most likely state per segment.34 This package was used in its default setting with human as the tissue type.
CONEXIC
CONEXIC23 is a computational algorithm that performs integrative analysis of CNV and GE in cancer. This algorithm is based on module network36 and combines matched GE and CNV of the samples to identify cancer driver genes. CONEXIC uses a Bayesian scoring function to detect the combination of modulators among the amplified or deleted regions, which explains the behaviors of GE modules across patient samples. The score measures how well a modulator can predict the behaviors of a GE module. CONEXIC searches for high-score modulators to recognize most probable driver genes in a stepwise manner: (1) selection of candidate drivers, (2) a single modulator step that builds an initial network between candidate drivers and gene modules, and (3) an iterative network learning step that improves the initial model. This method not only searches for important driver genes but also constructs regulatory networks. The concept of regulation networks or regulation program originates from Segal et al.36 This software package was used in its bootstrapping mode.
A score function was used to rank cancer driver genes in the CONEXIC method. The score function is a Bayesian function that maximizes the joint probability of data and model structure. Let D be the data and S represent the structure of the network. The scoring function is:
The first term is the likelihood of the data for a given model, and it has a normal gamma distribution. The normal gamma distribution function gives higher score to the data with lower variance. Therefore, it splits the data into two completely different distributions.
The second part is a priori on the structure of data, which is a penalty score on network complexity. The penalty function has two parts. The first part penalizes the number of leaves in each regulation program using exponential distribution over the total number of leaves. The second part is a network width penalty function that penalizes (1) the number of genes in the module for each modulator and (2) the number of distinct split values that a modulator has. Hence, the scoring function would be:
The parameters of the score are β, x, y, which can be determined by optimizing the log-likelihood of data through cross-validation test.23,37
IPA
IPA4 is a curated database that enables the search and visualization of experimentally confirmed molecular interactions and networks. Published molecular interactions can be explored from this web-based software application. This database gathers information from multiple resources, including experimental repositories or text collections of published literatures. In this study, only direct interactions among genes in human tissues were considered experimentally and confirmed molecular interactions in IPA analysis.
Genatomy
After constructing regulatory networks with CONEXIC and refining them using IPA, Genatomy was used for visualizing the networks. Genatomy is a software toolset for visualizing biological data such as GE, genotypes, and DNA copy number information.26 The software package was used based on its default settings. The CNV and GE values were normalized with the min and standard deviation to be in the range of [−1, 1].
Results
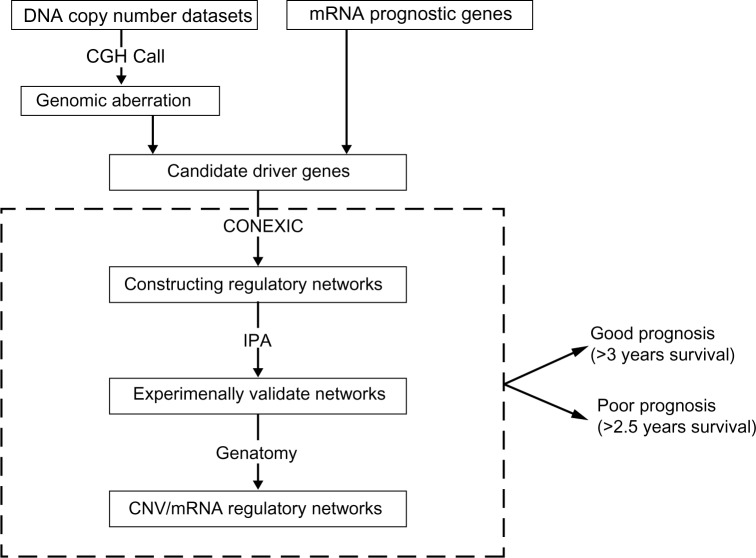
The scheme to perform integrative analysis of CNV and GE contains the following steps (Fig. 1). First, candidate driver genes were selected using DNA copy number information and mRNA prognostic genes. CGHcall34 was used to detect the aberrant regions of DNA copy numbers. Genes that showed consistent copy number aberrations in NSCLC compared with normal tissues in both datasets were selected. In addition, genes that had a significant association with patient survival time based on their copy number status were selected. The remaining candidate driver genes were mRNA prognostic genes identified in our previous studies. Second, after identifying candidate driver genes, CONEXIC was used to construct regulatory networks for good prognosis and poor prognosis NSCLC patients, respectively. Third, as the constructed regulatory networks were large, IPA was used to reduce the gene interactions to only experimentally validated ones. Finally, the Genatomy package was used to visualize the refined regulatory networks.
Figure 1.
Overview of integrative analysis of DNA copy number and GE.
Selecting candidate driver genes
The selection of candidate driver genes consists of two parts: first, DNA copy number information was used to find the aberrant regions and second, mRNA prognostic genes previously identified in our studies7,38,39 were included. To identify candidate driver genes based on DNA copy numbers, the CGHcall package was used. This method distinguishes three different states of CNV: loss (deletion), normal, and gain (amplification). Genes in the normal state have two copy numbers, in a loss state have less than two copies, and in a gain mode have three or more copy numbers. The CGHcall package was run on three data-sets (the dataset GSE31800 containing both AC and SQCC was analyzed separately according to histology). Based on results from CGHcall, genes that showed to be aberrant in more than 50 percent of the NSCLC tumor samples compared with normal tissues were selected for further analysis. Supplementary Tables S1–S3 list the genes with CNV in three corresponding datasets: SQCC and AC in the dataset (GSE31800), respectively, and SQCC in the TCGA dataset.31 To further select relevant genes with CNV in NSCLC initiation, genes that had consistent CNV among different datasets were pinpointed. There were 30 genes in common with CNV of type loss between AC and SQCC in the dataset GSE31800 (Table 2). Among these genes, NEUROG1 was found to be an important marker in early detection of colorectal cancer.40 LYN was a mediator of epithelial—mesenchymal transition and a therapeutic target of dasatinib in breast cancer.41 CPNE3, ABCB9, and AZIN1 were shown to be involved in promoting NSCLC metastasis or mediating chemoresponse.42–44 Table 3 shows the common genes among SQCC tumors in the TCGA dataset31 and the GSE31800 dataset. Only three genes (C3orf31, SELT, and C3orf52) were found to have consistent CNV with gain copy number aberration in both patient cohorts. Among them, C3orf52 was a prognosis gene of cancer in a US patent.45 Genes listed in Tables 2 and 3 showed a consistent CNV in multiple patient cohorts and histology of NSCLC, indicating their important role in NCLSC initiation.
Table 2.
Genes with consistent DNS CNVs between lung AC and SQCC in the dataset GSE31800. The percentage in this table stands for the percent of CNV in the corresponding patient cohort.
| GENE NAME | CHROMOSOME # | AC N=179 [ACCN GSE31800] | SQCC N=92 [ACCN GSE31800] | CNV TYPE |
|---|---|---|---|---|
| UBE1DC1 | 3q22.1 | 51% | 65% | Loss |
| CMTM6 | 3p22.3 | 51% | 65% | Loss |
| ULK4 | 3p22.1 | 51% | 65% | Loss |
| NKX2–5 | 5q34 | 78% | 78% | Loss |
| NEUROG1 | 5q23–q31 | 78% | 78% | Loss |
| LOC441150 | 6p21.1 | 78% | 76% | Loss |
| C6orf153 | 6p21.1 | 78% | 76% | Loss |
| C6orf134 | 6p21.33 | 78% | 76% | Loss |
| C6orf173 | 6q22.32 | 78% | 76% | Loss |
| C6orf194 | 6p22.1 | 78% | 76% | Loss |
| HIBADH | 7p15.2 | 68% | 67% | Loss |
| IFRD1 | 7q31.1 | 68% | 67% | Loss |
| PMPCB | 7q22.1 | 68% | 67% | Loss |
| TRIB1 | 8q24.13 | 51% | 68% | Loss |
| AZIN1 | 8q22.3 | 51% | 68% | Loss |
| CTSB | 8p22 | 70% | 79% | Loss |
| IKBKB | 8p11.2 | 70% | 79% | Loss |
| IMPAD1 | 8q12.1 | 70% | 79% | Loss |
| CPNE3 | 8q21.3 | 70% | 79% | Loss |
| TUSC3 | 8p22 | 70% | 79% | Loss |
| RIPK2 | 8q21 | 70% | 79% | Loss |
| LYN | 8q13 | 70% | 79% | Loss |
| ENTPD4 | 8p21.3 | 70% | 79% | Loss |
| ABCB9 | 12q24 | 67% | 66% | Loss |
| SPRYD4 | 12q13.3 | 67% | 66% | Loss |
| DDIT3 | 12q13.1 s | 67% | 66% | Loss |
| RAB22 A | 20q13.32 | 71% | 64% | Loss |
| NCOA6 | 20q11 | 71% | 64% | Loss |
| PRPF6 | 20q13.33 | 71% | 64% | Loss |
| STX16 | 20q13.32 | 71% | 64% | Loss |
Table 3.
Genes with consistent CNV in the TCGA dataset (31) and SQCC samples in dataset GSE31800. The percentage in this table stands for the percent of CNV in the corresponding patient cohort.
| GENE NAME | CHROMOSOME # | SQCC (n=201) (31) | SQCC (n=92) (GSE31800) | CNV TYPE |
|---|---|---|---|---|
| C3orf31 | 3p25.2 | 80% | 62% | Gain |
| SELT | 3q25.1 | 82% | 62% | Gain |
| C3orf52 | 3q13.2 | 80% | 64% | Gain |
Next, we sought to identify prognostic genes using three categories of CNV status (amplification, normal, and deletion). A Cox proportional hazards model was fit for the SQCC patients in the TCGA dataset.31 Genes with a significant (P < 0.05; Cox model) association between their CNV status and survival time in SQCC patients are listed in Table 4. Hazard ratios and their confidence intervals for the gain and loss categories are shown as well. A total of 30 genes had a significant association with patient survival time based on the gain status of DNA copy number, and 11 genes showed a strong association with survival time with their loss status. TADA3L is the only gene that had a strong association with survival time for both gain and loss DNA copy number value. TADA3L was associated with tumor suppressor gene p53 during the differentiation of hematopoietic stem and progenitor cells.46 As metastasis/relapse is the major treatment failure of lung cancer, aberrations in DNA copy numbers of genes in Table 4 are related to NSCLC metastasis and relapse.
Table 4.
Genes with a significant association between CNV and survival time in SQCC tumors in the TCGA dataset (31). Top genes are identified based on P-value. Hazard ratios and their confidence intervals are shown. Significant hazard ratios are marked with an asterisk (*).
| GENE NAME | P-VALUE | GAIN-HAZARD (95% CI) | LOSS-HAZARD (95% CI) |
|---|---|---|---|
| C14orf173 | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| BRMS1L | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| CHURC1 | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| NEK9 | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| CIDEB | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| SERPINA3 | 0.0007 | 2.7851 [1.61, 4.81]* | 1.3415 [0.77, 2.32] |
| C14orf172 | 0.0008 | 2.7625 [1.59, 4.77]* | 1.3195 [0.76, 2.28] |
| PTPRU | 0.0013 | 0.7796 [0.49, 1.23] | 0.2738 [0.12, 0.61]* |
| SEMA3F | 0.0014 | 0.4337 [0.24, 0.76]* | 1.1128 [0.54, 2.28] |
| TADA3L | 0.0015 | 0.3342 [0.19, 0.57]* | 0.4007 [0.18, 0.86]* |
| CCNB1IP1 | 0.0016 | 2.6560 [1.52, 4.62]* | 1.3451 [0.77, 2.34] |
| C14orf79 | 0.0016 | 2.6560 [1.52, 4.62]* | 1.3451 [0.77, 2.34] |
| UPB1 | 0.0016 | 0.3726 [0.20, 0.66]* | 0.7158 [0.42, 1.20] |
| SH3BP1 | 0.0016 | 0.3726 [0.20, 0.66]* | 0.7158 [0.42, 1.20] |
| TCF20 | 0.0016 | 0.3726 [0.20, 0.66]* | 0.7158 [0.42, 1.20] |
| MUSTN1 | 0.0020 | 0.4559 [0.25, 0.80]* | 1.1888 [0.57, 2.47] |
| ZC3H14 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| C14orf50 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| TDP1 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| C14orf24 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| KIAA0831 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| PTGER2 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| FOXN3 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| PRMT5 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| RIPK3 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| ZFP36L1 | 0.0021 | 2.5397 [1.46, 4.39]* | 1.2468 [0.71, 2.16] |
| LGR6 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| RFX5 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| CD1D | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| FLVCR1 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| MRPS14 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| DAB1 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| PUSL1 | 0.0022 | 0.9346 [0.59, 1.47] | 0.3007 [0.13, 0.66]* |
| CSDC2 | 0.0022 | 0.3736 [0.20, 0.67]* | 0.7728 [0.46, 1.29] |
| P2RXL1 | 0.0022 | 0.3736 [0.20, 0.67]* | 0.7728 [0.46, 1.29] |
| PNPLA3 | 0.0022 | 0.3736 [0.20, 0.67]* | 0.7728 [0.46, 1.29] |
| CSNK1E | 0.0022 | 0.3736 [0.20, 0.67]* | 0.7728 [0.46, 1.29] |
| CCDC117 | 0.0022 | 0.3736 [0.20, 0.67]* | 0.7728 [0.46, 1.29] |
| ATP8B2 | 0.0023 | 1.0134 [0.64, 1.59] | 0.3111 [0.13, 0.69]* |
| NPPB | 0.0023 | 1.0111 [0.64, 1.58] | 0.3108 [0.13, 0.69]* |
Additional candidate driver genes were from mRNA prognostic genes identified in our previous studies, showing a significant association with NSCLC patient survival and/or chemoresponse.7,38,39,47 These genes were further validated with Reverse transcription polymerase chain reaction (RT-PCR) assays of additional NSCLC patient samples (unpublished results). Supplementary Table S4 shows the complete list of candidate driver genes considered for constructing regulatory networks. Among these candidate driver genes, 30 genes had loss (Table 2) and 3 had gain CNV (Table 3) in the studied NSCLC cohorts. Forty genes had a strong association with SQCC survival time (Table 4), and the rest of the genes are mRNA prognostic genes identified in our previous studies.7,38,39,47 These candidate driver genes were used to construct regulatory networks.
Constructing CNV/mRNA regulatory networks
Having selected candidate driver genes as the first step, the second and third steps of the CONEXIC method were used to construct regulatory networks and rank the importance of driver genes based on DNA copy number and GE information.
In the second step of the CONEXIC method, the single modulator step, network initialization between candidate drivers and gene modules, was completed. Genome-wide CNV and GE datasets were filtered before this step. Specifically, genes with a variance above 0.2 in each patient cohort were considered for further analysis, because genes with a constant level of copy number or expression across all samples are unlikely to have a role in the regulatory networks.23 After the data filtering, the TCGA dataset31 was separated into good prognosis and poor prognosis groups. Patients who survived more than 3 years after surgery were grouped as good prognosis, whereas those who had survival time less than 2.5 years were grouped as poor prognosis. This single modulator step was performed with non-parametric bootstrapping for 10 times23 on good and poor prognosis groups, respectively. The bootstrapping method guarantees the robustness of the results. The modulators that were selected in at least 60% of the runs were picked. These modulators were used for a final run of the single modulator step. After performing the second step, the relation among genes was initialized and the basic regulatory networks were created. These networks were considered as the starting point for the third step, which is the network learning step.
The third step of the CONEXIC method was performed with non-parametric bootstrapping to ensure robustness of the results. This step was also completed for good prognosis and poor prognosis groups, separately. This step was also run 10 times, and the candidate driver genes that were selected in 60% of the runs were selected for the final run. Supplementary Table S5 shows the top-ranked genes (modulators) after running the final network learning step. As a result, each of these modulators is related to a large number of genes in the constructed module network.
As mentioned earlier, part of candidate driver genes were mRNA prognostic biomarkers identified from our previous studies, with potential clinical utilities for personalized lung cancer treatment.7,38,39,47 After constructing regulatory networks, we sought to investigate the functional roles of these mRNA prognostic genes. As the regulatory networks were constructed separately for good prognosis and poor prognosis patients, respectively, investigating important driver genes and regulatory modules could potentially reveal important mechanisms in NSCLC progression and metastasis and their impact on clinical outcome. Table 5 shows the regulatory role of top-ranked prognostic mRNA biomarkers in poor prognosis patients. Specifically, SAMD4B, APOA2, ATAD4, and VASH1 were top-ranked modulators (driver genes) in poor prognosis, with a potential functional involvement in rendering adverse clinical outcome in NSCLC patients. The other mRNA prognostic genes in Table 5 were not modulators, but they are in the modules of other driver genes such as CTSB. Table 6 shows the regulatory role of the mRNA prognostic genes in the good prognosis dataset. In particular, ADH1B, CCL19, FHL1, VASH1, and RB1 were top-ranked modulators (driver genes) in good prognosis group. Other mRNA prognostic genes were not modulators, but they were in the modules that were regulated by driver genes such as RB1 and VASH1. In the comparison of Tables 5 and 6, modulators in good prognosis and poor prognosis groups were vastly different, suggesting that NSCLC tumor progression and the ultimate clinical outcome are driven by very different molecular mechanisms.
Table 5.
mRNA prognostic biomarkers identified in our previous studies7,38,39 ranked as top driver genes in poor prognosis SQCC patients.
| GENE NAME | MODULATOR? (# MODULES)* | MODULATED BY | REFERENCE |
|---|---|---|---|
| SCLY | No | IFRD1 | 39 |
| TNFSF9 | No | CTSB | 38 |
| CD27 | No | STAT4 | 39 |
| DAG1 | No | SELT | N/A# |
| SAMD4B | Yes (3) | No modulator | N/A# |
| THBS1 | No | CTSB | 39 |
| XPO1 | No | PTGER2 | 39 |
| C8orf70 | No | PTGER2 | 39 |
| STK24 | No | CTSB | 39 |
| AKAP13 | No | CTSB | 38 |
| APOA2 | Yes (3) | No modulator | 7 |
| CCL19 | No | STAT4 | 7 |
| CLIC2 | No | STAT4 | 7 |
| COL14A1 | No | VASH1 | 7 |
| HMBOX1 | No | VASH1 | 7 |
| IRF3 | No | SAMD4B | 7 |
| ATAD4 | Yes (5) | No modulator | 7 |
| SLC39A8 | No | VASH1 | 7 |
| APIN1 | No | IFRD1 | 7 |
| TAF4 | No | SELT | 7 |
| TOMM34 | No | CTSB | 7 |
| VASH1 | Yes (2) | SAMD4B | 7 |
| VIPR2 | No | SERPINA3 | 7 |
| HFE | No | IFRD1 | 39 |
| HNF4A | No | CTSB | N/A# |
| STAT6 | No | SERPINA3 | 47 |
Notes:
If a gene is a modulator (driver gene), the number of modules regulated by it is listed in parentheses.
Unpublished mRNA prognostic biomarkers associated with NSCLC outcome.
Table 6.
mRNA prognostic biomarkers identified in our previous studies7,38,39 ranked as top driver genes in good prognosis SQCC patients.
| GENE NAME | MODULATOR? (# MODULES)* | MODULATED BY | REFERENCE |
|---|---|---|---|
| OGT | No | C6orf134 | 38 |
| CCDC99 | No | CCL19 | 39 |
| CD27 | No | CCL19 | 39 |
| DAG1 | No | C6orf134 | N/A# |
| XPO1 | No | PRMT5 | 39 |
| C8orf70 | No | RB1 | 39 |
| AKAP13 | No | RB1 | 38 |
| MSX2 | No | RB1 | 38 |
| ADH1B | Yes (4) | STAT4 | 7 |
| ANXA6 | No | VASH1 | 7 |
| CCL19 | Yes (4) | No Modulator | 7 |
| CLIC2 | No | ADH1B | 7 |
| COL14A1 | No | VASH1 | 7 |
| FHL1 | Yes (1) | VASH1 | 7 |
| ICA1 | No | RB1 | 7 |
| IRF3 | No | C6orf134 | 7 |
| IVD | No | VASH1 | 7 |
| SLC39A8 | No | ADH1B | 7 |
| SPIN1 | No | PRMT5 | 7 |
| TAF4 | No | C6orf134 | 7 |
| VASH1 | Yes (4) | ADH1B | 7 |
| HFE | No | C14orf50 | 39 |
| RB1 | Yes (7) | No Modulator | 7 |
| STAT6 | No | CCL19 | 47 |
| ZNF638 | No | C6orf134 | 38 |
| UBE1L2 | no | C6orf134 | 39 |
Notes:
If a gene is a modulator (driver gene), the number of modules regulated by it is listed in parentheses.
Unpublished mRNA prognostic biomarkers associated with NSCLC outcome.
Refinement and visualization of CNV/mRNA regulatory networks
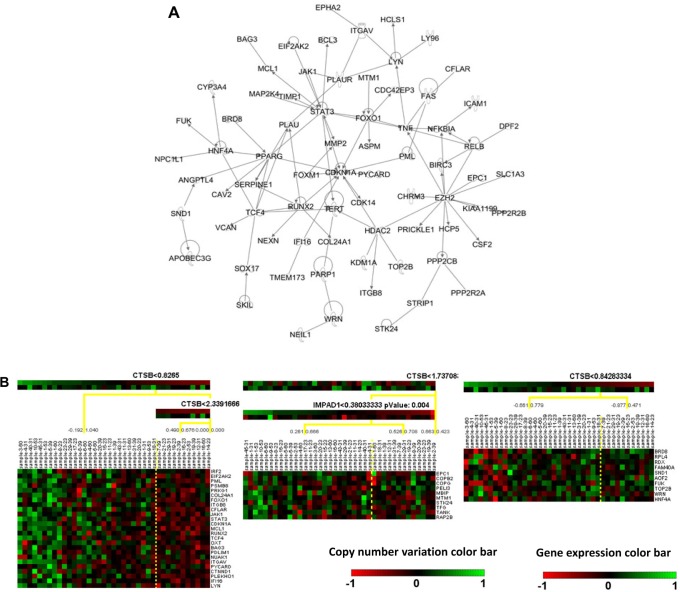
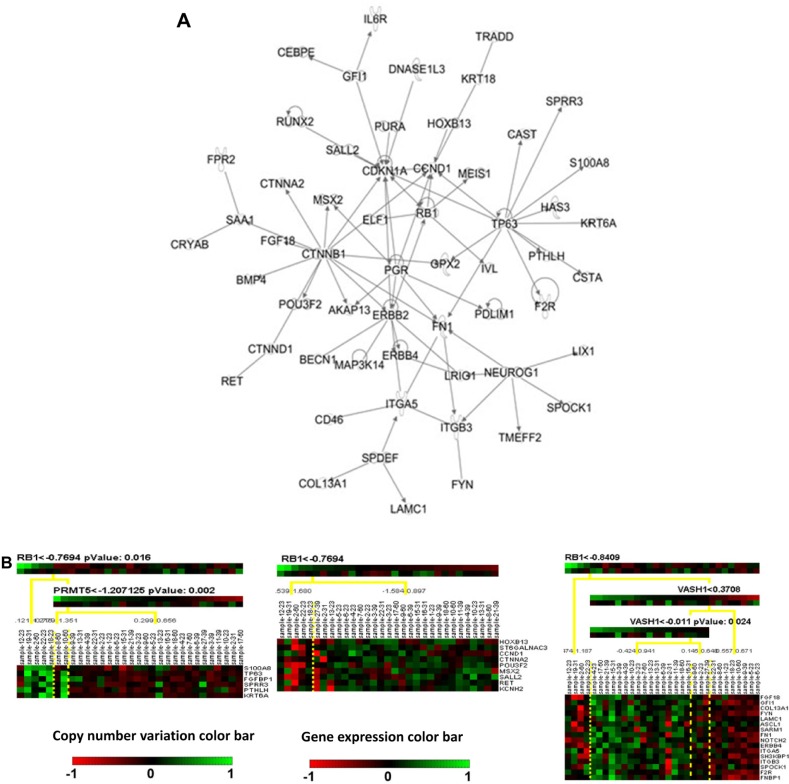
From the results of CONEXIC analysis, top-ranked driver genes were examined for further analysis. Specifically, CTSB was one of the top selected driver genes in poor prognosis patient group, and it was shown to modulate most mRNA prognostic genes, including TNSF9, THBS1, STK24, AKAP13, TOMM34, and HFE. Therefore, CTSB was selected for further analysis. For good prognosis group, RB1 was selected for further analysis because it is a prognostic biomarker gene by itself7 and modulated multiple mRNA prognostic biomarkers, including C8orf70, AKAP13, MSX2, and ICA1. In the regulatory network analysis, CTSB was related to 1,043 other genes in 10 different modules. RB1 modulated 681 genes in the 7 constructed regulatory modules. To refine the constructed regulatory networks, IPA was used to select experimentally validated molecular interactions in the module networks. Experimentally confirmed molecular interaction network for CTSB in poor prognosis SQCC patients is shown in Figure 2a, and the confirm network for RB1 in good prognosis SQCC patients is shown in Figure 3a, respectively. Next, Genatomy package was used to visualize the refined regulatory networks based on the text output from CONEXIC. Figure 2b shows the modules regulated by CTSB in poor prognosis SQCC patients, focusing on the modules involving previously identified mRNA prognostic genes. Similarly, Figure 3a shows RB1-modulated mRNA expression in good prognosis SQCC patients, focusing on the modules involving our identified mRNA biomarkers. These results reveal potential regulatory mechanisms of how CNV in a driver gene regulates the expression of a group of genes, among which are prognostic and predictive mRNA biomarkers associated with NSCLC metastasis and chemoresponse. The interactions among these genes in the computationally constructed regulatory networks were experimentally confirmed in the published literature with IPA.
Figure 2.
Regulatory networks of CTSB in poor prognosis SQCC patient group. (A) Experimentally validated network of CTSB in poor prognosis SQCC patients (31) with IPA analysis. (B) CNV/mRNA regulatory network of CTSB in poor prognosis SQCC patients (31).
Notes: The top color bar under the driver gene shows the mRNA expression and the below one shows CNV. Green color indicates overexpression/amplification and red color indicates under-expression/deletion.
Figure 3.
Regulatory networks of RB1 in good prognosis SQCC patient group. (A) Experimentally validated networks for RB1 in good prognosis SQCC patients (31) with IPA analysis. (B) CNV/mRNA regulatory network of RB1 in good prognosis SQCC patients (31).
Notes: Green color indicates overexpression/amplification, and red color indicates under-expression/deletion.
Discussion and Conclusion
Lung cancer is a complex disease involving numerous somatic mutations, amplifications, and deletions. Tumor relapse and metastasis is the major cause of treatment failure (ie, death) in lung cancer. Chromosomal CNVs are shown to be functionally important in regulating GE changes and genotypes in tumorigenesis and metastasis. While using GE changes alone has been shown to be a precise and feasible tool for clinical diagnostics and prognostics,48 it does not necessarily reveal molecular functional involvement and cancer mechanisms. Integrative analysis of GE and copy number is expected to identify cancer driver genes and prioritize them, as well as reveal disease mechanisms and provide insight into novel therapeutic targets. Regulatory network approaches have been developed for detecting GE affected by DNA CNVs, genetic biomarkers, or motif data. Lirnet,25 Geronimo,37 and CONEXIC23 are three major approaches based on module networks, which were originally conceived by Segal et al.36 CONEXIC was used in this study to construct CNV/mRNA regulatory networks because it models CNV together with GE, whereas Lirnet and Geronimo have not been used in such applications before. There are some other regulatory networks that were developed to model other molecular data types,49,50 but not with GE and DNA copy numbers.
The novelty of CONEXIC is that it does not require the correlation between CNV and GE in the search for driver genes. Many methods for identifying driver genes among aberrant regions focus on the genes with correlated CNV and GE.51,52 As a matter of fact, in many cases, the expression of a driver gene is poorly connected with the DNA copy number.23 The reason behind this is that CNV is only one of the many causes to induce up-regulation or down-regulation of a gene. In fact, the expression of many driver genes is less correlated with their copy numbers compared with passenger genes. In contrast, the expression of genes in a module is more related to the expression of the corresponding driver gene. The algorithm of CONEXIC was designed based on these biological contexts, which has the following advantages over other methods. First, by assessing both CNV and GE, CONEXIC provides a better way to find important areas in the aberrant chromosomal regions as potential candidate driver genes, comparing to the methods that consider only DNA copy number. Second, CONEXIC associates the expression of a driver gene to the expression of genes in the corresponding module in constructing regulatory networks.23 However, this could be considered as a limitation of CONEXIC, as it does not consider CNV in constructing regulatory networks in the second and third steps of the algorithm. One of the future improvements on this method would be considering two types of regulators (GE regulators and CNV regulators) and combining these two types of regulators in constructing regulatory networks.
In this study, we designed a framework based on CONEXIC to integrate CNV with GE to identify important driver genes in NSCLC and reveal experimentally confirmed regulatory networks in metastasis. In our approach, the candidate driver genes were selected and validated using multiple patient cohorts. Instead of using Genomic Identification of Significant Targets in Cancer (GISTIC) method53 as proposed in the original CONEXIC method, CGHcall34 was used to classify different regions of CNV based on mixture models. The selected candidate driver genes had either consistent CNV in different NSCLC patient cohorts and histology subtypes or a strong association with patient survival information based on their CNV status or mRNA expression levels. Next, regulatory networks were constructed for good prognosis and poor prognosis patient groups separately. The candidate driver genes were the same for both groups. After running CONEXIC with bootstrapping, the top-ranked driver genes and the constructed regulatory networks were very different in both groups, indicating that different molecular mechanisms render different clinical outcomes in NSCLC patients. As these computationally derived networks contained a large number of modules, IPA was used to select the genes and molecular interactions in the modules that were experimentally validated and reported in the literature. In this way, the gene modules were refined to the experimentally validated ones. Finally, Genatomy was used to visualize the refined regulatory networks. These experimentally confirmed regulatory networks reveal important CNV-regulated transcriptional activities in NSCLC metastasis. As many genes in the candidate drivers were NSCLC prognostic or chemoresponse predictive biomarkers identified in our previous studies, these results shed light on potential innovative therapeutic targets for NSCLC treatment.
Supplementary Files
Supplementary Tables 1–5. Tables S1–S3 list the genes with CNV in three corresponding datasets: SQCC and AC in the dataset (GSE31800), and SQCC in the TCGA dataset. Table S4 shows the complete list of candidate driver genes considered for constructing regulatory networks. Table S5 shows the top-ranked genes (modulators) after running the final network learning step.
Acknowledgments
We thank Julian Dymacek for editing the paper.
Footnotes
Author Contributions
Conceived and designed the experiments: NLG. Analyzed the data: SMI. Wrote the first draft of the manuscript: SMI. Contributed to the writing of the manuscript: NLG. Agree with manuscript results and conclusions: SMI, NLG. Jointly developed the structure and arguments for the paper: SMI, NLG. Made critical revisions and approved final version: NLG. Both authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: This study was funded by NIH R01/R56LM009500 (PI: Guo) and NCRR P20RR16440 and Supplement (PD: Guo). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants. Provenance: the authors were invited to submit this paper.
REFERENCES
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355(9202):479–85. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 4.Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379(9818):823–32. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 7.Wan YW, Beer DG, Guo NL. Signaling pathway-based identification of extensive prognostic gene signatures for lung adenocarcinoma. Lung Cancer. 2012;76(1):98–105. doi: 10.1016/j.lungcan.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker SG, Kramer BS, Srivastava S. Markers for early detection of cancer: statistical guidelines for nested case-control studies. BMC Med Res Methodol. 2002;2:4. doi: 10.1186/1471-2288-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emir B, Wieand S, Su JQ, Cha S. Analysis of repeated markers used to predict progression of cancer. Stat Med. 1998;17(22):2563–78. doi: 10.1002/(sici)1097-0258(19981130)17:22<2563::aid-sim952>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 10.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159(9):882–90. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 11.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 2005;26(4):178–82. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(suppl):S11–7. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 15.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99(20):12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotliarov Y, Steed ME, Christopher N, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006;66(19):9428–36. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bungaro S, Dell’Orto MC, Zangrando A, et al. Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer. 2009;48(1):22–38. doi: 10.1002/gcc.20616. [DOI] [PubMed] [Google Scholar]
- 18.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62(21):6240–45. [PubMed] [Google Scholar]
- 20.Kotliarov Y, Kotliarova S, Charong N, et al. Correlation analysis between single-nucleotide polymorphism and expression arrays in gliomas identifies potentially relevant target genes. Cancer Res. 2009;69(4):1596–603. doi: 10.1158/0008-5472.CAN-08-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsafrir D, Bacolod M, Selvanayagam Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66(4):2129–37. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 22.Haverty PM, Hon LS, Kaminker JS, Chant J, Zhang Z. High-resolution analysis of copy number alterations and associated expression changes in ovarian tumors. BMC Med Genomics. 2009;2:21. doi: 10.1186/1755-8794-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akavia UD, Litvin O, Kim J, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schafer M, Schwender H, Merk S, Haferlach C, Ickstadt K, Dugas M. Integrated analysis of copy number alterations and gene expression: a bivariate assessment of equally directed abnormalities. Bioinformatics. 2009;25(24):3228–35. doi: 10.1093/bioinformatics/btp592. [DOI] [PubMed] [Google Scholar]
- 25.Lee SI, Dudley AM, Drubin D, et al. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 2009;5(1):e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litvin O, Causton HC, Chen BJ, Pe’er D. Modularity and interactions in the genetics of gene expression. Proc Natl Acad Sci U S A. 2009;106(16):6441–6. doi: 10.1073/pnas.0810208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 28.Solinas-Toldo S, Lampel S, Stilgenbauer S, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20(4):399–407. [PubMed] [Google Scholar]
- 29.Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21(19):3763–70. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starczynowski DT, Lockwood WW, Delehouzee S, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. J Clin Invest. 2011;121(10):4095–105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wieringen WN, Unger K, Leday GG, et al. Matching of array CGH and gene expression microarray features for the purpose of integrative genomic analyses. BMC Bioinformatics. 2012;13(1):80. doi: 10.1186/1471-2105-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23(7):892–94. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 35.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21(22):4084–91. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 36.Segal E, Shapira M, Regev A, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34(2):166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 37.Lee SI, Pe’er D, Dudley AM, Church GM, Koller D. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci U S A. 2006;103(38):14062–7. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo NL, Wan YW, Tosun K, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14(24):8213–20. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan YW, Sabbagh E, Raese R, et al. Hybrid models identified a 12-gene signature for lung cancer prognosis and chemoresponse prediction. PLoS One. 2010;5(8):e12222. doi: 10.1371/journal.pone.0012222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herbst A, Rahmig K, Stieber P, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106(6):1110–8. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 41.Choi YL, Bocanegra M, Kwon MJ, et al. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res. 2010;70(6):2296–306. doi: 10.1158/0008-5472.CAN-09-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Vescovo V, Meier T, Inga A, Denti MA, Borlak J. A cross-platform comparison of affymetrix and Agilent microarrays reveals discordant miRNA expression in lung tumors of c-Raf transgenic mice. PLoS One. 2013;8(11):e78870. doi: 10.1371/journal.pone.0078870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Z, Zhong Z, Yang L, Wang S, Gong Z. MicroRNA-31 inhibits cisplatin-induced apoptosis in non-small cell lung cancer cells by regulating the drug transporter ABCB9. Cancer Lett. 2014;343(2):249–57. doi: 10.1016/j.canlet.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 44.Lin HC, Zhang FL, Geng Q, et al. Quantitative proteomic analysis identifies CPNE3 as a novel metastasis-promoting gene in NSCLC. J Proteome Res. 2013;12(7):3423–33. doi: 10.1021/pr400273z. [DOI] [PubMed] [Google Scholar]
- 45.Massague J, Zhang X, Padua D., inventors Gene signatures for the prognosis of cancer. US Patent. 2009. Patent US20110053804.
- 46.Fuhrken PG, Chen C, Apostolidis PA, Wang M, Miller WM, Papoutsakis ET. Gene ontology-driven transcriptional analysis of CD34+ cell-initiated megakaryocytic cultures identifies new transcriptional regulators of megakaryopoiesis. Physiol Genomics. 2008;33(2):159–69. doi: 10.1152/physiolgenomics.00127.2007. [DOI] [PubMed] [Google Scholar]
- 47.Ma Y, Ding Z, Qian Y, et al. Predicting cancer drug response by proteomic profiling. Clin Cancer Res. 2006;12(15):4583–9. doi: 10.1158/1078-0432.CCR-06-0290. [DOI] [PubMed] [Google Scholar]
- 48.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 49.Qi J, Michoel T, Butler G. An integrative approach to infer regulation programs in a transcription regulatory module network. J Biomed Biotechnol. 2012;2012:245968. doi: 10.1155/2012/245968. Epub April 11, 2012. (PMID:22577292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenfield A, Hafemeister C, Bonneau R. Robust data-driven incorporation of prior knowledge into the inference of dynamic regulatory networks. Bioinformatics. 2013;29(8):1060–7. doi: 10.1093/bioinformatics/btt099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Hassan NZ, Mokhtar NM, Kok ST, et al. Integrated analysis of copy number variation and genome-wide expression profiling in colorectal cancer tissues. PLoS One. 2014;9(4):e92553. doi: 10.1371/journal.pone.0092553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortiz-Estevez M, De Las RJ, Fontanillo C, Rubio A. Segmentation of genomic and transcriptomic microarrays data reveals major correlation between DNA copy number aberrations and gene-loci expression. Genomics. 2011;97(2):86–93. doi: 10.1016/j.ygeno.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 53.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1–5. Tables S1–S3 list the genes with CNV in three corresponding datasets: SQCC and AC in the dataset (GSE31800), and SQCC in the TCGA dataset. Table S4 shows the complete list of candidate driver genes considered for constructing regulatory networks. Table S5 shows the top-ranked genes (modulators) after running the final network learning step.