Abstract
Helicobacter pylori infection of gastric tissue results in an immune response dominated by Th1 cytokines and has also been linked with dysregulation of Sonic Hedgehog (SHH) signaling pathway in gastric tissue. However, since interactions between the cytokines and SHH during H. pylori infection are not well understood, any mechanistic understanding achieved through interpretation of the statistical analysis of experimental results in the context of currently known circuit must be carefully scrutinized. Here, we use mathematical modeling aided by restraints of experimental data to evaluate the consistency between experimental results and temporal behavior of H. pylori activated cytokine circuit model. Statistical analysis of qPCR data from uninfected and H. pylori infected wild-type and parietal cell-specific SHH knockout (PC-SHHKO) mice for day 7 and 180 indicate significant changes that suggest role of SHH in cytokine regulation. The experimentally observed changes are further investigated using a mathematical model that examines dynamic crosstalks among pro-inflammatory (IL1β, IL-12, IFNγ, MIP-2) cytokines, anti-inflammatory (IL-10) cytokines and SHH during H. pylori infection. Response analysis of the resulting model demonstrates that circuitry, as currently known, is inadequate for explaining of the experimental observations; suggesting the need for additional specific regulatory interactions. A key advantage of a computational model is the ability to propose putative circuit models for in-silico experimentation. We use this approach to propose a parsimonious model that incorporates crosstalks between NFĸB, SHH, IL-1β and IL-10, resulting in a feedback loop capable of exhibiting cyclic behavior. Separately, we show that analysis of an independent time-series GEO microarray data for IL-1β, IFNγ and IL-10 in mock and H. pylori infected mice further supports the proposed hypothesis that these cytokines may follow a cyclic trend. Predictions from the in-silico model provide useful insights for generating new hypothesis and design of subsequent experimental studies.
Introduction
Helicobacter pylori is a gram negative bacteria that resides in the stomach and duodenum of infected host [1], [2]. It is a significant risk factor for atrophic gastritis [3], [4], gastric ulcer [4], [5] and gastric cancer [4]–[6]. Improved treatment and early diagnosis of H. pylori associated diseases require a better understanding of the mechanisms by which this bacteria increases the risk for chronic inflammation and gastric cancer [7]. The host cell detects the presence of bacteria and produces an immune response to eliminate the bacteria. The final outcome depends on a balance between pro-inflammatory and anti-inflammatory cytokines produced during H. pylori infection [8], [9]. A strong pro-inflammatory response may allow eradication of the bacteria however at a cost of increased risk for gastritis [10] while anti-inflammatory cytokines may protect against gastritis, but H. pylori may continue to persist [11], [12]. Another gene that has been recently identified to play an important role in pathogenesis of chronic H. pylori infection [13], [14] and gastric cancer [17]–[20] is sonic hedgehog (SHH). In both human and mouse stomachs, it is expressed in the parietal cells [21]. Under normal conditions, SHH regulates differentiation of the gastric epithelium [21], [22] and T-cell [23], [24]. During chronic H. pylori infection, SHH-dependent proliferation of parietal cells plays a key role in gastric mucosal repair [13], [25]. It is also reported that SHH acts as a monocyte/macrophage chemoattractant during repair of the myocardium [26] and during H. pylori infection [15]. We and others have recently shown that SHH is involved in early immune response to H. pylori [15], [16]. We observed that parietal cell-specific SHH knock-out (PC-SHHKO) mice failed to develop gastritis, even after 6 months of H. pylori infection in contrast to infected control group (WT) which developed significant inflammatory response [15]. Zaatari et al have shown that overexpression of ShhWT induced gastritis while CMV-ShhF200H mice (carrying mutant SHH) did not develop gastritis. They also reported that SHH overexpression exacerbated the histologic severity observed with Helicobacter felis infection and increased the amount of myeloid cells recruited to the inflamed stomach as compared to that in non-transgenic mice [16].
Although recent studies have highlighted the immunoregulatory role of SHH in stomach [15], [16], a model studying temporal relationship between SHH and cytokines activated during H. pylori infection is still lacking, and it is unexplored what effect SHH may have in the context of regulating cytokine expression in the H. pylori infected stomach. Such temporal studies are not readily amenable to experimental approaches because of high cost and time associated with time series in-vivo experiments. In-vitro experiments of immune responses often face experimental limitations – for example, lack of a host immune cell response. Mathematical modeling is a powerful technique to complement such studies as it allows predicting dynamic behavior of the system under various perturbations and generating new hypotheses. Currently, there are only a handful of mathematical models that study the H. pylori - host immune response, and though they provide high-level or cellular-level details [27]–[30], they do not focus on quantifiable biomarkers involved.
An interaction map - a topological network of signaling pathways, activated by H. pylori, was manually curated from literature [31], [32] (Figure S1 in Methods S1). H. pylori virulence factors like CagA [33]–[35], VacA [33] and PGN [33] activate a cascade of signaling pathways in epithelial cells of host stomach. Virulence factor CagA (cytotoxin-associated gene A) activates ERK [25], [34], [36] and AKT [37] pathways which further stimulate nuclear translocation of NFĸB [37]–[39]. Effector molecule PGN (peptidogylcan) is sensed by intracellular receptor NOD1 (nucleotide-binding oligomerization domain 1) [25], [33] which activates NFĸB [25], [33], [35], [40], ERK, p38 and AP-1 [40]. Protein VacA (Vacuolating cytotoxin A) also stimulates ERK and p38 pathways [41] which activate transcription factor AP-1 [34], [42]–[44]. Both NFĸB and AP-1 positively regulate chemokine IL-8/MIP-2 transcription [34], [35], [45]. (MIP-2 is a functional homolog of human IL-8 in mouse [46]). H. pylori colonization of gastric epithelium eventually leads to recruitment of monocytes which secrete pro-inflammatory cytokines like IL-12, IL-1β, TNFα, IL6 [47]–[49] and IL8 [50], [51]. IL-12 and other H. pylori antigens synergistically stimulates release of IFNγ from natural killer cells [49], [52]–[54]. SHH, a crucial player in early immune response to H. pylori [15] has been reported as a putative target of NFĸB in gastric [55] and pancreatic cancer [56], [57]. Interestingly, Waghray et al have shown that IL-1β suppresses SHH expression in parietal cells by inhibiting acid secretion [58]. Along with the activation of pro-inflammatory cytokines, IL-10, an anti-inflammatory cytokine is also produced during H. pylori infection [47], [54], [59]. IL-10 has been reported to inhibit NFĸB activity, and subsequently IL-8 transcription in gastric epithelial cells during H. pylori infection [60] and also in macrophage [61] and T-cells [62]. In summary, different virulence factors of H. pylori activate various signaling pathways in the stomach tissue leading to increased activity or expression of NFĸB, chemokines and cytokines [63], [64]. The severity of inflammation and exact details of signaling pathways described above can vary depending on the strain of H. pylori and strain of mice used in the experiment [65], [66].
The role of SHH in H. pylori mediated inflammation was investigated experimentally using wild-type and parietal cell-specific SHH knock-out (PC-SHHKO) mice [15]. Here we investigate, using the experimental data and mathematical modelling technique, the putative dynamics of cytokine-SHH circuitry. Our analysis demonstrates an interaction between H. pylori infection and genotype of the mice, and positive regulation of the cytokines' expression involving SHH. A detailed examination of the current pathway databases (for example Strings, Pathway Commons), and molecular circuits (wiring diagram of interactions among genes and proteins) derived through manual curation of current literature, fail to identify the necessary positive regulation. Our mathematical model of the cytokine-SHH circuitry aims to understand temporal behavior of the network and to identify emergent patterns which may arise through interactions among components of the system and may not be apparent when the components are studied in isolation [67]–[69].
Materials and Methods
Animal Model
The mouse model with parietal cell-specific deletion of SHH (PC-SHHKO) was generated as previously described (C57Bl/6, 129/Sv background) [22] and HKCre mice expressing Cre transgene under the control of H+,K+-adenosine triphosphatase (ATPase) β subunit promoter (C57Bl/6, FVB/N background) were used as control [15]. HKCre (WT) and PC-SHHKO mice, aged 8 weeks were either infected with H. pylori or left uninfected. The uninfected group received 200 µl of Brucella broth over 3 consecutive days whereas the infected group was inoculated with 108 H. pylori SS1 (Sydney strain 1) bacteria per 200 µl of Brucella broth over 3 consecutive days. Mice (n = 3–4 per group) were sacrificed on day 7 and 180 post-infection and levels of gastric SHH and cytokines were assayed by quantitative reverse-transcriptase polymerase chain reaction. H. pylori colonization as measured by bacterial cultures and analysis of CFU/g Tissue (colony-forming units per gram tissue) for WT and PC-SHHKO infected mouse stomachs were shown to be equivalent [15].
Ethics Statement
All mouse studies were approved by the University of Cincinnati Institutional Animal Care and Use Committee (IACUC) that maintains an American Association of Assessment and Accreditation of Laboratory Animal Care (AAALAC) facility.
Quantitative real-time RT-PCR (qPCR)
Total RNA was isolated from stomachs of uninfected and infected WT and PC-SHHKO mice. The High Capacity cDNA Reverse Transcription Kit was used for cDNA synthesis from 100 ng of RNA following the recommended protocol (Applied Biosystems). Pre-designed real-time PCR assays were purchased for the following genes (Applied Biosystems): SHH (Mm00436528_m1), IFNγ (Mm01168134_m1), IL-1β (Mm01336189_m1) and mouse GAPDH (20X) (4352932-0803020), MIP2 (Mm00436450_m1). PCR amplifications were performed in a total volume of 20 µl, containing 20X TaqMan Expression Assay primers, 2X TaqMan Universal Master Mix (Applied Biosystems, TaqMan Gene Expression Systems) and cDNA template. Each PCR amplification was performed in duplicate wells in a StepOne Real-Time PCR System (Applied Biosystems), using the following conditions: 50°C 2 minutes, 95°C 10 minutes, 95°C 15 seconds (denature) and 60°C 1 minute (anneal/extend) for 40 cycles. IL-12 expression was quantified using specific primers as previously published for IL-12: Forward- 5′- GGA AGC ACG GCA GAA TA-3′ and Reverse- 5′- AAC TTG AGG GAG AAG TAG GAA TGG -3′ [70] using the SYBR Green PCR Master Mix and protocol (Applied Biosystems). GAPDH was used as an internal control. See Methods S1.
Statistical Analysis
For each sample, dCT value for target gene was calculated by subtracting CT value of calibrator gene (GAPDH) from CT value of target gene. To test if expression of target gene in gastric mucosa was affected by H. pylori and if this effect was same in all genotypes, a two-way ANOVA (Analysis of variance) test was performed, comparing dCT values from all four condition: uninfected wild type, infected wild type, uninfected PC-SHHKO and infected PC-SHHKO. Bonferroni test was performed as post ANOVA test to assess the effect of H. pylori on target genes in each genotype. dCT values from uninfected mice were compared with dCT values from H. pylori infected mice in both WT and PC-SHHKO conditions. For graphical presentation of qPCR results, for each gene in WT and PC-SHHKO conditions, ddCT was calculated by subtracting dCT value of H. pylori infected group from dCT of uninfected group. The data were plotted as 2−ddCT (mean fold change) [12]. A P-value <0.05 was considered statistically significant. Interaction test was also performed to study the interaction between infection status and genotype. Since CT value is inversely proportional to gene's expression level, negative dCT values were used for interaction test. The interaction plot displays levels of treatment (absence and presence of H. pylori) on the x-axis and mean of negative dCT values for each treatment on the y-axis. A separate line connects the means corresponding to each level of the trace factor – genotype (WT and PC-SHH-KO). The qPCR data was analyzed using R statistical software. Link to the R script used for analyzing the data and generating graphs: http://rpubs.com/marwahsi/20168.
Mathematical model
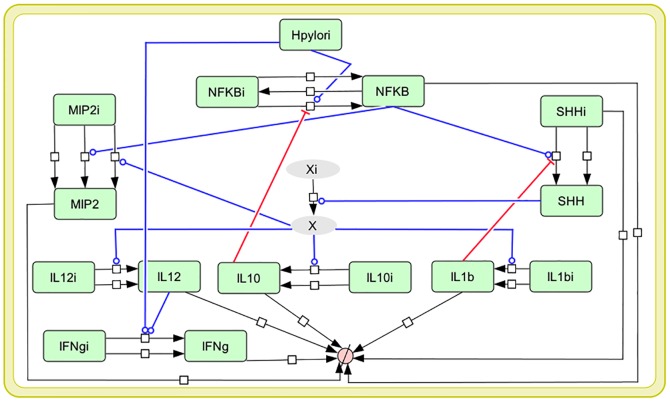
A comprehensive interaction map was developed by manual curation of published literature, capturing key signaling pathways activated in gastric epithelium and in macrophages by H. pylori virulence factors. For our mathematical model, a selected subset of biomolecules from the interaction map was included. The molecules were selected for inclusion if: a) they were reported in the literature to be regulated with H. pylori SS1 strain (as SS1 strain was used in our experiments) [63], [64] or b) their experimentally measured levels showed variations in our data. Mathematical modelling of very large networks can be impractical, primarily due to unavailability of parameter values, especially in case of higher organisms [31], [71]–[73]. In order to overcome these challenges, the complex interaction map was pruned to a simpler reduced network (Figure 1) focusing on cytokines and SHH, for which experimental trends were available [74]. The reduced model can capture key characteristics of the larger network and conserve the regulatory mechanisms present in the system [71]–[73]. Our reduced model compresses the details of a complex pathway into a “black box”, which can be scaled up, by adding detailed components as needed. The reduced model was further enriched with a new “influence” link between SHH and cytokines as predicted by our experimental data. It must be noted that this link may not be a direct interaction between SHH and cytokines and rather may involve an indirect mechanism. This unknown mechanism is currently shown by an intermediate species “X” in the model which links SHH to all the cytokines. Equations based on Michaelis-Menten and mass action kinetics were used in the reduced model in order to construct a dynamic system that describes the evolution of the biomolecules over time. The sampling frequency (control equaling time zero, day 7 equaling early, and day 180 equaling late) was unlikely to be sufficient for obtaining detailed dynamics of concentration and oscillation frequencies. Therefore, unit-free measures were used to express “time” and “amount” in order to focus on the qualitative behavior of the cytokine dynamics. The kinetic parameters initially selected for the model, represent a range of biologically feasible values [75]–[77]. Subsequent computational optimization (iterative trial and error) was used to select the parameter set that best satisfies the trends observed in experimental data and are within biologically relevant limits. This approach is similar to the methodology used for parameter estimation for computational models when no experimental values are available [76], [78], [79]. In addition to the reactions activated through H. pylori signaling, each model species is activated by a small “constitutive” flux which accounts for some basal levels of the species formed through a pathway either not represented in the model or currently unknown. Each model species is connected to a source and sink. The source represents inactive form of the protein. The sink accounts for downstream signaling and half-life of the protein. Complete list of model assumptions are available in Table S1 in Methods S1. Ordinary Differential Equations (ODE) were simulated to characterize the system using modeling tool CellDesigner [80]. Knock-out (KO) of different genes was modeled by setting the concentration of its source to zero. See Methods S1 for model equations and parameter values (Table S2, S3 and S4 in Methods S1).
Figure 1. Diagram of mathematical model of cytokine-SHH network during H. pylori infection.
This reduced network derived from interaction map, represents the key cytokines activated as host's immune response to H. pylori. Blue arrows represent activation whereas while red arrows depict inhibition. Model species with suffix “i” represent the inactive form. The link between SHH and cytokines, as predicted by our experimental data is modelled through unknown model species “X” (grey colored). Detailed interaction network of host immune response to H. pylori is available in MethodsS1.
GEO Data
A time-series microarray dataset – GSE37938, available through GEO (Gene Expression Omnibus) database was used as an auxiliary data set in order to analyze the temporal behavior of the genes present in our model. The microarray data set contains six-week-old female BALB/c mice. The mice were uninfected or infected with SS1 strain of H. pylori for 2,7,14 & 28 days. A total of 71 samples (35 infected +36 uninfected) were collected from 3 cell types: chief cell, parietal cell and mucous producing pit cell [81]. We extracted and analyzed time-series data for IL-10, IFNγ, IL-1β from GSE37938 dataset. Data was not available for SHH and MIP-2 while IL-12 data was not used for analysis due to large number of missing values. For a given probe, median of its expression values was calculated for each time point. Probes with two or more missing values for more than one time point, were not considered for further analysis. The median values of a probe from both uninfected and H. pylori infected mice were plotted against time to study their trajectory in the two conditions. To address the problem of probe selection in case of multiple probes representing the same gene, median value was taken for probes with same Genbank ID (GB_list ID) while probes with different Genbank ID were analyzed separately. Plots are shown only for Genbank IDs which are represented by at least three probes. Link to the R script used for extracting and analyzing the data: http://rpubs.com/marwahsi/8932.
Results
SHH positively regulates cytokine expression during H. pylori infection
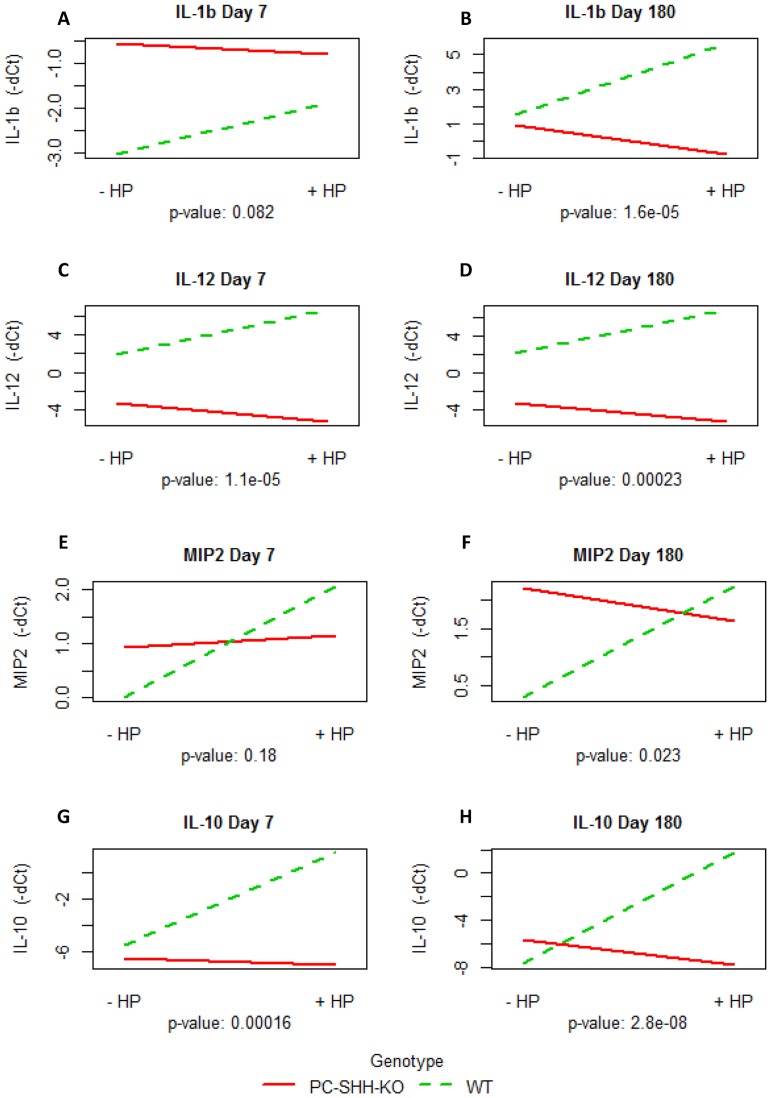
To determine if gastric SHH regulates expression of cytokines activated during H. pylori infection, WT and PC-SHHKO mice were infected with Brucella broth or H. pylori, and cytokine expression was measured by qPCR in tissues collected from mouse stomachs, 7 and 180 days post inoculation. Interaction plots shown in Figure 2 (and Figure S2 in Methods S1) examines if H. pylori affects cytokine expression and if this effect is different in WT and PC-SHHKO mice. Non-parallel lines that do not cross or crossing lines, imply an interaction effect between the two factors (genotype and infection status) [82]. An interaction effect between genotype and infection was observed for expression of all cytokines (Figure 2). Parallel lines would have implied that the effect of H. pylori on cytokine expression is the same in WT and PC-SHHKO mice. On day 7, the interaction effect was statistically significant for IL-12 and IL-10 but not for IL-1β and MIP-2. For day 180, statistically significant interaction effect between genotype and infection was observed for all cytokines.
Figure 2. Interaction between infection status and genotype.
RNA was extracted from stomachs of uninfected (-HP) and H. pylori-infected (+HP) wild type (WT) and parietal cell specific SHH knock out (PC-SHH-KO) mice 7 and 180 days post-inoculation. Expression of genes was measured by qPCR and interaction test was performed. Parallel lines imply that H. pylori has same effect on gene's expression in WT and PC-SHHKO mice whereas intersecting or non-parallel lines indicate an interaction between genotype and infection. The graphs show interaction plot between infection status and genotype for (A) IL1β on day 7, (B) IL1β on day 180, (C) IL-12 on day 7, (D) IL-12 on day 180, (E) MIP-2 on day 7 (F) MIP-2 on day 180 (G) IL-10 on day 7 and (H) IL-10 on day 180. P-value for interaction between infection and genotype were calculated by two-way ANOVA test. Y-axis: mean of negative dCT value of cytokine, X-axis: infection status, trace-factor: genotype.
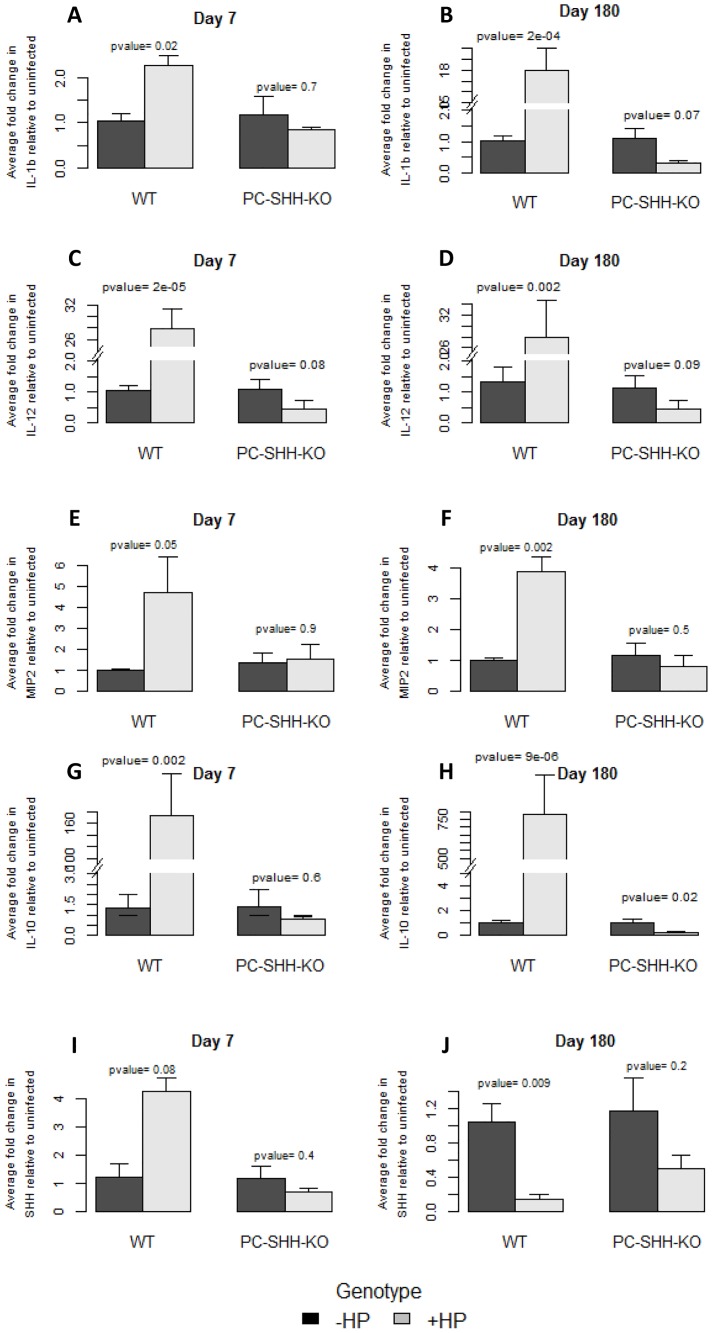
Two-way ANOVA test was performed, comparing dCT values from all four conditions (WT+ Brucella Broth, WT + H. pylori, PC-SHHKO + Brucella Broth, PC-SHHKO + H. pylori), followed by Bonferroni test to compare specific groups (Figure 3 and Figure S2 in Methods S1). All cytokines showed a statistically significant increase with H. pylori in WT mice, on both day 7 and day 180 (except MIP-2 on day 7, which increased with a P-value of 0.05). However, this increase in cytokines' expression with H. pylori was not observed in PC-SHHKO mice. These observations suggest the potential role of SHH in positive regulation of expression of IL-12, IL-1β, IL-10, IFNγ and MIP-2 during H. pylori infection. An unexpected trend was observed for IL-10 on day 180 in PC-SHHKO mice - IL-10 expression was significantly lower in H. pylori infected mice as compared with uninfected mice on day 180, although it did not change significantly with H. pylori on day 7.
Figure 3. Effect of H. pylori on SHH and cytokines' expression in WT and PC-SHHKO mouse stomachs, day 7 and day 180 post-inoculation.
RNA was extracted from stomachs of uninfected and H. pylori-infected wild type (WT) and parietal cell-specific SHH knock out (PC-SHH-KO) mice 7 and 180 days post-inoculation. Expression of genes was measured by qPCR and two-way ANOVA test was performed, followed by Bonferroni test to compare uninfected (-HP) with H. pylori infected group (+HP) in each genotype. The graphs show average fold change in expression of IL-1β (A, B), IL-12 (C, D), MIP-2 (E, F), IL-10 (G, H) and SHH (I, J) upon H. pylori infection relative to uninfected condition. Bars represent the mean ± SEM, n = 3-4 per group.
Mathematical Model Behavior
An important goal of model building is to ensure that the model can recapitulate the experimental trends. SHH knock-out condition was tested in the model that is curated using current pathway databases and literature. This model lacks the putative positive regulation link (kcat9 = 0) suggested by our qPCR analysis. The model exhibited no change in levels of IL-12, IL-10, IL-1β, IFNγ or MIP-2 upon simulating SHH KO condition (Figure S3 in Methods S1). The visual results of simulation were mathematically verified by examining the eigenvalues of the Jacobian matrix. All eigenvalues were real and negative, which indicated a constant-level of all constituents at equilibrium. The experimental observation of diminished concentration of IL-10 on day 180 highlights the need to account for a missing regulatory link between SHH and cytokines.
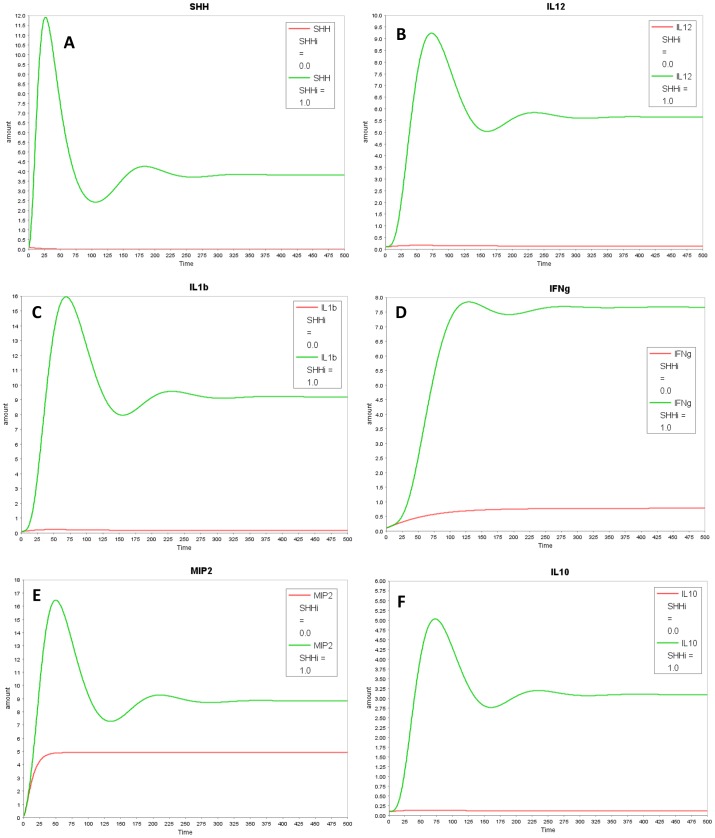
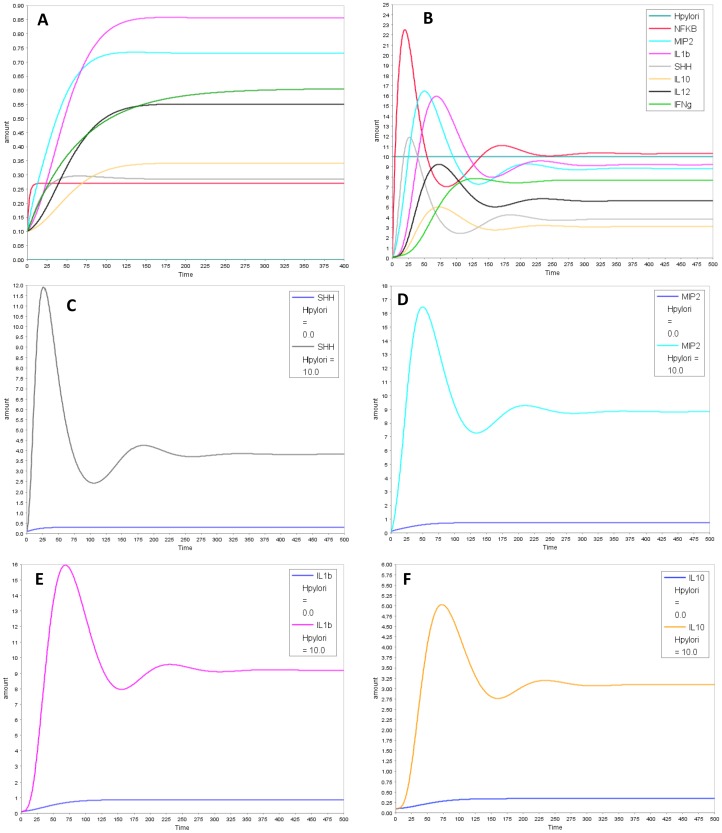
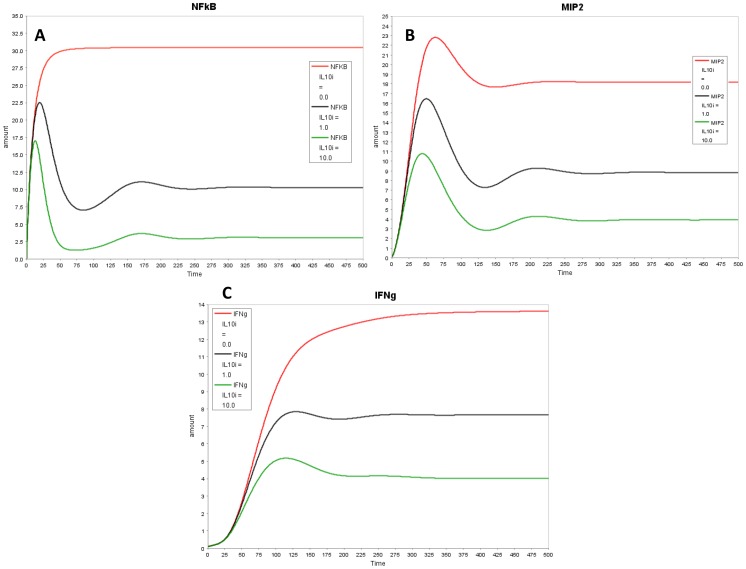
In our model with the putative link, all cytokines: IL-12, IL-1β, IFNγ, IL-10 and MIP-2 decreased with SHH knock-out (Figure 4), aligning the model closer to the experimental results. The decrease in MIP-2 was not considerable as MIP-2 is also formed through a parallel pathway via NFĸB. Next, we assessed the effect of H. pylori on the temporal profile for all model species in-silico (Figure 5). The simulation results align qualitatively with the experimental trends of cytokines. All cytokines increased with switching H. pylori “on” in the model. Temporal profile for each model species is compared in the presence and absence of H. pylori in Figure 5 (MIP-2, SHH, IL-1β and IL-10) and Figure S4 in Methods S1 (IL-12, IFNγ). Dynamic modeling of the reduced network shows that with H. pylori stimulation, there is increase in NFĸB which transcribes MIP-2 and SHH. SHH activates IL-1β, IL-12 and IL-10 through unknown mechanism (shown through “X” in the model). IL-10 in turn inhibits NFĸB, forming a negative feedback loop. This inhibition decreases NFĸB which lowers SHH and eventually decreases IL-10 and other cytokines. The decrease in IL-10 relieves the inhibition on NFĸB, allowing it to rise again. These interactions between NFĸB, SHH and IL-10 form a negative feedback loop and give rise to damped oscillations in the model (Figure 5B). Next, we simulated IL-10 overexpression and knock-out conditions. Overexpression of IL-10 led to decrease in NFĸB and in all the cytokines examined while knock-out of IL-10 in the model relieved the inhibition on NFĸB and resulted in increase in NFĸB and in pro-inflammatory cytokines downstream (Figure 6 and Figure S5 in Methods S1).
Figure 4. In-silico SHH KO results show a decrease in cytokines as comared to WT.
SHH KO condition was simulated by setting SHHi to zero. Graph A–F shows profiles of (A) SHH (B) IL-1β (C) IL12 (D) IFNγ (E) MIP2 (F) IL10. Wild type condition (SHHi = 1) is shown in green and in-silico SHH KO condition (SHHi = 0) is represented in red.
Figure 5. Temporal profiles of model species in uninfected and infected conditions.
Simulation results comparing temporal profiles of model species in (A) absence and (B) presence of H. pylori. Graph C–F show temporal profiles of (C) SHH (D) MIP-2 (E) IL-1β and (F) IL-10 in absence and presence of H. pylori.
Figure 6. In-silico IL-10 knock-out and overexpression.
Effect of IL-10 knock-out and overexpression on (A) NFkB (B) MIP-2 and (C) IFNγ. Wild type condition (IL10i = 1) is shown in black, in-silico IL-10 knok-out (IL10i = 0) in red and in-silico IL-10 overexpression (IL10i = 10) in green.
Temporal Analysis of GEO Data
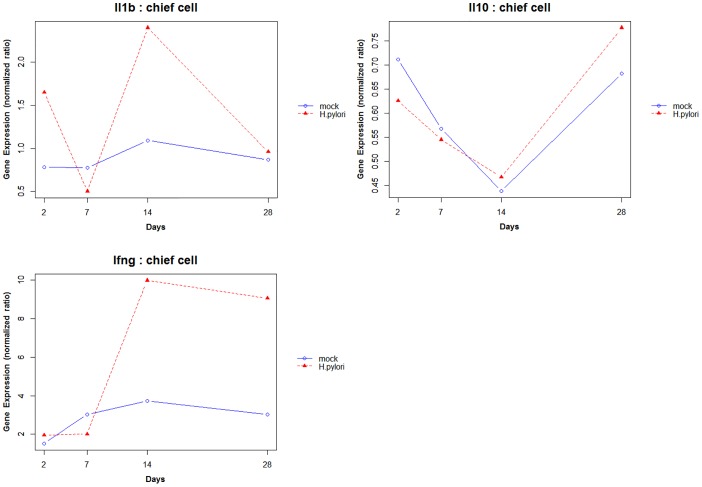
We also analyzed a microarray time-series dataset, available through GEO database. This data allowed us to study the temporal profile of IL-1β, IL-10 and IFNγ for both mock-infected and H. pylori infected mice. Figure 7 shows the trajectory of the three cytokines in chief cell for day 2, 7, 14 and 28. This time based analysis of the data suggests that these cytokines show a cyclic behavior as opposed to a linear trend. Figure S6 in Methods S1 shows the profiles for these cytokines in parietal and pit cells.
Figure 7. Trajectories of cytokines in mock-infected and H. pylori infected mice from GEO microarray dataset.
(A) IL-1β, (B) IL-10 and (C) IFNγ for day 2, 7, 14 and 28 from chief cell of mock-infected and H. pylori infected mice. The temporal profiles indicate that these cytokines potentially display a cyclic expression pattern in response to H. pylori infection.
Sensitivity and Stability Analysis
Our weakly non-linear system, modelled using a set of ordinary differential equations (ODE), can be viewed as a linear time-invariant system in the neighborhood of equilibrium and can be further studied using standard methods of analysis. In particular, the eigen-structure of the system can be used to algebraically identify the system behavior. For example, oscillations, a common result of negative feedback loops in biology, can be deduced from the complex eigenvalues. The damped oscillatory nature of our model, for example, depends on the negative inhibition of NFĸB by IL-10 and it is deduced from the complex eigenvalues (with negative real part) of the system's response. Removal of this inhibitory link in the model results in steady state behavior (Figure S7 in Methods S1) – which can be mathematically identified with the vanishing imaginary part of the eigenvalues. A sensitivity analysis is also performed to find the range of parameters across which the model displays damped oscillations. These results indicate that the cyclic behavior of the model remains viable within a large, biologically feasible, parameter region and is not limited to specific parameters and concentration values. Jacobian matrix and eigenvalues for the model were calculated using simulation tool – Copasi [83]. The parameters and concentrations that have the maximum influence on the imaginary part of eigenvalues were selected and varied over a wide range of values. The results presented in Table S5 in Methods S1 show that the model exhibits oscillatory trend even when original value of key parameters are increased or decreased by 50%. Further details of sensitivity analysis are provided in MethodsS1.
Discussion
H. pylori is associated with high risk of gastric diseases including chronic gastritis, gastric ulcer and stomach cancer [84]. Experimental evidence has shown that H. pylori activates pro-inflammatory Th1-dependent cytokines such as IL-12, IFNγ, IL-1β, TNFα [85]–[88]. Often, immune system succeeds in decreasing H. pylori numbers but the bacteria is not completely eradicated [86], [89] and a constant low level of infection and chronic inflammation may ensue. In recent years, SHH has emerged as an important player in acute immune response to H. pylori [15]. But the relationship between SHH and cytokines remains unexplored. Our analysis shows that SHH positively regulates expression of cytokines like IL-12, IL-1β, MIP-2, IL-10, however the mechanism remains unclear.
We developed a simple mathematical model of the cytokines and SHH activated during H. pylori infection. Our model captures the positive regulation of cytokines by SHH through an intermediate model species ‘X’. We speculate the following mechanism through which SHH may play a key role in regulating the cytokines during H. pylori infection. We have recently shown that SHH acts as a macrophage chemoattractant in response to H. pylori infection [15]. It has been reported by independent studies that monocytes and macrophages treated with H. pylori secrete IL-1β, IL-12, IL-10, TNFγ, IL-6 [47]–[49]. Thus the positive influence on the expression of cytokines by SHH, predicted by our qPCR data may be mediated through recruitment of macrophage by SHH to the gastric epithelium. IL-12 secreted in turn activates release of IFNγ from natural killer cells [52], [53]. In future, it will be interesting to experimentally test if treatment of H. pylori stimulated macrophages, with SHH can further enhance the expression of above mentioned cytokines. Results from such experiments can help to identify unknown molecule ‘X’ in the current model and reduce the existing knowledge gap between SHH and cytokines activated during H. pylori infection.
A key advantage of in-silico modeling is that it facilitates the investigation of perturbations on a scale and extent that would be challenging and expensive to achieve on the bench top in an expedient manner. To understand the role of the anti-inflammatory cytokine IL-10 in our model circuit, we simulated overexpression and knock-out IL-10 conditions in the model. Our in-silico outcomes qualitatively matched published results of similar experiments. Robinson et al studied the effect of IL-10 in gastric epithelial cells (AGS cell line) [60] where they showed that addition of recombinant human IL-10 to co-cultures of H. pylori strain 60190 with AGS cells caused decrease in nuclear NFĸB and IL-8. Similar decreasing trends in NFĸB and MIP-2 (murine homologue of human IL-8) were observed with increase of IL-10 in our model (Figure 6 A, B). Knock-out of IL-10 in our model relieved the inhibition on NFĸB and resulted in increase in NFĸB and in pro-inflammatory cytokines downstream. Figure 6 C shows increase in pro-inflammatory cytokine IFNγ with IL-10 KO as compared to that in WT, thus suggesting the potential protective role of IL-10 against H. pylori mediated gastritis. Bodger et al have also suggested that IL-10 secretion during H. pylori infection may serve a protective role, reducing local tissue damage caused by inflammation [90]. Ismail et al have shown that Helicobacter felis infection in IL-10 knock-out in mice resulted in a higher inflammation score and severe gastritis as compared to that in infected wild type mice [11]. They also cultured splenocytes from control uninfected and H. felis-infected WT and IL-10 KO mice, with sonicated H. felis Ag and the culture supernatants were evaluated for the concentration of IFN-γ. Splenocytes from H. felis-infected WT mice produced low levels of IFN-γ while splenocytes from H. felis-infected IL-10−/− mice produced large amounts of IFN-γ [11]. Future experiments evaluating the effect of IL-10 on pro-inflammatory cytokines (like IL-12, IL-1β, IFNγ, IL6, TNFα) and SHH can enhance our current understanding about the role of IL-10 in gastric mucosa during H. pylori infection.
The model also helps to bring out emergent properties of the network, which can guide future experimental studies and enhance our current understanding of the system. Our model suggests that NFĸB, SHH and the cytokines engage in a feedback loop which can result in damped oscillations. Temporal analysis of cytokines from a microarray Geo dataset also indicates that IL-1β, IFNγ and IL-10 from H. pylori infected mice, show a cyclic behavior rather than a linear trend. However, time course experiments capturing dynamic trajectories of expression of genes involved in the network are required to validate this hypothesis. Based on our preliminary results from analysis of Geo dataset, we propose that experiments capturing expression of cytokines and other genes at different time points (on scale of days) can inform about time dependent variation in expression of genes and also help to study the phase of correlations among pairs of genes and understand their relationships. We use the mathematical model as a tool to gain insights into cytokine and SHH relationship during H. pylori infection and as a hypothesis generating tool to predict host responses that may be associated with gastric disease or clinical treatments that may provide a better outcome.
Supporting Information
Supporting table. Table S6, Raw qPCR data.
(CSV)
Supporting files. Figure S1, Interaction Map of signaling pathways activated in host stomach in response to H. pylori. H. pylori virulence factors (CagA, VacA and PGN, shown in orange) activate cascade of signaling pathways in host gastric epithelium that leads to nuclear translocation of NFĸB. NFĸB further activates IL8/MIP-2 and SHH. Immune response to the bacteria involves recruitment of monocytes to gastric epithelium where they secrete cytokines like IL-12, IL-1β, TNFα, IL6, IL10 and IL8. Blue arrows show activation while red lines represent inhibition. The network was built using Cytoscape using information based on current literature. However, the current knowledge does not inform about any role of SHH in regulation of cytokines as suggested by our analysis. Figure S2, Effect of H. pylori infection on IFNγ expression on day 180 in wild-type (WT) and parietal cell-specific SHH KO (PC-SHH-KO) mice. RNA was extracted from stomachs of uninfected (-HP) and H. pylori-infected (+HP) wild type and parietal cell specific SHH knock-out mice 180 days post-inoculation and expression of IFNγ was measured by qPCR. (A) Interaction plot between infection status and genotype. P-value for interaction between infection and genotype was calculated by two-way ANOVA test. Y-axis: Negative dCT value of IFNγ, X-axis: infection status, trace-factor: genotype. (B) Fold change in expression of IFNγ relative to uninfected condition in WT and PC-SHHKO mice. Two-way ANOVA test was performed, followed by Bonferroni test to compare uninfected (-HP) with infected group (+HP) in each genotype. Bars represent the mean ± SEM, n = 4 per group. Figure S3, In-silico SHH KO results in model lacking the predicted link show no change in cytokines as comared to WT. SHH KO condition was simulated by setting SHHi to zero. Graph A-F shows profiles of (A) SHH (B) IL-1β (C) IL-12 (D) IFNγ (E) MIP2 (F) IL10. Wild type condition (SHHi = 1) is shown in yellow and in-silico SHH KO condition (SHHi = 0) is represented in red. Cytokines show no change (orange color is observed as a result of overlap of red and yellow lines). Figure S4, In-silico temporal profiles of (A) IL-12 and (B) IFNγ in absence and presence of H. pylori. Figure S5, In-silico IL-10 knock-out and overexpression. Effect of IL-10 knock-out and overexpression on (A) IL-1β (B) IL-12 and (C) SHH. Wild type condition (IL10i = 1) is shown in black, in-silico IL-10 knok-out (IL10i = 0) in red and in-silico IL-10 overexpression (IL10i = 10) in green. Figure S6, Trajectory of IL-1β, IL-10 and IFNγ for day 2, 7, 14 and 28 for parietal and pit cell from mock-infected and H.pylori infected mice. The temporal profiles indicate that these cytokines show non-uniform behavior than a linear trend. Figure S7, Temporal profiles of model species in absence of negative feedback on NFĸB by IL-10. The damped oscillatory nature of the model depends on the negative inhibition of NFĸB by IL-10. Removal of this inhibitory link in the model results in steady state behavior. Table S1, Model Assumptions. Table S2, Mathematical equations used in the model. Table S3, Species' parameters used in the model. Table S4, Kinetic parameters used in the model. Table S5, Sensitivity Analysis of model parameters for damped oscillations. Key parameters and their range for which the model shows damped oscillations.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by start-up funds from the Department of Molecular and Cellular Physiology, University of Cincinnati (Hamid R. Eghbalnia). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blaser MJ (1990) Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis 161: 626–633. [DOI] [PubMed] [Google Scholar]
- 2. Jhala NC, Siegal GP, Klemm K, Atkinson BF, Jhala DN (2003) Infiltration of Helicobacter pylori in the gastric mucosa. Am J Clin Pathol 119: 101–107 10.1309/YDTX-KE06-XHTH-FNP2 [DOI] [PubMed] [Google Scholar]
- 3. Kuipers EJ, Uyterlinde AM, Peña AS, Roosendaal R, Pals G, et al. (1995) Long-term sequelae of Helicobacter pylori gastritis. Lancet 345: 1525–1528. [DOI] [PubMed] [Google Scholar]
- 4.Sipponen P, Hyvärinen H (1993) Role of Helicobacter pylori in the pathogenesis of gastritis, peptic ulcer and gastric cancer. Scand J Gastroenterol Suppl 196: 3–6. [DOI] [PubMed]
- 5. Blaser MJ, Chyou PH, Nomura A (1995) Age at establishment of Helicobacter pylori infection and gastric carcinoma, gastric ulcer, and duodenal ulcer risk. Cancer Res 55: 562–565. [PubMed] [Google Scholar]
- 6. Huang JQ, Sridhar S, Chen Y, Hunt RH (1998) Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114: 1169–1179. [DOI] [PubMed] [Google Scholar]
- 7. Noto JM, Khizanishvili T, Chaturvedi R, Piazuelo MB, Romero-Gallo J, et al. (2013) Helicobacter pylori promotes the expression of Krüppel-like factor 5, a mediator of carcinogenesis, in vitro and in vivo. PLoS One 8: e54344 10.1371/journal.pone.0054344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zambon C-F, Basso D, Navaglia F, Belluco C, Falda A, et al. (2005) Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine 29: 141–152 10.1016/j.cyto.2004.10.013 [DOI] [PubMed] [Google Scholar]
- 9. Ernst P (1999) Review article: the role of inflammation in the pathogenesis of gastric cancer. Aliment Pharmacol Ther 13 Suppl 113–18. [DOI] [PubMed] [Google Scholar]
- 10. Sawai N, Kita M, Kodama T, Tanahashi T, Yamaoka Y, et al. (1999) Role of gamma interferon in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Infect Immun 67: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ismail HF, Fick P, Zhang J, Lynch RG, Berg DJ (2003) Depletion of neutrophils in IL-10(−/−) mice delays clearance of gastric Helicobacter infection and decreases the Th1 immune response to Helicobacter. J Immunol 170: 3782–3789. [DOI] [PubMed] [Google Scholar]
- 12. Algood HMS, Cover TL (2006) Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses. Clin Microbiol Rev 19: 597–613 10.1128/CMR.00006-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katoh Y, Katoh M (2005) Hedgehog signaling pathway and gastric cancer. Cancer Biol Ther 4: 1050–1054. [DOI] [PubMed] [Google Scholar]
- 14. Katoh Y, Katoh M (2006) Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review). Int J Mol Med 18: 1019–1023. [PubMed] [Google Scholar]
- 15. Schumacher MA, Donnelly JM, Engevik AC, Xiao C, Yang L, et al. (2012) Gastric Sonic Hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology 142: 1150–1159.e6 10.1053/j.gastro.2012.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El-Zaatari M, Kao JY, Tessier A, Bai L, Hayes MM, et al. (2013) Gli1 deletion prevents Helicobacter-induced gastric metaplasia and expansion of myeloid cell subsets. PLoS One 8: e58935 10.1371/journal.pone.0058935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S-Y, Han HS, Lee KY, Hwang TS, Kim JH, et al. (2007) Sonic hedgehog expression in gastric cancer and gastric adenoma. Oncol Rep 17: 1051–1055. [PubMed] [Google Scholar]
- 18. Myung K, Sang J, Sik H, Kyun D, Sun H, et al. (2010) Late reactivation of sonic hedgehog by Helicobacter pylori results in population of gastric epithelial cells that are resistant to apoptosis: Implication for gastric carcinogenesis. Cancer Lett 287: 44–53 10.1016/j.canlet.2009.05.032 [DOI] [PubMed] [Google Scholar]
- 19. Wan J, Zhou J, Zhao H, Wang M, Wei Z, et al. (2014) Sonic hedgehog pathway contributes to gastric cancer cell growth and proliferation. Biores Open Access 3: 53–59 10.1089/biores.2014.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin J, Donnelly JM, Houghton J, Zavros Y (2010) The role of sonic hedgehog reemergence during gastric cancer. Dig Dis Sci 55: 1516–1524 10.1007/s10620-010-1252-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, et al. (2001) Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology 121: 317–328. [DOI] [PubMed] [Google Scholar]
- 22. Xiao C, Ogle SA, Schumacher MA, Orr-Asman MA, Miller ML, et al. (2010) Loss of parietal cell expression of Sonic hedgehog induces hypergastrinemia and hyperproliferation of surface mucous cells. Gastroenterology 138: 550–e1 – 10.1053/j.gastro.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lowrey JA, Stewart GA, Lindey S, Hoyne GF, Dallman MJ, et al. (2002) Sonic hedgehog promotes cell cycle progression in activated peripheral CD4(+) T lymphocytes. J Immunol 169: 1869–1875. [DOI] [PubMed] [Google Scholar]
- 24. Crompton T, Outram SV, Hager-Theodorides AL (2007) Sonic hedgehog signalling in T-cell development and activation. Nat Rev Immunol 7: 726–735 10.1038/nri2151 [DOI] [PubMed] [Google Scholar]
- 25. Katoh M (2007) Dysregulation of stem cell signaling network due to germline mutation, SNP, Helicobacter pylori infection, epigenetic change and genetic alteration in gastric cancer. Cancer Biol Ther 6: 832–839. [DOI] [PubMed] [Google Scholar]
- 26. Dunaeva M, Voo S, van Oosterhoud C, Waltenberger J (2010) Sonic hedgehog is a potent chemoattractant for human monocytes: diabetes mellitus inhibits Sonic hedgehog-induced monocyte chemotaxis. Basic Res Cardiol 105: 61–71 10.1007/s00395-009-0047-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blaser MJ, Kirschner D (1999) Dynamics of Helicobacter pylori colonization in relation to the host response. Proc Natl Acad Sci U S A 96: 8359–8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kirschner DE, Blaser MJ (1995) The dynamics of Helicobacter pylori infection of the human stomach. J Theor Biol 176: 281–290 10.1006/jtbi.1995.0198 [DOI] [PubMed] [Google Scholar]
- 29.Bisset K, Alam MM, Bassaganya-Riera J, Carbo A, Eubank S, et al.. (2012) High-Performance Interaction-Based Simulation of Gut Immunopathologies with ENteric Immunity Simulator (ENISI). 2012 IEEE 26th Int Parallel Distrib Process Symp: 48–59. doi:10.1109/IPDPS.2012.15.
- 30. Carbo A, Bassaganya-Riera J, Pedragosa M, Viladomiu M, Marathe M, et al. (2013) Predictive computational modeling of the mucosal immune responses during Helicobacter pylori infection. PLoS One 8: e73365 10.1371/journal.pone.0073365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franke R, Müller M, Wundrack N, Gilles E-D, Klamt S, et al. (2008) Host-pathogen systems biology: logical modelling of hepatocyte growth factor and Helicobacter pylori induced c-Met signal transduction. BMC Syst Biol 2: 4 10.1186/1752-0509-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghosh S, Matsuoka Y, Asai Y, Hsin K-Y, Kitano H (2011) Software for systems biology: from tools to integrated platforms. Nat Rev Genet 12: 821–832 10.1038/nrg3096 [DOI] [PubMed] [Google Scholar]
- 33. Polk DB, Peek RM (2010) Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 10: 403–414 10.1038/nrc2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fox JG, Wang TC (2007) Inflammation, atrophy, and gastric cancer. J Clin Invest 117: 60–69 10.1172/JCI30111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. (2004) Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5: 1166–1174 10.1038/ni1131 [DOI] [PubMed] [Google Scholar]
- 36. Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, et al. (2002) SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295: 683–686 10.1126/science.1067147 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, et al. (2009) Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 5: 23–34 10.1016/j.chom.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 38. Nozawa Y, Nishihara K, Peek RM, Nakano M, Uji T, et al. (2002) Identification of a signaling cascade for interleukin-8 production by Helicobacter pylori in human gastric epithelial cells. Biochem Pharmacol 64: 21–30. [DOI] [PubMed] [Google Scholar]
- 39. Backert S, Naumann M (2010) What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori. Trends Microbiol 18: 479–486 10.1016/j.tim.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 40. Allison CC, Kufer TA, Kremmer E, Kaparakis M, Ferrero RL (2009) Helicobacter pylori induces MAPK phosphorylation and AP-1 activation via a NOD1-dependent mechanism. J Immunol 183: 8099–8109 10.4049/jimmunol.0900664 [DOI] [PubMed] [Google Scholar]
- 41. Nakayama M, Kimura M, Wada A, Yahiro K, Ogushi K, et al. (2004) Helicobacter pylori VacA activates the p38/activating transcription factor 2-mediated signal pathway in AZ-521 cells. J Biol Chem 279: 7024–7028 10.1074/jbc.M308898200 [DOI] [PubMed] [Google Scholar]
- 42. Naito Y, Yoshikawa T (2002) Molecular and cellular mechanisms involved in Helicobacter pylori-induced inflammation and oxidative stress. Free Radic Biol Med 33: 323–336. [DOI] [PubMed] [Google Scholar]
- 43. Yamada H, Aihara T, Okabe S (2001) Mechanism for Helicobacter pylori stimulation of interleukin-8 production in a gastric epithelial cell line (MKN 28): roles of mitogen-activated protein kinase and interleukin-1beta. Biochem Pharmacol 61: 1595–1604. [DOI] [PubMed] [Google Scholar]
- 44. Seo JH, Lim JW, Kim H, Kim KH (2004) Helicobacter pylori in a Korean isolate activates mitogen-activated protein kinases, AP-1, and NF-kappaB and induces chemokine expression in gastric epithelial AGS cells. Lab Invest 84: 49–62 10.1038/sj.labinvest.3700010 [DOI] [PubMed] [Google Scholar]
- 45. Aihara M, Tsuchimoto D, Takizawa H, Azuma A, Wakebe H, et al. (1997) Mechanisms involved in Helicobacter pylori-induced interleukin-8 production by a gastric cancer cell line, MKN45. Infect Immun 65: 3218–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Remick DG, Green LB, Newcomb DE, Garg SJ, Bolgos GL, et al. (2001) CXC chemokine redundancy ensures local neutrophil recruitment during acute inflammation. Am J Pathol 159: 1149–1157 10.1016/S0002-9440(10)61791-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fehlings M, Drobbe L, Moos V, Renner Viveros P, Hagen J, et al. (2012) Comparative analysis of the interaction of Helicobacter pylori with human dendritic cells, macrophages, and monocytes. Infect Immun 80: 2724–2734 10.1128/IAI.00381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mai UE, Perez-Perez GI, Wahl LM, Wahl SM, Blaser MJ, et al. (1991) Soluble surface proteins from Helicobacter pylori activate monocytes/macrophages by lipopolysaccharide-independent mechanism. J Clin Invest 87: 894–900 10.1172/JCI115095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bimczok D, Smythies LE, Waites KB, Grams JM, Stahl RD, et al. (2013) Helicobacter pylori infection inhibits phagocyte clearance of apoptotic gastric epithelial cells. J Immunol 190: 6626–6634 10.4049/jimmunol.1203330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bliss CM, Golenbock DT, Keates S, Linevsky JK, Kelly CP (1998) Helicobacter pylori lipopolysaccharide binds to CD14 and stimulates release of interleukin-8, epithelial neutrophil-activating peptide 78, and monocyte chemotactic protein 1 by human monocytes. Infect Immun 66: 5357–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Jonge R, Kusters JG, Timmer MS, Gimmel V, Appelmelk BJ, et al. (2001) The role of Helicobacter pylori virulence factors in interleukin production by monocytic cells. FEMS Microbiol Lett 196: 235–238. [DOI] [PubMed] [Google Scholar]
- 52. Lindgren A, Pavlovic V, Flach C-F, Sjöling A, Lundin S (2011) Interferon-gamma secretion is induced in IL-12 stimulated human NK cells by recognition of Helicobacter pylori or TLR2 ligands. Innate Immun 17: 191–203 10.1177/1753425909357970 [DOI] [PubMed] [Google Scholar]
- 53. Yun CH, Lundgren A, Azem J, Sjöling A, Holmgren J, et al. (2005) Natural killer cells and Helicobacter pylori infection: bacterial antigens and interleukin-12 act synergistically to induce gamma interferon production. Infect Immun 73: 1482–1490 10.1128/IAI.73.3.1482-1490.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pellicanò A, Sebkova L, Monteleone G, Guarnieri G, Imeneo M, et al. (2007) Interleukin-12 drives the Th1 signaling pathway in Helicobacter pylori-infected human gastric mucosa. Infect Immun 75: 1738–1744 10.1128/IAI.01446-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim J-H, Choi YJ, Lee SH, Shin HS, Lee IO, et al. (2010) Effect of Helicobacter pylori infection on the sonic hedgehog signaling pathway in gastric cancer cells. Oncol Rep 23: 1523–1528. [DOI] [PubMed] [Google Scholar]
- 56. Singh AP, Arora S, Bhardwaj A, Srivastava SK, Kadakia MP, et al. (2012) CXCL12/CXCR4 protein signaling axis induces sonic hedgehog expression in pancreatic cancer cells via extracellular regulated kinase- and Akt kinase-mediated activation of nuclear factor κB: implications for bidirectional tumor-stromal interactions. J Biol Chem 287: 39115–39124 10.1074/jbc.M112.409581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakashima H, Nakamura M, Yamaguchi H, Yamanaka N, Akiyoshi T, et al. (2006) Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res 66: 7041–7049 10.1158/0008-5472.CAN-05-4588 [DOI] [PubMed] [Google Scholar]
- 58.Waghray M, Zavros Y, Saqui-Salces M, El-Zaatari M, Alamelumangapuram CB, et al. (2010) Interleukin-1beta promotes gastric atrophy through suppression of Sonic Hedgehog. Gastroenterology 138: : 562–72, 572.e1–2. doi:10.1053/j.gastro.2009.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haeberle H a, Kubin M, Bamford KB, Garofalo R, Graham DY, et al. (1997) Differential stimulation of interleukin-12 (IL-12) and IL-10 by live and killed Helicobacter pylori in vitro and association of IL-12 production with gamma interferon-producing T cells in the human gastric mucosa. Infect Immun 65: 4229–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robinson K, Kenefeck R, Pidgeon EL, Shakib S, Patel S, et al. (2008) Helicobacter pylori-induced peptic ulcer disease is associated with inadequate regulatory T cell responses. Gut 57: 1375–1385 10.1136/gut.2007.137539 [DOI] [PubMed] [Google Scholar]
- 61. Wang P, Wu P, Siegel MI, Egan RW, Billah MM (1995) Interleukin (IL)-10 inhibits nuclear factor kappa B (NF kappa B) activation in human monocytes. IL-10 and IL-4 suppress cytokine synthesis by different mechanisms. J Biol Chem 270: 9558–9563. [DOI] [PubMed] [Google Scholar]
- 62. Romano MF, Lamberti A, Petrella A, Bisogni R, Tassone PF, et al. (1996) IL-10 inhibits nuclear factor-kappa B/Rel nuclear activity in CD3-stimulated human peripheral T lymphocytes. J Immunol 156: 2119–2123. [PubMed] [Google Scholar]
- 63. Kundu P, De R, Pal I, Mukhopadhyay AK, Saha DR, et al. (2011) Curcumin alleviates matrix metalloproteinase-3 and -9 activities during eradication of Helicobacter pylori infection in cultured cells and mice. PLoS One 6: e16306 10.1371/journal.pone.0016306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Algood HMS, Gallo-Romero J, Wilson KT, Peek RM, Cover TL (2007) Host response to Helicobacter pylori infection before initiation of the adaptive immune response. FEMS Immunol Med Microbiol 51: 577–586 10.1111/j.1574-695X.2007.00338.x [DOI] [PubMed] [Google Scholar]
- 65. Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, et al. (1997) A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112: 1386–1397. [DOI] [PubMed] [Google Scholar]
- 66. Thompson LJ, Danon SJ, Wilson JE, O'Rourke JL, Salama NR, et al. (2004) Chronic Helicobacter pylori infection with Sydney strain 1 and a newly identified mouse-adapted strain (Sydney strain 2000) in C57BL/6 and BALB/c mice. Infect Immun 72: 4668–4679 10.1128/IAI.72.8.4668-4679.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Franks NR, Sneyd J, Theraulaz G, Bonabeau E (2001) Self-Organization in Biological Systems.
- 68. Ideker T, Galitski T, Hood L (2001) A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2: 343–372 10.1146/annurev.genom.2.1.343 [DOI] [PubMed] [Google Scholar]
- 69. Bhalla US, Iyengar R (1999) Emergent properties of networks of biological signaling pathways. Science 283: 381–387. [DOI] [PubMed] [Google Scholar]
- 70. Lehmann J, Bellmann S, Werner C, Schröder R, Schütze N, et al. (2001) IL-12p40-dependent agonistic effects on the development of protective innate and adaptive immunity against Salmonella enteritidis. J Immunol 167: 5304–5315. [DOI] [PubMed] [Google Scholar]
- 71. Koschorreck M, Conzelmann H, Ebert S, Ederer M, Gilles ED (2007) Reduced modeling of signal transduction - a modular approach. BMC Bioinformatics 8: 336 10.1186/1471-2105-8-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anderson J, Chang Y-C, Papachristodoulou A (2011) Model decomposition and reduction tools for large-scale networks in systems biology. Automatica 47: 1165–1174 10.1016/j.automatica.2011.03.010 [DOI] [Google Scholar]
- 73. Maurya MR, Bornheimer SJ, Venkatasubramanian V, Subramaniam S (2005) Reduced-order modelling of biochemical networks: application to the GTPase-cycle signalling module. Syst Biol (Stevenage) 152: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brazhnik P, Tyson JJ (2006) Cell cycle control in bacteria and yeast: a case of convergent evolution? Cell Cycle 5: 522–529. [DOI] [PubMed] [Google Scholar]
- 75. Novak B, Pataki Z, Ciliberto A, Tyson JJ (2001) Mathematical model of the cell division cycle of fission yeast. Chaos 11: 277–286 10.1063/1.1345725 [DOI] [PubMed] [Google Scholar]
- 76. Ciliberto A, Petrus MJ, Tyson JJ, Sible JC (2003) A kinetic model of the cyclin EyCdk2 developmental timer in Xenopus laevis embryos. Biophys Chem 104: 573–589 10.1016/S0301-4622 [DOI] [PubMed] [Google Scholar]
- 77. Chen KC, Calzone L, Csikasz-nagy A, Cross FR, Novak B, et al. (2004) Integrative Analysis of Cell Cycle Control in Budding Yeast. Mol Biol Cell 15: 3841–3862 10.1091/mbc.E03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang T, Brazhnik P, Tyson JJ (2009) Computational analysis of dynamical responses to the intrinsic pathway of programmed cell death. Biophys J 97: 415–434 10.1016/j.bpj.2009.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, et al. (2000) Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A 97: 11250–11255 10.1073/pnas.210171597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Funahashi A, Morohashi M, Kitano H, Tanimura N (2003) CellDesigner: a process diagram editor for gene-regulatory and. 1: 159–162. [Google Scholar]
- 81. Mueller A, Merrell DS, Grimm J, Falkow S (2004) Profiling of microdissected gastric epithelial cells reveals a cell type—specific response to Helicobacter pylori infection. Gastroenterology 127: 1446–1462 10.1053/j.gastro.2004.08.054 [DOI] [PubMed] [Google Scholar]
- 82.Reinard JC (2006) Communication Research Statistics p. 218.
- 83. Hoops S, Sahle S, Gauges R, Lee C, Pahle J, et al. (2006) COPASI–a COmplex PAthway SImulator. Bioinformatics 22: 3067–3074 10.1093/bioinformatics/btl485 [DOI] [PubMed] [Google Scholar]
- 84. Lacy BE, Rosemore J (2001) Helicobacter pylori: ulcers and more: the beginning of an era. J Nutr 131: 2789S–2793S. [DOI] [PubMed] [Google Scholar]
- 85. Lohoff M, Röllinghoff M, Sommer F (2000) Helicobacter pylori gastritis: a Th1 mediated disease? J Biotechnol 83: 33–36. [DOI] [PubMed] [Google Scholar]
- 86. Moss SF, Blaser MJ (2005) Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol 2: 90–7 ; quiz 1 p following 113. 10.1038/ncponc0081 [DOI] [PubMed] [Google Scholar]
- 87. Luzza F, Parrello T, Sebkova L, Pensabene L, Imeneo M, et al. (n.d.) Expression of proinflammatory and Th1 but not Th2 cytokines is enhanced in gastric mucosa of Helicobacter pylori infected children. Dig Liver Dis 33: 14–20. [DOI] [PubMed] [Google Scholar]
- 88. Sommer F, Faller G, Konturek P, Kirchner T, Hahn EG, et al. (1998) Antrum- and corpus mucosa-infiltrating CD4(+) lymphocytes in Helicobacter pylori gastritis display a Th1 phenotype. Infect Immun 66: 5543–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mohammadi M, Nedrud J, Redline R, Lycke N, Czinn SJ (1997) Murine CD4 T-cell response to Helicobacter infection: TH1 cells enhance gastritis and TH2 cells reduce bacterial load. Gastroenterology 113: 1848–1857. [DOI] [PubMed] [Google Scholar]
- 90.Bodger K, Wyatt JI, Heatley RV (1997) Gastric mucosal secretion of interleukin- 10: relations to histopathology, Helicobacter pyloni status, and tumour necrosis factor-a secretion: 739–744. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting table. Table S6, Raw qPCR data.
(CSV)
Supporting files. Figure S1, Interaction Map of signaling pathways activated in host stomach in response to H. pylori. H. pylori virulence factors (CagA, VacA and PGN, shown in orange) activate cascade of signaling pathways in host gastric epithelium that leads to nuclear translocation of NFĸB. NFĸB further activates IL8/MIP-2 and SHH. Immune response to the bacteria involves recruitment of monocytes to gastric epithelium where they secrete cytokines like IL-12, IL-1β, TNFα, IL6, IL10 and IL8. Blue arrows show activation while red lines represent inhibition. The network was built using Cytoscape using information based on current literature. However, the current knowledge does not inform about any role of SHH in regulation of cytokines as suggested by our analysis. Figure S2, Effect of H. pylori infection on IFNγ expression on day 180 in wild-type (WT) and parietal cell-specific SHH KO (PC-SHH-KO) mice. RNA was extracted from stomachs of uninfected (-HP) and H. pylori-infected (+HP) wild type and parietal cell specific SHH knock-out mice 180 days post-inoculation and expression of IFNγ was measured by qPCR. (A) Interaction plot between infection status and genotype. P-value for interaction between infection and genotype was calculated by two-way ANOVA test. Y-axis: Negative dCT value of IFNγ, X-axis: infection status, trace-factor: genotype. (B) Fold change in expression of IFNγ relative to uninfected condition in WT and PC-SHHKO mice. Two-way ANOVA test was performed, followed by Bonferroni test to compare uninfected (-HP) with infected group (+HP) in each genotype. Bars represent the mean ± SEM, n = 4 per group. Figure S3, In-silico SHH KO results in model lacking the predicted link show no change in cytokines as comared to WT. SHH KO condition was simulated by setting SHHi to zero. Graph A-F shows profiles of (A) SHH (B) IL-1β (C) IL-12 (D) IFNγ (E) MIP2 (F) IL10. Wild type condition (SHHi = 1) is shown in yellow and in-silico SHH KO condition (SHHi = 0) is represented in red. Cytokines show no change (orange color is observed as a result of overlap of red and yellow lines). Figure S4, In-silico temporal profiles of (A) IL-12 and (B) IFNγ in absence and presence of H. pylori. Figure S5, In-silico IL-10 knock-out and overexpression. Effect of IL-10 knock-out and overexpression on (A) IL-1β (B) IL-12 and (C) SHH. Wild type condition (IL10i = 1) is shown in black, in-silico IL-10 knok-out (IL10i = 0) in red and in-silico IL-10 overexpression (IL10i = 10) in green. Figure S6, Trajectory of IL-1β, IL-10 and IFNγ for day 2, 7, 14 and 28 for parietal and pit cell from mock-infected and H.pylori infected mice. The temporal profiles indicate that these cytokines show non-uniform behavior than a linear trend. Figure S7, Temporal profiles of model species in absence of negative feedback on NFĸB by IL-10. The damped oscillatory nature of the model depends on the negative inhibition of NFĸB by IL-10. Removal of this inhibitory link in the model results in steady state behavior. Table S1, Model Assumptions. Table S2, Mathematical equations used in the model. Table S3, Species' parameters used in the model. Table S4, Kinetic parameters used in the model. Table S5, Sensitivity Analysis of model parameters for damped oscillations. Key parameters and their range for which the model shows damped oscillations.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.