Abstract
We investigated MET mRNA expression status using RNA in situ hybridization (ISH) technique in primary and metastatic lesions of 535 surgically resected gastric carcinoma (GC) cases. We compared the results with those of immunohistochemistry and silver in situ hybridization, and examined the association with clinicopathologic characteristics and prognosis. Among 535 primary GCs, 391 (73.1%) were scored 0, 87 (16.3%) were scored 1, 38 (7.1%) were scored 2, 12 (2.2%) were scored 3 and 7 (1.3%) were scored 4 by RNA ISH. High MET mRNA expression (score ≥3) was associated with lymph node metastasis (P = .014), distant metastasis (P = .001), and higher TNM stage (P<.001). MET mRNA expression was correlated with protein expression (r = 0.398; P<.001) and gene copy number (r = 0.345; P<.001). The patients showing high-MET mRNA in primary or metastatic lesions had shorter overall survival than those showing low-MET mRNA (primary tumors, P = .002; metastatic lymph nodes, P<.001). The patients showing positive conversion of MET mRNA status in metastatic lymph node had shorter overall survival than those with no conversion (P = .011). Multivariate analysis demonstrated that high MET mRNA expression in metastatic lymph node was an independent prognostic factor for overall survival (P = .007). Therefore, this study suggests that MET mRNA expression assessed by RNA ISH could be useful as a potential marker to identify MET oncogene-addicted GC.
Introduction
During the past decade, receptor tyrosine kinase (RTK) pathways have proven to be attractive drug targets for anticancer therapy [1], and the MET pathway is one of these promising targets. MET is a proto-oncogene located on the 7q31 locus and encodes an RTK for hepatocyte growth factor (HGF) [2], [3]. The tight regulation of the HGF/MET pathway that is observed in development and regeneration is lost in cancer, and such deregulation occurs through multiple mechanisms [1]. Aberrant MET activation plays important roles in cancer cell survival, growth, angiogenesis, and metastasis in various cancers including lung, breast, kidney, and gastrointestinal tract malignancies [4]. However, although patient stratification according to MET expression or activity is important for therapeutic success, the methods for assessing the level of MET expression or activity have not been established [4].
For gastric carcinoma (GC), aberrant MET activation has been thought to be related to a gene dosage effect [5], and MET gene amplification (GA) or protein overexpression has been associated with aggressive tumor characteristics and/or worse clinical outcome [6]–[14]. Furthermore, MET-amplified or -overexpressed GC showed response to treatment with several inhibitors of the HGF/MET signaling pathway in preclinical studies [15] and phase I clinical trials [12], [16]. Hence, MET inhibition has the potential as another successful therapeutic strategy following human epidermal growth factor receptor 2 (HER2)-targeted therapy in advanced GC. However, previous studies have used various methods to identify MET-positive GC and have shown discrepancy in the prevalence of MET overexpression or amplification: MET overexpression ranged 18% to 73.7% in studies using immunohistochemistry (IHC) [7]–[9], [13], [14], [17], MET gene copy number (GCN) gain ranged 10% to 21.2% in studies using quantitative real-time polymerase chain reaction (qPCR) [10], [11], and MET GA ranged 2% to 3.9% in studies using fluorescence in situ hybridization (FISH) [12], [18] or silver in situ hybridization (SISH) [13]. Of these methods, IHC is widely used in clinical practice and the most likely screening method for detection of MET-positive GC. However, further exploration is still needed to find a predictive biomarker or assay methodology for MET inhibition therapy.
In this study, we performed an RNA in situ hybridization (ISH) assay using paired DNA oligonucleotide probes and preamplifier-amplifier-label probes for visualization [19]. This method uses formalin-fixed, paraffin-embedded (FFPE) tissues and allows single-molecule visualization under a bright-field microscope. In our previous study, we proved that HER2 mRNA expression evaluated by RNA ISH was well correlated with protein overexpression and GA evaluated by IHC and FISH in 211 GC cases [20]. Also, we showed the correlation between MET GCN and protein expression in a previous study [13]. Here, we evaluated MET mRNA expression using RNA ISH method, and compared the results with those of IHC and SISH in a large series of GC. In addition, clinicopathologic parameters and clinical outcomes of GC patients according to MET mRNA expression status were evaluated.
Materials and Methods
Patients and tissue specimens
We collected archival tissue samples of GC patients who consecutively underwent gastrectomy at Seoul National University Hospital, Seoul, Korea, from January 2004 through December 2005. Finally, 535 samples of primary GC and 199 samples of synchronous regional metastatic lymph node (LN) from 535 patients were available for this study. The clinicopathologic characteristics of the patients were examined by reviewing medical charts and pathologic records (Table 1). TNM stage was classified according to the system of the American Joint Committee on Cancer Staging Manual, 7th edition. Clinical outcomes were followed up from the date of surgery until death or 60 months.
Table 1. Demographic and clinical characteristics of 535 gastric carcinoma patients.
| Characteristics | |
| Median age (range), y | 60 (24–87) |
| Gender, n (%) | |
| Male | 368 (68.8) |
| Female | 167 (31.2) |
| Tumor location, n (%) | |
| Upper third | 53 (16.3) |
| Middle third | 90 (27.6) |
| Lower third | 170 (52.1) |
| Tumor histology and differentiation, n (%) | |
| Tubular/Papillary ADC, WD | 37 (6.9) |
| Tubular/Papillary ADC, MD | 179 (33.5) |
| Tubular/Papillary ADC, PD | 206 (38.5) |
| Signet ring cell carcinoma | 83 (15.5) |
| Others | 30 (5.6) |
| Lauren classification, n (%) | |
| Intestinal | 238 (44.5) |
| Diffuse | 209 (39.1) |
| Mixed/indeterminate | 88 (16.4) |
| Radicality. n (%) | |
| R0 | 499 (93.3) |
| R1/R2 | 36 (6.7) |
| Adjuvant chemotherapy, n (%) | |
| No | 234 (43.7) |
| Yes | 301 (56.3) |
| TNM stage, n (%) | |
| I | 170 (31.8) |
| II | 142 (26.5) |
| III | 175 (32.7) |
| IV | 48 (9.0) |
Abbreviations: ADC, adenocarcinoma; MD, moderately differentiated; PD, poorly differentiated; TNM, Tumor-Node-Metastasis; WD, well differentiated.
All tissue samples were fixed in 10% buffered formalin for 24–48 hours and then embedded in paraffin. Core tissues (2 mm in diameter) were taken using a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). For the primary GCs, the invasion front of each primary tumor was selected. Metastatic LNs were subjected to the tissue array but the cases with micrometastasis were excluded. Total 22 tissue microarray blocks which contained up to 60 cores were constructed.
Ethical statement
All human specimens were obtained during therapeutic surgery. The participants did not provide the written consent to participate in this study. The retrospective study was performed using the samples over the shelves after the pathologic diagnosis, and all of the samples were anonymized before the study. Our IRB (Seoul National University Hospital) approved this retrospective study under the condition of the anonymization (Reference: H-1006-035-320).
RNA ISH
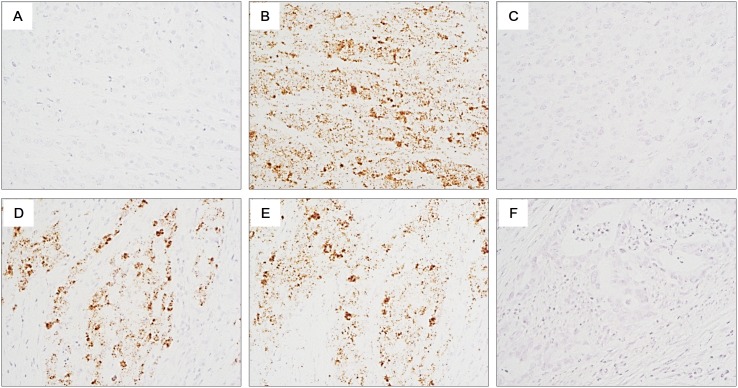
For in situ detection of MET mRNA, the RNAscope FFPE 2.0 assay kit (Advanced Cell Diagnostics, Hayward, CA, USA) was used according to the manufacturer’s instructions. Briefly, 2- to 3-µm thick FFPE tissue sections were deparaffinized, heated, treated by protease, and hybridized with probe at 40°C for 2 hours (the reference sequence, NM_001127500; probe region, 1236–2257). After washing and amplification, 3, 3′-diaminobenzidine was added for detection of target RNA. Nuclei were counterstained with hematoxylin. Positive staining was indicated by brown punctate dots in the nucleus and/or cytoplasm. MET mRNA expression levels were categorized into 5 grades according to the manufacturer’s scoring guideline: score 0, no staining or <1 dot per cell; score 1, 1–3 dots per cell (visible at 20–40×); score 2, 4–10 dots per cell and no or very few dot clusters (visible at 20–40×); score 3, >10 dots per cell and <10% positive cells have dot clusters (visible at 20×); score 4, >10 dots per cell and >10% positive cells have dot clusters (visible at 20×) (Figure 1). The probes for UBC (ubiquitin C) and dapB (a bacterial gene) were used as the positive and negative control, respectively. Samples were considered adequate when the UBC mRNA signals were easily visible under a 10x objective lens and the dapB signal was not visible.
Figure 1. Representative figures of RNA in situ hybridization (ISH).
(A–C) a negative case showing MET RNA ISH score 0: (A) MET mRNA, (B) UBC mRNA, and (C) dapB mRNA. (D–F) a positive case showing MET RNA ISH score 4: (D) MET mRNA, (E) UBC mRNA, and (F) dapB mRNA (original magnification: ×400).
IHC and SISH
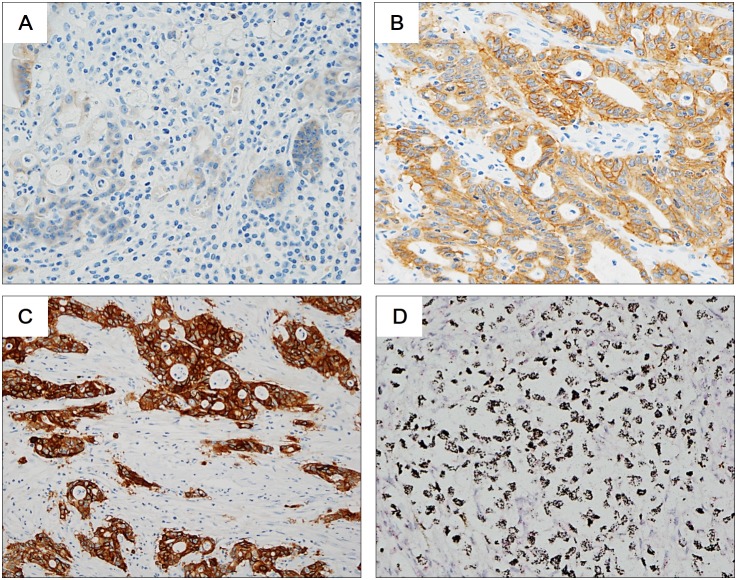
Immunohistochemical staining for MET was performed with anti-total MET (SP44) rabbit monoclonal primary antibodies (Ventana Medical Systems, Tucson, AZ, USA). An automatic immunostainer (BenchMark XT, Ventana Medical Systems) was used according to the manufacturer’s instructions. MET immunostaining was scored with the HercepTest scoring guidelines for GC (DAKO, Glostrup, Denmark): score 0, no membrane staining or membrane staining in <10% of tumor cells; score 1, faint/barely perceptible partial membrane staining in >10% of tumor cells; score 2, weak to moderate staining of the entire membrane in >10% of tumor cells; score 3, strong staining of the entire membrane in >10% of tumor cells (Figure 2).
Figure 2. Representative figures of MET immunohistochemistry (IHC) and silver in situ hybridization.
(A) IHC score 1, (B) IHC score 2, (C) IHC score 3, and (D) gene amplification (original magnification: ×400).
Dual-color SISH assay was performed with INFORM MET DNA probe and INFORM Chromosome 7 probe (Ventana Medical Systems) on a Ventana BenchMark XT following the manufacturer’s protocols. Signals were enumerated in 40 tumor nuclei per core, and MET gene status was classified into 6 groups using the University of Colorado Cancer Center criteria for epidermal growth factor receptor gene [21] (Figure 2).
Statistical analysis
The χ2 test or Fisher’s exact test was used to test the association between MET status and clinicopathologic factors. The Student’s t-test was used to compare means of continuous variables. The Spearman correlation test was used to assess the relationship between RNA ISH results and IHC or SISH results. The Kaplan-Meier method was used to estimate overall survival (OS), and OS differences between the groups with different MET status were compared by using the log-rank test. Multivariate survival analysis was performed using the Cox proportional hazards ratio model. Data analysis was conducted by using SPSS version 20.0 (SPSS, Chicago, IL, USA), and the results were considered significant when P<.05.
Results
1. MET mRNA status assessed by RNA ISH
Of 535 primary tumors, 391 (73.1%), 87 (16.3%), 38 (7.1%), 12 (2.2%) and 7 (1.3%) showed RNA ISH score 0, 1, 2, 3, and 4, respectively. When we compared the results of RNA ISH with clinicopathologic data including survival, and analyzed results of RNA ISH with IHC and SISH data, the groups of score 3 and 4 showed distinct features. Therefore, we regarded the score 3 and 4 as high-MET mRNA group, and total 19 cases (3.5%) belonged to high-MET mRNA.
High-MET mRNA was associated with older age (P = .002), larger tumor size (P = .006), LN metastasis (P = .014), lymphatic invasion (P<.001), increased number of metastatic lymph nodes (P<.001), distant metastasis (P = .001), and higher TNM stage (P<.001) when compared to low-MET mRNA (Table 2). However, high-MET mRNA did not show any association with gender, tumor location, Lauren classification, and invasion depth (Table 2).
Table 2. Clinicopathologic characteristics of gastric carcinoma patients according to MET mRNA expression status.
| Characteristics | MET mRNA status by RNA ISH | ||
| Low-MET mRNA | High-MET mRNA | P-value | |
| (score 0–2) | (score 3–4) | ||
| n = 516 (96.4%) | n = 19 (3.6%) | ||
| Mean age, y | 57.9 | 70 | .002 |
| Mean tumor size, cm | 5.64 | 7.62 | .006 |
| Gender, n (%) | .639 | ||
| Male | 354 (69.6) | 14 (73.7) | |
| Female | 162 (31.4) | 5 (26.3) | |
| Lauren classification, n (%) | .832 | ||
| Intestinal | 230 (44.6) | 8 (42.1) | |
| Diffuse/mixed | 286 (55.4) | 11 (57.9) | |
| Tumor invasion, n (%) | .392 | ||
| EGC | 113 (21.9) | 2 (10.5) | |
| AGC | 403 (78.1) | 17 (89.5) | |
| LN metastasis, n (%) | .014 | ||
| Absent | 277 (41.5) | 3 (15.8) | |
| Present | 391 (58.5) | 16 (84.2) | |
| Distant metastasis, n (%) | .001 | ||
| Absent | 612 (91.6) | 12 (63.2) | |
| Present | 56 (8.4) | 7 (36.8) | |
| TNM stage, n (%) | <.001 | ||
| I | 191 (28.6) | 1 (5.3) | |
| II | 201 (30.1) | 3 (15.8) | |
| III | 220 (32.9) | 8 (42.1) | |
| IV | 56 (8.4) | 7 (36.8) | |
Abbreviations: AGC, advanced gastric carcinoma; EGC, early gastric carcinoma; ISH, in situ hybridization; LN, lymph node; TNM, Tumor-Node-Metastasis.
Of 199 synchronous metastatic LNs, 119 (59.8%), 45 (22.6%), 22 (11.1%), 4 (2.0%) and 9 (4.5%) showed RNA ISH score 0, 1, 2, 3, and 4, respectively. Therefore, 13 cases (6.5%) were high-MET mRNA. Among 199 pairs of primary and metastatic lesions, 186 (93.5%) showed concordant MET mRNA status and 13 (6.5%) did not. Of these 13 discordant cases, negative conversion was found in 50% (7/14) of high-MET mRNA primary tumors, and positive conversion was found in 3.2% (6/185) of low-MET mRNA primary tumors (Table 3).
Table 3. Comparison of MET mRNA status between primary tumors and synchronous metastatic lymph nodes.
| Primary tumor | |||
| Low-METmRNA | Low-METmRNA | High-METmRNA | |
| (score 0–1) | (score 2) | (score 3–4) | |
| Metastatic lymph node | |||
| Low-MET mRNA (score 0–1) | 145 | 13 | 6 |
| Low-MET mRNA (score 2) | 17 | 4 | 1 |
| High-MET mRNA (score 3–4) | 3 | 3 | 7 |
2. MET protein and GCN status assessed by IHC and SISH
Using IHC, 236 (44.1%), 171 (32%), 113 (21.1%) and 15 (2.8%) primary tumors were scored 0, 1, 2 and 3, respectively. IHC score 3 showed distinct clinicopathologic features, and this MET overexpression group was significantly associated with older age (P = .005), larger tumor size (P = .009), invasion depth (P = .05), LN metastasis (P = .018), lymphatic invasion (P = .026), increased number of metastatic lymph nodes (P<.001), distant metastasis (P = .007), and higher TNM stage (P = .001). However, MET overexpression did not show any association with gender, tumor location, and Lauren classification (Table S1).
Of 199 synchronous metastatic LNs, 46 (23.1%), 92 (46.2%), 46 (23.1%) and 15 (7.5%) showed IHC score 0, 1, 2 and 3, respectively. Among 199 pairs of primary and metastatic lesions, 187 (94.0%) showed concordant MET protein status and 12 (6.0%) did not. Of these 12 discordant cases, negative conversion was found in 36.4% (4/11) of primary tumors with MET overexpression, and positive conversion was found in 4.3% (8/188) of primary tumors without MET overexpression.
Using SISH, MET GA was observed in 2.6% (14/535) of primary tumors. MET GA showed significant association with larger tumor size (P = .038), lymphatic invasion (P = .009), increased number of metastatic lymph nodes (P<.001), distant metastasis (P = .005), and higher TNM stage (P = .002). However, MET GA did not show any association with gender, tumor location, Lauren classification, invasion depth and LN metastasis (Table S2).
3. Correlation of MET status evaluated by RNA ISH, IHC and SISH
The correlation between MET mRNA and protein status assessed by RNA ISH and IHC is presented in Table 4. In 535 primary tumors, there was a positive correlation between MET mRNA and protein expression (r = 0.398, P<.001). All 7 cases with RNA ISH score 4 showed MET protein overexpression. Among 12 cases with RNA ISH score 3, 5 cases (41.7%) showed IHC score 3. The cases with RNA ISH score 2 exhibited variable IHC scores. The cases with RNA ISH score 0 or 1 did not show MET protein overexpression except for 1 case. Among 10 cases showing discrepancy (i.e., positive in RNA ISH and negative in IHC or vice versa), 8 showed IHC score 2 or RNA ISH score 2. In 199 metastatic LNs, there was a good positive correlation between MET mRNA and protein expression (r = 0.462, P<.001) (Table S3).
Table 4. Correlation of MET mRNA assessed by RNA in situ hybridization with protein and gene copy number assessed by immunohistochemistry and silver in situ hybridization.
| RNA ISH score, n (%) | |||||
| 0 (n = 391) | 1 (n = 87) | 2 (n = 38) | 3 (n = 12) | 4 (n = 7) | |
| IHC score | |||||
| 0 (n = 236) | 205 (52.4) | 26 (29.9) | 5 (13.2) | 0 (0) | 0 (0) |
| 1 (n = 171) | 131 (33.5) | 26 (29.9) | 13 (34.2) | 1 (8.3) | 0 (0) |
| 2 (n = 113) | 54 (13.8) | 35 (40.2) | 18 (47.4) | 6 (50.0) | 0 (0) |
| 3 (n = 15) | 1 (0.3) | 0 (0) | 2 (5.3) | 5 (41.7) | 7 (100) |
| SISH | |||||
| DS (n = 221) | 190 (48.6) | 27 (31.0) | 4 (10.5) | 0 (0) | 0 (0) |
| LT (n = 130) | 96 (24.6) | 22 (25.3) | 12 (31.6) | 0 (0) | 0 (0) |
| HT (n = 1) | 1 (0.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| LP (n = 111) | 75 (19.2) | 23 (26.4) | 10 (26.3) | 3 (25.0) | 0 (0) |
| HP (n = 58) | 29 (7.4) | 15 (17.2) | 10 (26.3) | 3 (25.0) | 1 (14.3) |
| GA (n = 14) | 0 (0) | 0 (0) | 2 (5.3) | 6 (50.0) | 6 (85.7) |
Abbreviations: DS, disomy; GA, gene amplification; HP, high polysomy; HT, high trisomy; IHC, immunohistochemistry; ISH, in situ hybridization; LP, low polysomy; LT, low trisomy; SISH, silver in situ hybridization.
In addition, there was a positive correlation between MET mRNA expression and MET GCN (r = 0.345; P<.001) (Table 4). Among the 7 cases with RNA ISH score 4, 6 (85.7%) showed MET GA and only one case showed HP (14.3%). The 12 cases with RNA ISH score 3 showed GA (50%) or polysomy (50%) by SISH. The cases with RNA ISH score 2 showed various SISH patterns including GA (5.3%). None of the cases with RNA ISH score 0 or 1 showed GA.
Table 5 summarizes the results of RNA ISH, IHC and SISH of primary GC. Among the 535 cases, 513 cases (95.9%) were negative by both RNA ISH and IHC, and only 22 cases (4.1%) showed positive results by either RNA ISH or IHC. These 22 cases exhibited MET GA (54.5%) or polysomy (45.5%) by SISH, and disomy or trisomy was never observed. In terms of SISH, among the 14 cases showing GA, 11 cases (78.6%) exhibited high expression of both mRNA and protein. Among the 58 cases showing HP, however, only 7 cases (12.1%) exhibited high expression of either mRNA or protein. Among the 111 cases showing LP, only 3 cases (2.7%) exhibited high mRNA expression. All 352 cases showing disomy or trisomy exhibited negative results by both RNA ISH and IHC.
Table 5. Simultaneous comparison of MET status evaluated by RNA in situ hybridization, immunohistochemistry and silver in situ hybridization.
| RNA ISH scores | IHC scores | SISH patterns, n (%) | |||
| Non-GA (n = 521) | GA (n = 14) | ||||
| DS, TS (n = 352) | LP (n = 111) | HP (n = 58) | |||
| 0/1 | 0/1 | 291 (82.7) | 73 (65.8) | 24 (41.4) | 0 (0) |
| 0/1 | 2 | 45 (12.8) | 25 (22.5) | 19 (32.8) | 0 (0) |
| 0/1 | 3 | 0 (0) | 0 (0) | 1 (1.7) | 0 (0) |
| 2 | 0/1 | 9 (2.6) | 6 (5.4) | 3 (5.2) | 0 (0) |
| 2 | 2 | 7 (2.0) | 4 (3.6) | 5 (8.6) | 2 (14.3) |
| 2 | 3 | 0 (0) | 0 (0) | 2 (3.4) | 0 (0) |
| 3/4 | 0/1 | 0 (0) | 0 (0) | 1 (1.7) | 0 (0) |
| 3/4 | 2 | 0 (0) | 3 (2.7) | 2 (3.4) | 1 (7.1) |
| 3/4 | 3 | 0 (0) | 0 (0) | 1 (1.7) | 11 (78.6) |
Abbreviations: DS, disomy; GA, gene amplification; HP, high polysomy; IHC, immunohistochemistry; ISH, in situ hybridization; LP, low polysomy; SISH, silver in situ hybridization; TS, trisomy.
4. Prognostic implications of MET status in primary and metastatic lesions
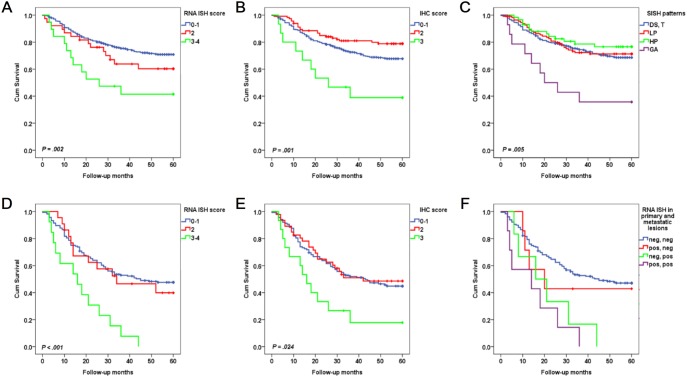
High-MET mRNA in primary tumors or metastatic LNs was significantly associated with poor OS (primary tumors, P = .002, Figure 3A; metastatic LNs, P<.001, Figure 3D). In the primary tumors with low-MET mRNA, the patients with positive conversion showed worse OS than those with no conversion (P = .011, Figure 3F). In the primary tumors with high-MET mRNA, the patients with negative conversion showed better OS than those with no conversion, but there was no statistical significance (P = .137, Figure 3F).
Figure 3. Kaplan-Meier curves for overall survival (OS) according to MET status.
In 535 primary GC, (A) high-MET mRNA was associated with poor OS compared to low-MET mRNA (P = .002), (B) MET overexpression was associated with poor OS compared to no overexpression (P = .001), and (C) MET gene amplification was associated with poor OS compared to no amplification (P = .005). In 199 metastatic lymph nodes, (D) high-MET mRNA was associated with poor OS compared to low-MET mRNA (P<.001), and (E) MET overexpression was associated with poor OS compared to no overexpression (P = .024). (F) In 199 matched primary tumors and metastatic LNs, concordantly positive and positive conversion groups were associated with poor OS compared to concordantly negative group (concordantly negative vs. negative conversion, P = .640; concordantly negative vs. positive conversion, P = .011; concordantly negative vs. concordantly positive, P<.001; concordantly positive vs. negative conversion, P = .137; concordantly positive vs. positive conversion, P = .382; negative conversion vs. positive conversion, P = .260).
MET overexpression in primary tumors or metastatic LNs was significantly associated with poor OS (primary tumors, P = .001, Figure 3B; metastatic LNs, P = .024, Figure 3E). In the primary tumors without MET overexpression, positive conversion trended toward prediction of poor OS, but there was no statistical significance (P = .393). In the primary tumors with MET overexpression, negative conversion showed a trend of better OS than no conversion, but it did not reach statistical significance (P = .132). In addition, MET GA in primary tumors was also significantly associated with poor OS (P = .005, Figure 3C).
In multivariate analysis, high-MET mRNA in metastatic LNs was an independent negative prognostic factor for OS, after adjusting for age (<60 y vs. ≥60 y), Lauren classification (intestinal type vs. diffuse or mixed type), and TNM stage (I–II vs. III–IV). The hazard ratio was 2.27 (P = .007) (Table 6). However, MET overexpression in metastatic LNs was not a statistically significant prognostic factor by multivariate analysis, although the hazard ratio was 1.76 (P = .067). In addition, MET GA, high-MET mRNA or protein overexpression in primary tumor was not a statistically significant prognostic factor by multivariate analysis (data not shown).
Table 6. Univariate and multivariate Cox proportional hazards ratio model for the predictors of overall survival in gastric carcinoma (n = 199).
| Characteristics | Univariate analysis | Multivariate analysis | ||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age | 1.47 (1.00–2.15) | .05 | 1.55 (1.05–2.30) | .029 |
| (<60 y vs. ≥60 y) | ||||
| Lauren classification | 1.36 (0.93–2.00) | .117 | 1.34 (0.90–1.98) | .149 |
| (Intestinal vs. diffuse/mixed) | ||||
| TNM stage | 12.8 (4.06–40.4) | <.001 | 11.8 (3.73–37.4) | <.001 |
| (Stage I–II vs. III–IV) | ||||
| RNA ISH score of metastatic LN | 3.18 (1.77–5.70) | <.001 | 2.27 (1.26–4.09) | .007 |
| (0–2 vs. 3–4) | ||||
Abbreviations: CI, confidence interval; HR, hazard ratio; ISH, in situ hybridization; LN, lymph node.
Discussion
In the present study, we demonstrated that high MET mRNA expression was significantly associated with adverse clinicopathologic features and poor prognosis in a large series of GC patients using RNA ISH method. In addition, RNA ISH results were well correlated with those of SISH and IHC. The previous studies evaluating MET mRNA levels in GC used the Northern blot assay [8], [22] or reverse transcription-polymerase chain reaction (RT-PCR) [8], [14], [23]–[25]. However, most of the studies had small sample sizes, and only a few of them investigated its clinical implications [22], [24] or performed comparison with DNA or protein status [14], [25]: Kuniyasu et al. firstly studied MET mRNA expression using the Northern-blot analysis, and they reported that expression of 6.0-kb transcript was closely correlated with tumor stage and LN metastasis [22]. Amemiya et al. reported that Stage IV GC patients with liver metastasis showed higher MET expression at both mRNA and protein levels than stage IV GC patients without liver metastasis using RT-PCR and IHC [24].
Recently, we reported that high levels of HER2 mRNA was well correlated with protein overexpression and GA by comparing the results of 4 different in situ-based methodologies (RNA ISH, IHC, FISH, and SISH) in 211 GC cases [20]. Likewise, in this study for MET status, the results of RNA ISH showed fairly good correlation with those of IHC and SISH. These results support that RNA ISH can be a reliable assay for FFPE tissue samples, although further validation studies are needed.
We demonstrated that MET GA, high MET mRNA and protein overexpression evaluated by SISH, RNA ISH and IHC were highly concordant, and high MET status at the DNA, mRNA, and protein were significantly associated with poor prognosis. These findings support that MET overexpression is mainly due to increased MET GCN and this mechanism contributes to aggressive behavior of MET oncogene-addicted GC. Nevertheless, there were some cases showing inconsistency among the MET GCN, mRNA and protein levels. We speculate that technical problems (e.g., sensitivity and specificity of the probe or antibody, and poor mRNA quality of FFPE tissues) and intratumoral heterogeneity of MET status may be the main causes of this discrepancy. However, some biological mechanisms can also be related to this discrepancy. For example, MET overexpression without GA can occur through transcriptional activation via HGF-dependent autocrine/paracrine loops or other signaling pathways [23], [26]. On the contrary, MET GA may not increase the gene product. Asaoka et al. reported that a few GC cell lines harboring MET GA expressed the protein as same level as other cell lines without GA, but their tyrosine residues at the kinase domain were more phosphorylated [27]. The mechanisms of MET activation and the role of HGF in GC remain to be elucidated.
It is well known that MET plays a role in metastatic progression of cancer. Several studies showed that MET GA or overexpression was associated with LN metastasis [7], [13], [22] or distant metastasis [13], in GC patients. In addition, it was shown that the administration of MET inhibitor reduced peritoneal dissemination of GC in a xenograft model [23]. Furthermore, Di Renzo et al. found that cancer cells carrying MET activating mutations were selected during metastatic spread of head and neck squamous cell carcinomas by comparing the gene sequence between primary tumor and metastatic lymph node [28]. However, direct comparison of MET status between primary tumor and metastasis has not been performed in a large series of GC. When we compared the MET expression status between 199 matched primary tumors and metastatic LNs, and the overall concordance was 93.5% and 94% by RNA ISH and IHC, respectively. These results suggest that MET expression status of GC is relatively constant during metastasis to regional LNs. However, positive conversion of MET mRNA status was significantly associated with poor prognosis by univariate and multivariate analysis. Therefore, these results suggest that the evaluation of MET status in metastatic lesions may be important to predict prognosis and to identify additional candidates for MET-targeted therapy.
In situ-based RNA analysis has several advantages over the ‘grind and bind’ analysis such as RT-PCR [19] and is applicable for both clinical practice and retrospective research. Moreover, RNA ISH is more favorable than IHC when there is no suitable antibody or when the target molecule is the secreted protein. In regard to MET, this advantage can be useful because the HGF-producing cells can be visualized in a tissue section using the HGF probe. However, vulnerable mRNA stability during the tissue processing and higher cost than that of IHC are the disadvantages of RNA ISH method. We hope that further technical improvement will resolve these limitations.
In this retrospective study, MET mRNA status evaluated by RNA ISH is well correlated with protein and GCN assessed by IHC and SISH, respectively, and MET GA is highly concordant with high expression of either mRNA or protein. In survival analysis, high expression of MET mRNA in primary or metastatic lesions, and positive conversion of MET mRNA status are significantly associated with poor prognosis. In addition, MET mRNA status in metastatic LNs is an independent prognostic factor by multivariate analysis. Our findings indicate that MET mRNA can be an alternative marker to identify the MET oncogene-addicted GC.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are very grateful to Ms. Hyun Ju Park for her technical support.
Funding Statement
This work was supported by the National Research Foundation of Korea (NRF) A3 Foresight program (http://www.nrf.re.kr). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Peters S, Adjei AA (2012) MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol 9: 314–326. [DOI] [PubMed] [Google Scholar]
- 2. Dean M, Park M, Le Beau MM, Robins TS, Diaz MO, et al. (1985) The human met oncogene is related to the tyrosine kinase oncogenes. Nature 318: 385–388. [DOI] [PubMed] [Google Scholar]
- 3. Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, et al. (1991) Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251: 802–804. [DOI] [PubMed] [Google Scholar]
- 4. Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G (2012) Targeting MET in cancer: rationale and progress. Nat Rev Cancer 12: 89–103. [DOI] [PubMed] [Google Scholar]
- 5. Rege-Cambrin G, Scaravaglio P, Carozzi F, Giordano S, Ponzetto C, et al. (1992) Karyotypic analysis of gastric carcinoma cell lines carrying an amplified c-met oncogene. Cancer Genet Cytogenet 64: 170–173. [DOI] [PubMed] [Google Scholar]
- 6. Kuniyasu H, Yasui W, Kitadai Y, Yokozaki H, Ito H, et al. (1992) Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun 189: 227–232. [DOI] [PubMed] [Google Scholar]
- 7. Nakajima M, Sawada H, Yamada Y, Watanabe A, Tatsumi M, et al. (1999) The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer 85: 1894–1902. [DOI] [PubMed] [Google Scholar]
- 8. Huang TJ, Wang JY, Lin SR, Lian ST, Hsieh JS (2001) Overexpression of the c-met protooncogene in human gastric carcinoma–correlation to clinical features. Acta Oncol 40: 638–643. [DOI] [PubMed] [Google Scholar]
- 9. Drebber U, Baldus SE, Nolden B, Grass G, Bollschweiler E, et al. (2008) The overexpression of c-met as a prognostic indicator for gastric carcinoma compared to p53 and p21 nuclear accumulation. Oncol Rep 19: 1477–1483. [PubMed] [Google Scholar]
- 10. Graziano F, Galluccio N, Lorenzini P, Ruzzo A, Canestrari E, et al. (2011) Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol 29: 4789–4795. [DOI] [PubMed] [Google Scholar]
- 11. Lee J, Seo JW, Jun HJ, Ki CS, Park SH, et al. (2011) Impact of MET amplification on gastric cancer: possible roles as a novel prognostic marker and a potential therapeutic target. Oncol Rep 25: 1517–1524. [DOI] [PubMed] [Google Scholar]
- 12. Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, et al. (2011) MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 29: 4803–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee HE, Kim MA, Lee HS, Jung EJ, Yang HK, et al. (2012) MET in gastric carcinomas: comparison between protein expression and gene copy number and impact on clinical outcome. Br J Cancer 107: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha SY, Lee J, Kang SY, Do IG, Ahn S, et al. (2013) MET overexpression assessed by new interpretation method predicts gene amplification and poor survival in advanced gastric carcinomas. Mod Pathol 26: 1632–1641. [DOI] [PubMed] [Google Scholar]
- 15. Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, et al. (2006) Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A 103: 2316–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Catenacci DV, Henderson L, Xiao SY, Patel P, Yauch RL, et al. (2011) Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer Discov 1: 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaji M, Yonemura Y, Harada S, Liu X, Terada I, et al. (1996) Participation of c-met in the progression of human gastric cancers: anti-c-met oligonucleotides inhibit proliferation or invasiveness of gastric cancer cells. Cancer Gene Ther 3: 393–404. [PubMed] [Google Scholar]
- 18. Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, et al. (1998) Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest 78: 1143–1153. [PubMed] [Google Scholar]
- 19. Wang F, Flanagan J, Su N, Wang LC, Bui S, et al. (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim MA, Jung JE, Lee HE, Yang HK, Kim WH (2013) In situ analysis of HER2 mRNA in gastric carcinoma: comparison with fluorescence in situ hybridization, dual-color silver in situ hybridization, and immunohistochemistry. Hum Pathol 44: 487–494. [DOI] [PubMed] [Google Scholar]
- 21. Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, et al. (2005) Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 97: 643–655. [DOI] [PubMed] [Google Scholar]
- 22. Kuniyasu H, Yasui W, Yokozaki H, Kitadai Y, Tahara E (1993) Aberrant expression of c-met mRNA in human gastric carcinomas. Int J Cancer 55: 72–75. [DOI] [PubMed] [Google Scholar]
- 23. Toiyama Y, Yasuda H, Saigusa S, Matushita K, Fujikawa H, et al. (2012) Co-expression of hepatocyte growth factor and c-Met predicts peritoneal dissemination established by autocrine hepatocyte growth factor/c-Met signaling in gastric cancer. Int J Cancer 130: 2912–2921. [DOI] [PubMed] [Google Scholar]
- 24. Amemiya H, Kono K, Itakura J, Tang RF, Takahashi A, et al. (2002) c-Met expression in gastric cancer with liver metastasis. Oncology 63: 286–296. [DOI] [PubMed] [Google Scholar]
- 25. Janjigian YY, Tang LH, Coit DG, Kelsen DP, Francone TD, et al. (2011) MET expression and amplification in patients with localized gastric cancer. Cancer Epidemiol Biomarkers Prev 20: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park WS, Oh RR, Kim YS, Park JY, Shin MS, et al. (2000) Absence of mutations in the kinase domain of the Met gene and frequent expression of Met and HGF/SF protein in primary gastric carcinomas. Apmis 108: 195–200. [DOI] [PubMed] [Google Scholar]
- 27. Asaoka Y, Tada M, Ikenoue T, Seto M, Imai M, et al. (2010) Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem Biophys Res Commun 394: 1042–1046. [DOI] [PubMed] [Google Scholar]
- 28. Di Renzo MF, Olivero M, Martone T, Maffe A, Maggiora P, et al. (2000) Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene 19: 1547–1555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)