Abstract
The purpose of this report was to evaluate the expression patterns of selected glutathione transferase genes (gst1, gst18, gst23 and gst24) in the tissues of two maize (Zea mays L.) varieties (relatively resistant Ambrozja and susceptible Tasty Sweet) that were colonized with oligophagous bird cherry-oat aphid (Rhopalosiphum padi L.) or monophagous grain aphid (Sitobion avenae L.). Simultaneously, insect-triggered generation of superoxide anion radicals (O2 •−) in infested Z. mays plants was monitored. Quantified parameters were measured at 1, 2, 4, 8, 24, 48 and 72 h post-initial aphid infestation (hpi) in relation to the non-infested control seedlings. Significant increases in gst transcript amounts were recorded in aphid-stressed plants in comparison to the control seedlings. Maximal enhancement in the expression of the gst genes in aphid-attacked maize plants was found at 8 hpi (gst23) or 24 hpi (gst1, gst18 and gst24) compared to the control. Investigated Z. mays cultivars formed excessive superoxide anion radicals in response to insect treatments, and the highest overproduction of O2 •− was noted 4 or 8 h after infestation, depending on the aphid treatment and maize genotype. Importantly, the Ambrozja variety could be characterized as having more profound increments in the levels of gst transcript abundance and O2 •− generation in comparison with the Tasty Sweet genotype.
Introduction
Maize (Zea mays L.) has increasingly emerged as a pivotal model plant species (Poaceae family, Panicoideae subfamily) that is widely used in a variety of genetic and ecotoxicological experiments [1]–[3]. During the last decade, its world production and utilization in many sectors of industrial production was substantially increased; therefore, it is important to get better insight into the complex mechanisms underlying maize tolerance towards a vast array of biotic and abiotic stressors [4]–[5]. Among the numerous insects attacking Z. mays plants, destructive influence of cereal aphids (Hemiptera, Aphidoidea) colonization should be underlined [6]–[8]. These phloem feeding parasites are involved in severe exploitation of the host systems, resulting in a broad range of detrimental effects, such as mechanical injuries of the stylet-penetrated tissues, local chlorosis or necrosis, deformations of organs, biomass reduction, significant disturbances of cellular homeostasis and transmission of pathogenic viruses. The harmfulness of the aphid attack is linked to the suppression of photosynthesis, diminution in chlorophyll content, intensive removal of water and photosynthates from the sieve elements [9]–[12]. Recently, there has been evidence showing that the severity of aphid-induced damages is largely associated with the composition of species-specific elicitors present in the salivary secretions injected into the host tissues [13]–[14]. Importantly, an aphid-triggered oxidative burst in tissues of host systems colonized by these hemipterans has scarcely been reported [6], [15]. On the other hand, it has been documented that cereal aphids evoked a significant decrease in ascorbate content in triticale and deterioration of the antioxidative capacity toward DPPH (1,1-diphenyl-2-picrylhydrazyl) radicals in maize plants [6], [16]. It should be noted that cellular redox imbalance in plant cells due to a chronic overproduction of various reactive oxygen species (ROS) may result in profound oxidative damages of lipids, polysaccharides, proteins and nucleic acids [15]–[16].
Cytosolic glutathione transferases (GSTs, E.C.2.5.1.18) embrace a multifunctional superfamily of enzymes participating in many physiological processes involved in plant growth and development, shoot regeneration and adaptability to adverse environmental stimuli [17]. Plant GSTs catalyze the nucleophilic substitution or addition reactions of endogenous substrates and xenobiotics with glutathione molecules, leading to the synthesis of less toxic compounds with greater solubility in water, which secondarily improves their vacuolar sequestration [18]–[19]. Additionally, glutathione transferases are involved in scavenging of excessive amounts of ROS generated in plant tissues under oxidative stress conditions, and they participate in the signal transduction pathways, cellular responses to auxins and cytokinins, as well as metabolic turnover of cinnamic acid and anthocyanins [20]–[21]. According to Dixon et al. [22], AtGSTZ1-1 from Arabidopsis thaliana L. possesses maleylacetone isomerase activity and participates in tyrosine degradation. Furthermore, GSTs display glutathione-peroxidase activity associated with the reduction of hydroperoxides [23]. Some authors have proposed that the activation of glutathione transferases in plants exposed to different stressors is associated with an increased ability to neutralize the lipid hydroperoxides synthesised in oxidatively damaged membranes [24]–[25]. It has been previously reported that GST isoforms overexpressed in transgenic plants markedly augment tolerance levels to herbicide treatment and oxidative stress [26]–[27]. Consistent with these observations, tau-GST from Lycopersicon esculentum Mill., elevated resistance to hydrogen peroxide-stimulated stress and repressed Bax-stimulated apoptosis in transformed yeast cells [28]. Likewise, upregulation of several plant glutathione transferases in catalase-deficient mutants were reported [29].
There are numerous studies indicating a rapid and substantial increase in the activity of various plant GST isozymes and differential regulation of gst genes influenced by multifarious external factors (e.g. heavy metals, herbicides, drought, low and high temperatures, UV radiation, exogenous application of chemical inducers of oxidative stress, insect infestation and fungal or viral infection) [30]–[32]. However, there is a lack of published data concerning expression profiling of the gst genes and superoxide anion radical (O2 •−) production in the seedlings of maize varieties exposed to cereal aphid colonization. It may be assumed that mono- and oligophagous aphids differentially affect the transcriptional activity of gst genes and O2 •−generation in tissues of maize genotypes, exhibiting diverse resistance levels to the aphid infestation. To verify this hypothesis, the relative quantification of four gst genes (gst1, gst18, gst23 and gst24) was performed and the amount of O2 •− was measured in the seedlings of Z. mays Ambrozja (susceptible) and Tasty Sweet (relatively resistant) varieties infested by monophagous grain aphid (Sitobion avenae F.) or oligophagous bird cherry-oat aphid (Rhopalosiphum padi L.). The study was also aimed at assessing whether the scale of aphid-triggered changes in the levels of the analysed parameters may be dependent on the insect density.
Methods
Plant material
The seeds of two investigated Z. mays varieties (Ambrozja and Tasty Sweet) were acquired from local commercial grain suppliers: Reheza (Moszna, Poland) and PNOS S.A. (Ożarów Mazowiecki, Poland). Before performing the bioassays, intact maize seeds without any visible damages were surface sterilized as described previously [32]. Subsequently, portions of plant material (5 seeds of each cultivar per plate; four replicates) were subjected to potato dextrose agar (PDA) plate screening in order to confirm the absence of mycoflora, according to the method of Adejumo et al. [33]. Ambrozja genotype has previously been classified as relatively resistant, whereas Tasty Sweet is susceptible to the cereal aphids' infestation [6]. Maize seeds were sown in round plastic pots (10×9 cm; diameter × height) filled with general-purpose horticultural substrate and no additional fertilization was applied. Seedlings were grown in a climate chamber at 22±2°C/16±2°C (day/night) with a light intensity of 100 µM m−2 s−1, a long-day photoperiod (L16: D8) and a relative humidity of 65±5%. It is important to note that only health maize seedlings of similar height were included during the experiments.
Aphids
Wingless parthenogenetic females of R. padi and S. avenae aphids were collected from the field crops within the Siedlce district, Poland (52°09′54″N, 22°16′17″E). The authors state that no specific permissions were required for the sampling of aphids in this location, and confirm that the field studies did not involve endangered or protected species. The collected females were transferred to the seedlings of common wheat (Triticum aestivum L.) cv. Tonacja in the Department of Biochemistry and Molecular Biology, University of Natural Sciences and Humanities (Siedlce, Poland). New wheat seedlings were provided every week, and the aphids were reared for a year in the climate chamber under the conditions described above. Adult apterous females of the cereal aphids used in the leaf infestation experiments originated from the mother stock cultures of parthenogenetic individuals.
Infestation experiments
Leaves of 14-day-old maize seedlings (Ambrozja and Tasty sweet cultivars) were colonized with 10, 20, 40, or 60 adult wingless females of the relevant cereal aphids (R. padi or S. avenae) per plant. The control groups of seedlings were not infested with hemipterans. The levels of relative expression of the selected gst genes (gst1, gst18, gst23 and gst24) and O2 •− generation in Z. mays seedling leaves were determined 1, 2, 4, 8, 24, 48, and 72 h after initial insect infestation (hpi). Maize plants infested with aphids and the non-infested (control) plants were isolated in gauze-covered plastic cylinders (20×50 cm; diameter × height). At the end of each variant of biotests, the aphids were removed from the plants and, subsequently, the seedling leaves were excised and used immediately for further analytic procedures.
Determination of superoxide anion radical generation in the maize seedlings
The formation of O2 •− was measured by the reduction of nitroblue tetrazolium (NBT), according to the method of Chaitanya and Naithani [34] with necessary modifications. Freshly collected Z. mays seedling leaves were cut into small pieces, and 0.5 g of the plant material was homogenized in 5 cm3 of ice-cold phosphate buffer (100 mM, pH 7.2) with 1 mM diethyldithiocarbamate (superoxide dismutase inhibitor). The homogenate was filtered through four layers of nylon mesh and centrifuged at 19 000×g for 20 min at 4°C. A portion of the supernatant (0.2 cm3) was combined with 0.8 cm3 of the phosphate buffer and 0.1 cm3 of 25 mM NBT (Sigma-Aldrich, Poland), and then, the mixture was incubated at 25°C for 5 min. Absorbance values of the sample before the incubation (A 0) and after the incubation period (A S) were determined at 540 nm using an Epoch UV-Vis microplate spectrophotometer (BioTek, USA). The amount of O2 •− in Z. mays seedling leaves was calculated using the following formula: ΔA 540 = A S – A 0, and it was expressed as ΔA 540 (min−1 g−1) fresh weight.
Isolation of total RNA and cDNA synthesis
The insect-infested and non-infested seedling leaves of both investigated Z. mays genotypes were collected and homogenized immediately in liquid nitrogen by employing a sterile ceramic mortar and pestle. Total RNA was extracted with the application of Spectrum Plant Total RNA Kit (Sigma Aldrich, Poland) and, subsequently, trace amounts of genomic DNA were degraded using the On-Column DNase I Digestion Set (Sigma Aldrich, Poland). The quantitative-qualitative evaluation of the RNA samples was conducted with the use of an Epoch UV-Vis microplate spectrophotometer (BioTek, USA). High-quality RNA preparates (A 260/280 >2.0; A 260/230 >1.8) were exclusively accepted for the reverse-transcription reaction. Synthesis of complementary DNA (cDNA) was performed with the use of RevertAid Premium First Strand cDNA Synthesis Kit (Fermentas, Poland). It should be noted that the protocol scheme with oligo(dT)18 primers was applied. Additionally, two negative controls (NTC – no template control, and NRT – no reverse transcriptase) were included.
Gene expression quantification
The relative expression of the target gst genes in foliar tissues of the aphid-infested and non-infested (control) Z. mays seedlings was estimated using the quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR). The glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene was used as the internal reference [6]. Transcriptional activity of four gst genes (gst1, gst18, gst23 and gst24) was measured with the application of TaqMan Gene Expression Assays (Life Technologies, Poland). The selection of target genes was based on their regulation under specific stress conditions (gst1 has widely been described as a molecular marker of oxidative stress in maize tissues and gst23 has been thought to be associated with multiple disease resistance, whereas expression of gst18 and gst24 genes was markedly altered under fungal infections) [19], [32]. Reference sequences and unique assay names (IDs) of the quantified gst transcripts are listed in Table S1. The reaction mixtures (20 mm3 final volume) contained 10 mm3 2× TaqMan Fast Universal PCR Master Mix, 1 mm3 20× TaqMan gene expression assay solution, 4 mm3 template (cDNA) and 5 mm3 RNase-free deionised water. Detection of the fluorescence signals was carried out on the StepOne Plus Real-Time PCR System equipped with StepOnePlus Software v2.3 (Life Technologies, USA). Amplification plots were obtained under the following thermal cycling conditions: initial activation of AmpliTaq Gold DNA polymerase at 95°C (20 s) and, subsequently, 40 cycles of 95°C (1 s) and 60°C (20 s). Relative gene expression was estimated according to the comparative C T (ΔΔC T) method [35], and the results are reported as the mean n-fold change ± standard deviation (SD) in the specific transcript amount of the aphid-stressed plants compared to the relevant non-infested control plants. Three biological and three technical replicates were included for each tested sample.
Statistical analysis
The data are presented as the mean ± SD of three independent experiments. Each group of aphid-stressed and non-infested maize plants consisted of ten seedlings of a similar height. Factorial analysis of variance (ANOVA) was applied to assess the effects of four experimental indicators (maize cultivar, hemipteran species, insect abundance and aphid exposure period) as well as their interdependence. Afterwards, a post-hoc Tukey's test was performed (p values less than 0.05 were considered significant). Statistical analyses were carried out with the implementation of STATISTICA 10 software (StatSoft, Poland).
Results
Effects of cereal aphids colonization on O2 •− generation in Z. mays seedlings
Both R. padi and S. avenae aphids accelerated O2 •− production in the colonized Ambrozja and Tasty Sweet maize cultivars compared with the relevant control plants (Table 1, 2). Bird cherry-oat aphid infestation led to a greater increase in O2 •− amounts than grain aphid attack. For example, at 4 hpi, colonization of Tasty Sweet or Ambrozja plants with bird cherry-oat aphids at the highest density (60 per seedling) led to 65 and 209% increases in the O2 •− levels relative to the control, respectively, whereas infestation of these cultivars with the same number of S. avenae and insect exposure time led to 49 and 117% increases in O2 •−, respectively. Ambrozja seedlings that were attacked with either aphid species were characterized by significantly higher production of O2 •− than the insect-stressed Tasty Sweet plants (Table 1, 2). The lowest initial number of both aphid species (10 per seedling) resulted in slight increments in the superoxide anion radicals content in the leaves of both maize varieties in relation to the control. Plants treated with higher numbers of aphids showed proportionally greater levels of O2 •− accumulation. Consequently, the largest differences in aphid-stimulated production of O2 •− between the two maize cultivars were observed at the highest insect density (60 per seedling). For example, R. padi–stressed Tasty Sweet plants generated 2–26% more O2 •− (depending on duration of aphid colonization) than seedlings attacked by S. avenae, whereas R. padi–stressed Ambrozja plants had 4–91% greater rates of O2 •− formation than S. avenae–infested seedlings. Additionally, slightly more superoxide anion radicals production was found in the non-infested Ambrozja seedlings than in Tasty Sweet plants (Table 1, 2). Importantly, the duration of aphid infestation had a strong influence on the generation of O2 •− in leaves of both Z. mays genotypes. Comparative analysis of all treatments revealed that the lowest level of O2 •− generation occurred at 1 hpi (2–9% increase, depending on the aphid infestation level) relative to the control. Maximal O2 •− formation was observed at 4 hpi in Tasty Sweet seedlings infested with 60 individuals of bird cherry-oat aphid or grain aphid, and in Ambrozja seedlings colonized with 20–60 R. padi or 40–60 S. avenae aphids per plant. For the other tested bioassay variants, the highest O2 •− generation occurred after 8 hpi compared to the non-stressed seedlings. Prolonged aphid feeding resulted in a progressive decrease in the amount of analysed ROS in comparison to maximal changes observed after 4–8 h of aphid colonization. Furthermore, factorial analysis of variance (ANOVA) revealed significant effects of the experimental indicators and their interactions on levels of O2 •− production in the maize seedlings (Table 3).
Table 1. Levels of O2 •− generation (ΔA 540 min−1 g−1 fresh weight) in leaves of the maize seedlings colonized with R. padi.
| Time intervals of aphid infestation (hpi) | Aphid abundance (per plant) | |||||
| 0 | 10 | 20 | 40 | 60 | ||
| Ambrozja genotype | ||||||
| 0 | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a | |
| 1 | 0.47±0.02b | 0.47±0.02b | 0.47±0.02b | 0.49±0.03ab | 0.51±0.04a | |
| 2 | 0.48±0.04b | 0.48±0.04b | 0.49±0.03b | 0.52±0.04ab | 0.58±0.03a | |
| 4 | 0.47±0.03d | 0.50±0.04d | 0.81±0.06c | 1.23±0.08b | 1.45±0.10a | |
| 8 | 0.49±0.04d | 0.65±0.04c | 0.70±0.05c | 0.91±0.06ab | 1.03±0.06a | |
| 24 | 0.48±0.04d | 0.57±0.05c | 0.62±0.05bc | 0.69±0.04b | 0.87±0.07a | |
| 48 | 0.48±0.03cd | 0.55±0.04c | 0.57±0.03bc | 0.63±0.03b | 0.81±0.06a | |
| 72 | 0.49±0.04c | 0.53±0.03c | 0.54±0.04bc | 0.60±0.04ab | 0.76±0.05a | |
| Tasty Sweet genotype | ||||||
| 0 | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | |
| 1 | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.43±0.02a | 0.44±0.02a | |
| 2 | 0.44±0.03a | 0.44±0.03a | 0.45±0.02a | 0.47±0.03a | 0.49±0.04a | |
| 4 | 0.43±0.02bc | 0.44±0.03bc | 0.47±0.03b | 0.50±0.05b | 0.71±0.08a | |
| 8 | 0.43±0.02d | 0.50±0.04cd | 0.53±0.05b | 0.62±0.06a | 0.66±0.06a | |
| 24 | 0.45±0.04bc | 0.48±0.03b | 0.51±0.04ab | 0.54±0.05a | 0.61±0.05a | |
| 48 | 0.44±0.03bc | 0.45±0.04bc | 0.50±0.04b | 0.51±0.05ab | 0.58±0.05a | |
| 72 | 0.43±0.03b | 0.43±0.03b | 0.46±0.03ab | 0.48±0.03ab | 0.55±0.05a | |
Values are the means ± standard deviation (SD) of three independent experiments (10 plants per repeat); hpi-hours post-initial insect infestation; the different letters in rows denote significant differences according to Tukey's test (P≤0.05).
Table 2. Levels of O2 •− generation (ΔA 540 min−1 g−1 fresh weight) in leaves of the maize seedlings colonized with S. avenae.
| Time intervals of aphid infestation (hpi) | Aphid abundance (per plant) | ||||
| 0 | 10 | 20 | 40 | 60 | |
| Ambrozja genotype | |||||
| 0 | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a | 0.47±0.03a |
| 1 | 0.47±0.02a | 0.47±0.02a | 0.47±0.02a | 0.48±0.03a | 0.49±0.03a |
| 2 | 0.48±0.04ab | 0.48±0.04ab | 0.48±0.04ab | 0.50±0.04ab | 0.54±0.04a |
| 4 | 0.47±0.03c | 0.49±0.03c | 0.52±0.05c | 0.88±0.07ab | 1.02±0.08a |
| 8 | 0.49±0.04c | 0.60±0.04b | 0.67±0.05b | 0.65±0.04b | 0.85±0.06a |
| 24 | 0.48±0.04b | 0.54±0.03b | 0.63±0.05ab | 0.62±0.05ab | 0.76±0.05a |
| 48 | 0.48±0.03bc | 0.52±0.04b | 0.59±0.03ab | 0.59±0.03ab | 0.68±0.04a |
| 72 | 0.49±0.04bc | 0.51±0.03b | 0.54±0.04b | 0.55±0.03b | 0.63±0.05a |
| Tasty Sweet genotype | |||||
| 0 | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a |
| 1 | 0.42±0.02a | 0.42±0.02a | 0.42±0.02a | 0.43±0.02a | 0.43±0.03a |
| 2 | 0.44±0.03a | 0.44±0.03a | 0.44±0.03a | 0.45±0.02a | 0.48±0.04a |
| 4 | 0.43±0.02b | 0.45±0.03b | 0.45±0.02b | 0.47±0.03b | 0.64±0.05a |
| 8 | 0.43±0.02b | 0.48±0.03ab | 0.51±0.05a | 0.58±0.04a | 0.55±0.04a |
| 24 | 0.45±0.04ab | 0.47±0.02ab | 0.49±0.04a | 0.51±0.04a | 0.52±0.05a |
| 48 | 0.44±0.03ab | 0.44±0.03ab | 0.47±0.03a | 0.49±0.03a | 0.50±0.04a |
| 72 | 0.43±0.03ab | 0.43±0.04ab | 0.44±0.03ab | 0.46±0.02a | 0.48±0.03a |
Values are means ± standard deviation (SD) of three independent experiments (10 plants per repeat); hpi-hours post-initial insect infestation; different letters in rows denote significant differences according to Tukey's test (P≤0.05).
Table 3. Factorial ANOVA results for tested indicators (Z. mays cultivar, hemipteran species, insect abundance and aphid exposure period) and interdependence between these parameters affecting O2 •− formation in the maize seedlings.
| Tested factors and interactions | Df | F | p |
| Maize cultivar (C) | 1 | 175.2 | ≤0.001 |
| Hemipteran species (S) | 2 | 87.9 | ≤0.001 |
| Insect abundance (A) | 3 | 68.2 | ≤0.001 |
| Aphid exposure period (EP) | 7 | 52.7 | ≤0.001 |
| S × C | 2 | 19.6 | ≤0.001 |
| S × A | 6 | 14.5 | ≤0.001 |
| C × A | 3 | 27.1 | ≤0.001 |
| S × EP | 14 | 14.9 | ≤0.001 |
| C × EP | 7 | 18.5 | ≤0.001 |
| A × EP | 21 | 10.6 | ≤0.001 |
| S × C × A | 6 | 12.4 | ≤0.001 |
| S × C × EP | 14 | 9.9 | ≤0.001 |
| S × A × EP | 42 | 8.7 | ≤0.001 |
| C × A × EP | 21 | 4.9 | ≤0.001 |
| S × C × A × EP | 42 | 3.7 | ≤0.008 |
Df-degrees of freedom; p-values less than 0.05 were considered significant; F-ratio is defined as the variance between samples/the variance within samples.
Transcriptional activity of gst1 gene in the aphid-stressed maize seedlings
The conducted biotests demonstrated that short-term feeding of the examined cereal aphids (R. padi or S. avenae) did not influence the amount of gst1 mRNA transcript in the seedlings of Ambrozja and Tasty Sweet maize genotypes (Figure 1). Two hours after initial infestation, the low abundance of aphids (10–20 individuals per plant) did not alter the gene expression, but a higher number of insects (40–60 per seedlings) stimulated a slight increment in transcriptional activity of the target gene (from 5% increase in Tasty Sweet plants colonized with 60 S. avenae to a 26% increase in Ambrozja plants infested with the same number of R. padi aphids). After 4 and 8 hpi, the levels of gst1 transcript gradually enhanced in both maize varieties colonized with the tested aphid species, with the exception of two aphid treatments (10 and 20 insects per plant) at 4 hpi when there were no changes in the relative expression of the analysed gene in Tasty Sweet genotype. The highest accumulation of the gst1 transcript amount in the aphid-infested maize seedlings of both Z. mays genotypes occurred at 24 hpi and 60 aphids per plant (4.3–5.5-fold elevations in Ambrozja, and 2.4–3.1-fold increases in Tasty Sweet seedlings, depending on the aphid species). However, extended insect colonization (48–72 hpi) resulted in a gradually lower gene expression in relation to the levels recorded at 24 hpi. Generally, R. padi infestation led to a more profound increase in the transcriptional activity of the gst1 gene in comparison with S. avenae (e.g. 120% higher increase in Ambrozja and 69% increment in Tasty Sweet plants, at 24 hpi and 60 aphids per plant). The results of factorial ANOVA confirmed a significant influence of the analysed indicators and their interactions on expression of the gst1 gene in the maize seedlings (Table 4).
Figure 1. Influence of the tested cereal aphids on gst1 gene expression in the seedlings of Ambrozja and Tasty Sweet maize cultivars.
Values signify the mean n-fold changes in the gst1 transcript abundance in the aphid-stressed Z. mays plants in comparison with the non-infested group of seedlings. Error bars represent the standard deviation (± SD). For each maize-aphid treatment, three independent biological replicates were accomplished. The obtained gene expression data were normalized to the gapdh gene. The different letters above the SD bars designate significant differences among compared plants at P≤0.05 based on the Tukey's test. I-10, I-20, I-40 and I-60 are the levels of aphid infestation (10, 20, 40 and 60 insects per plant, accordingly).
Table 4. Factorial ANOVA results for tested indicators (Z. mays cultivar, hemipteran species, insect abundance and aphid exposure period) and interactions between these parameters affecting gst1 and gst18 transcript amounts in the maize seedlings.
| Tested factors and interactions | Df | F | p | F | p |
| gst1 gene | gst18 gene | ||||
| Maize cultivar (C) | 1 | 986.8 | ≤0.001 | 916.9 | ≤0.001 |
| Hemipteran species (S) | 2 | 852.4 | ≤0.001 | 1645.2 | ≤0.001 |
| Insect abundance (A) | 3 | 1447.0 | ≤0.001 | 1362.5 | ≤0.001 |
| Aphid exposure period (EP) | 7 | 1078.3 | ≤0.001 | 1573.8 | ≤0.001 |
| S × C | 2 | 561.4 | ≤0.001 | 965.2 | ≤0.001 |
| S × A | 6 | 374.2 | ≤0.001 | 1258.9 | ≤0.001 |
| C × A | 3 | 309.2 | ≤0.001 | 724.5 | ≤0.001 |
| S × EP | 14 | 407.4 | ≤0.001 | 1419.8 | ≤0.001 |
| C × EP | 7 | 275.1 | ≤0.001 | 1160.4 | ≤0.001 |
| A × EP | 21 | 223.2 | ≤0.001 | 583.9 | ≤0.001 |
| S × C × A | 6 | 79.5 | ≤0.001 | 185.3 | ≤0.001 |
| S × C × EP | 14 | 77.8 | ≤0.001 | 306.0 | ≤0.001 |
| S × A × EP | 42 | 58.0 | ≤0.001 | 163.2 | ≤0.001 |
| C × A × EP | 21 | 45.2 | ≤0.001 | 142.5 | ≤0.001 |
| S × C × A × EP | 42 | 13.8 | ≤0.001 | 40.5 | ≤0.001 |
Df-degrees of freedom; p-values less than 0.05 were considered significant; F-ratio is defined as the variance between samples/the variance within samples.
Amount of gst18 transcript in the insect-injured Z. mays seedlings
The performed analyses revealed that the transcriptional activity of the gst18 gene in tissues of both maize cultivars remained unaffected after 1 or 2 h of aphid colonization (Figure 2). The 4 h infestation with a higher density of insects (40–60 per seedling) resulted in slightly enhanced levels of gene expression (16–112% increment), whereas a lower abundance (10–20 aphids per plant) did not evoke any alternations compared to the control. Eight hours after the initial infestation, the transcriptional activity of the gst18 gene in seedlings of the investigated Z. mays cultivars gradually increased in proportion to the number of hemipterans per plant (21–82% increase in Tasty Sweet and 27–440% increase in Ambrozja variety). It is important to note, that the highest stimulation of target gene expression occurred at 24 hpi. Colonization of maize plants with R. padi aphids at this time point led to 1.4–4.1-fold and 2.1–6.2-fold elevations in the transcript abundance in Tasty Sweet and Ambrozja seedlings, accordingly, whereas the grain aphid attack resulted in 1.3–3.4-fold and 1.7–5.5-fold increases in the corresponding maize cultivars. During the next two periods of aphid infestation the scale of upregulation of the gst18 gene in both maize genotypes was less pronounced (1.2–4.9-fold increments at 48 hpi; 1.1–4.2-fold elevations at 72 hpi, depending on the aphid treatments). Importantly, R. padi-colonized maize plants were characterized with greater amounts of the target transcript (20–97% Tasty Sweet and 61–163% Ambrozja) in relation to S. avenae-attacked seedlings. Furthermore, it was evidenced that the aphid-infested Ambrozja plants responded with much greater increases in the gst18 gene expression compared to the infested Tasty Sweet genotype (e.g. 50–258% greater increments at 60 insects per plant). Statistical analysis confirmed the considerable impact of tested parameters and their interrelation on expression of the analysed gene in the investigated Z. mays plants (Table 4).
Figure 2. Influence of the tested cereal aphids on gst18 gene expression in the seedlings of Ambrozja and Tasty Sweet maize cultivars.
Values signify the mean n-fold changes in the gst18 transcript abundance in the aphid-stressed Z. mays plants in comparison with the non-infested group of seedlings. Error bars represent the standard deviation (± SD). For each maize-aphid treatment, three independent biological replicates were accomplished. The obtained gene expression data were normalized to the gapdh gene. The different letters above the SD bars designate significant differences among compared plants at P≤0.05 based on the Tukey's test. I-10, I-20, I-40 and I-60 are the levels of aphid infestation (10, 20, 40 and 60 insects per plant, accordingly).
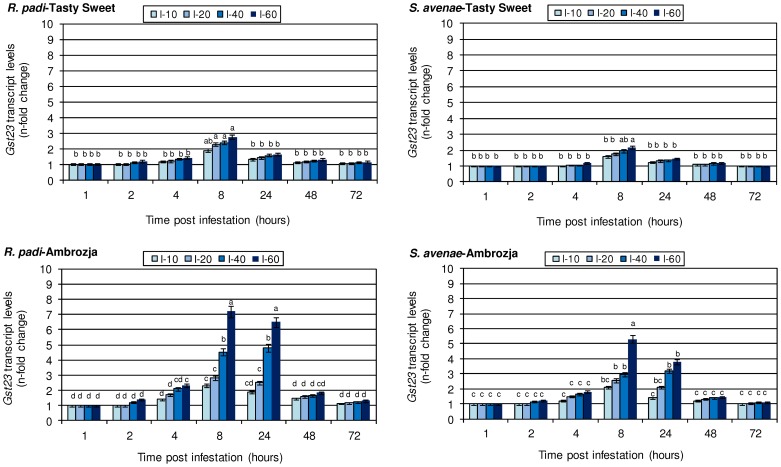
Relative expression of gst23 gene in maize plants colonized with cereal aphids
Results concerning the expression levels of the gst23 gene in the aphid-infested seedlings of Z. mays are depicted in figure 3. It has been found that feeding S. avenae or R. padi for 1 h did not evoke any disturbances in the transcriptional activity of the target gene in tissues of the investigated maize cultivars. Insect feeding for 2 h did not result in any changes in the gst1 gene expression in S. avenae-infested Tasty Sweet plants, regardless of the number of aphids per plant. Likewise, the tested cereal aphids (10–20 insects per seedling) did not affect the transcriptional activity of the analysed gene in both maize genotypes (Ambrozja or Tasty Sweet). However, higher numbers of aphids (40–60 per plant) led to an elevation in the gst23 transcript abundance, ranging from 10% in Tasty Sweet plants colonized by 40 R. padi aphids to 42% increase in Ambrozja seedlings infested by 60 insects per plant. Further extension of colonization period (4 hpi) resulted in a continuous increase (3–132%) in gst23 gene expression in the maize tissues compared with the relevant control plants. The maximal induction of the target gene in aphid-attacked maize plants occurred at 8 hpi. At this time point, 10–60 R. padi per plant evoked 1.9–2.8-fold and 2.3–7.2-fold increases in the levels of transcript accumulation in Tasty Sweet and Ambrozja plants, respectively. Infestation with S. avenae (10–60 aphids per seedling) caused 1.6–2.2-fold and 2.1–5.3-fold elevations in Tasty Sweet and Ambrozja varieties, respectively. Prolonged exposure to aphids (24–72 hpi) could be linked to a progressively lower upregulation of the gst23 gene in comparison with the changes observed at 8 hpi. Interestingly, long-term colonization (72 hpi) by the grain aphid did not influence the analysed transcript amount in Tasty Sweet plants in relation to the relative non-infested seedlings. Comparative analyses revealed that the bird cherry-oat aphid caused a more noticeable augmentation of gst23 gene expression (11–170%, depending on the aphid treatment and maize cultivar) in comparison with S. avenae aphids. Moreover, elevation of the transcriptional activity of the gst23 gene in maize plants occurred in parallel with increasing aphid densities per plant. A markedly higher transcript amount was found in the insect-stressed Ambrozja seedlings compared to Tasty Sweet plants. For example, after 8 h infestation, 60 R. padi aphids stimulated 2.8- and 7.2-fold increments in Tasty Sweet and Ambrozja plants, respectively, whereas feeding the same number of S. avenae individuals led to 2.2- and 5.3-fold increases in Tasty Sweet and Ambrozja varieties, respectively. The results of factorial ANOVA analysis proved that there was a significant impact of the investigated parameters and their interdependence on the transcriptional activity of the gst23 gene in the maize seedlings (Table 5).
Figure 3. Influence of the tested cereal aphids on gst23 gene expression in the seedlings of Ambrozja and Tasty Sweet maize cultivars.
Values signify the mean n-fold changes in the gst23 transcript abundance in the aphid-stressed Z. mays plants in comparison with the non-infested group of seedlings. Error bars represent the standard deviation (± SD). For each maize-aphid treatment, three independent biological replicates were accomplished. The obtained gene expression data were normalized to the gapdh gene. The different letters above the SD bars designate significant differences among compared plants at P≤0.05 based on the Tukey's test. I-10, I-20, I-40 and I-60 are the levels of aphid infestation (10, 20, 40 and 60 insects per plant, accordingly).
Table 5. The factorial analysis of variance of tested indicators (Z. mays cultivar, hemipteran species, insect abundance and aphid exposure period) and interactions between these parameters affecting gst23 and gst24 transcript amounts in the maize seedlings.
| Tested factors and interactions | Df | F | p | F | p |
| gst23 gene | gst24 gene | ||||
| Maize cultivar (C) | 1 | 1142.5 | ≤0.001 | 573.2 | ≤0.001 |
| Hemipteran species (S) | 2 | 748.3 | ≤0.001 | 1229.2 | ≤0.001 |
| Insect abundance (A) | 3 | 1325.2 | ≤0.001 | 495.5 | ≤0.001 |
| Aphid exposure period (EP) | 7 | 1409.8 | ≤0.001 | 1050.9 | ≤0.001 |
| S × C | 2 | 721.4 | ≤0.001 | 147.6 | ≤0.001 |
| S × A | 6 | 438.9 | ≤0.001 | 120.8 | ≤0.001 |
| C × A | 3 | 166.3 | ≤0.001 | 29.5 | ≤0.001 |
| S × EP | 14 | 844.6 | ≤0.001 | 275.0 | ≤0.001 |
| C × EP | 7 | 275.0 | ≤0.001 | 50.6 | ≤0.001 |
| A × EP | 21 | 196.5 | ≤0.001 | 47.3 | ≤0.001 |
| S × C × A | 6 | 45.9 | ≤0.001 | 16.7 | ≤0.001 |
| S × C × EP | 14 | 69.4 | ≤0.001 | 7.4 | ≤0.004 |
| S × A × EP | 42 | 53.1 | ≤0.001 | 12.8 | ≤0.001 |
| C × A × EP | 21 | 27.7 | ≤0.001 | 3.6 | ≤0.006 |
| S × C × A × EP | 42 | 10.5 | ≤0.001 | 1.5 | ≤0.017 |
Df-degrees of freedom; p-values less than 0.05 were considered significant; F-ratio is defined as the variance between samples/the variance within samples.
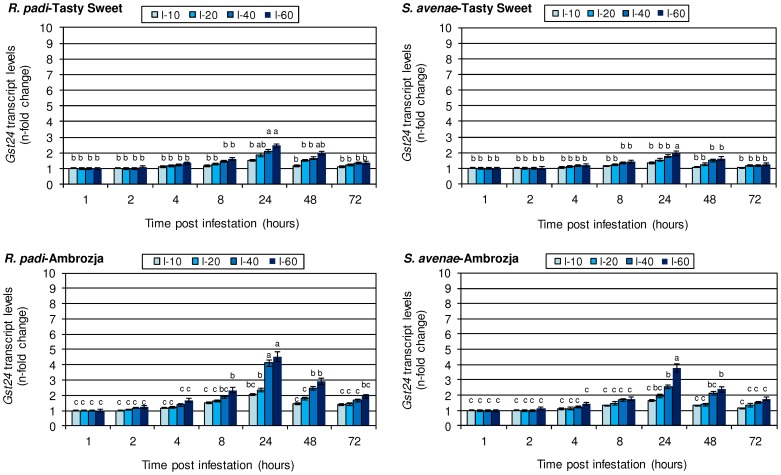
Abundance of gst24 transcript in Z. mays seedlings infested with the cereal aphids
Relative expression data of the gst24 gene in aphid-colonized maize seedlings are presented in figure 4. Transcriptional activity of the target gene in tissues of both tested Z. mays cultivars infested with R. padi or S. avenae remained at the same levels after 1 hpi, when compared to the respective control seedlings. In maize plants exposed to insect infestation for 2 h, only subtle accumulation of the gst24 transcript was recorded (3–10% increase in Tasty Sweet seedlings, and 6–24% elevation in Ambrozja plants). Prolonged aphid colonization (4–8 hpi) was associated with a steady enhancement in the expression of the analysed gene from 5% elevation in S. avenae-infested Tasty Sweet plants to 133% increment in R. padi-attacked Ambrozja seedlings, compared to the controls. The highest enhancement in the transcript amount for tissues of the aphid-infested maize plants was found at 24 hpi (e.g. 60 R. padi aphids influenced 2.5-fold and 4.5-fold increases in Tasty Sweet and Ambrozja seedlings, respectively, whereas the same abundance of S. avenae affected 2.0- and 3.8-fold increments in the relevant maize genotypes). It should be emphasized that insect infestation for 48 and 72 h resulted in a gradually decreasing upregulation of gst24 gene expression in Z. mays seedlings of the investigated cultivars in relation to the changes demonstrated after 24 h. Furthermore, the aphid-attacked Ambrozja plants responded to a higher elevation in the transcriptional activity of the target gene when compared with Tasty Sweet variety (e.g. 12–205% larger increase at the highest level of aphid infestation). It was additionally demonstrated that there was a higher abundance of the target mRNA transcript in R. padi-infested maize cultivars in comparison with S. avenae-stressed seedlings. It is important to underline that the scale of alternations in the gene expression was proportional to densities of the tested hemipterans on the seedlings of the investigated maize varieties. The maximal abundance of bird cherry-oat aphids (60 insects per plant) led to 7–50% and 9–71% higher increments of gst24 gene expression in the Tasty Sweet and Ambrozja plants, respectively, relative to the number of grain aphids. The statistical analysis evidenced significant effects of the tested variables and their interconnections in terms of gst24 gene expression in Z. mays plants (Table 5).
Figure 4. Influence of the tested cereal aphids on gst24 gene expression in the seedlings of Ambrozja and Tasty Sweet maize cultivars.
Values signify the mean n-fold changes in the gst24 transcript abundance in the aphid-stressed Z. mays plants in comparison with the non-infested group of seedlings. Error bars represent the standard deviation (± SD). For each maize-aphid treatment, three independent biological replicates were accomplished. The obtained gene expression data were normalized to the gapdh gene. The different letters above the SD bars designate significant differences among compared plants at P≤0.05 based on the Tukey's test. I-10, I-20, I-40 and I-60 are the levels of aphid infestation (10, 20, 40 and 60 insects per plant, accordingly).
Discussion
Monophagous Sitobion avenae F. (grain aphid) and oligophagous Rhopalosiphum padi L. (bird cherry-oat aphid) become serious pest species building up numerous colonies on many maize varieties grown in Poland, especially during warm and moist vegetative seasons [36]–[38]. Despite many research groups conducting extensive studies on the complex plant-aphid interactions, the participation of these hemipterans in the generation of oxidative stress and the functioning of the antioxidant defence network in the host systems still remain to be unraveled. To the best of our knowledge, this is the first report evaluating the impact of R. padi or S. avenae infestations on the expression profiles of the four genes encoding glutathione transferase isozymes (GSTF1, GST18, GST23 and GST24), as well as the levels of superoxide anion radical generation in the seedlings of susceptible (Tasty Sweet) and relatively resistant (Ambrozja) maize genotypes.
Aphid salivary glands produce a battery of hydrolytic enzymes that participate in the cleavage of primary and secondary cell walls, plasma membranes, and a variety of intracellular compounds. Additionally, salivary secretions of these hemipterans contain various elicitors, metabolic regulators, and phytotoxic constituents that trigger cascades of local and/or systemic defensive reactions as well as the processes of premature senescing, apoptosis, or necrosis within the colonized plant systems [39]–[41]. Studies have documented that proteinaceous effectors (Mp10 and Mp42) from M. persicae are capable of enhancing the defence systems in Nicotiana benthamiana Dom. plants, whereas two elicitors of Macrosiphum euphorbiae Thom., Me10 and Me23, possess the ability to suppress the host reactions in order to facilitate prolonged phloem feeding [13]-[14]. Aphid saliva infiltration and profound ultrastructural damages induced by insect mouthparts in the host tissues may be linked to excessive ROS release in the attacked organs. Superoxide anion radical is one of the major and most deleterious reactive oxygen species generated in plant cells both in the normal physiological state and in response to adverse environmental stimuli. It was found that the seedlings of both maize varieties colonized with R. padi or S. avenae aphids responded an early overproduction of O2 •– in comparison to the non-stressed control. The maximal enhancement in the superoxide anion radical generation in Z. mays seedlings was noted after 4–8 h of aphid feeding. Interestingly, a more marked elevation in O2 •− amounts occurred in the seedlings of Ambrozja (relatively resistant) plants in relation to Tasty Sweet (susceptible) cultivar. These observations are coherent with the results obtained by Mai et al. [15] who ascertained that Pisum sativum L. plants infested with the pea aphid (Acyrthosiphon pisum Harr.) possessed substantially higher amounts of O2 •− relative to the insect-free control. Furthermore, the most significant increase in excessive O2 •− formation was found at the highest infestation level (30 aphids per seedling). According to these authors, the prolonged aphid feeding resulted in the progressive increase in O2 •− levels within the attacked plants (e.g. 1.46- and 1.81-fold increments in relation to the reference plants at 24 and 96 hpi, accordingly). Moreover, it was reported that Russian wheat aphid (Diuraphis noxia Mordv.) markedly augmented the biosynthesis of hydrogen peroxide in resistant wheat plants in relation to the aphid-susceptible line. The oxidative burst in plants is associated with a dramatic increase in superoxide anion radicals' production at early stages of the exposure to various biotic stressing factors [28]. This phenomenon is linked with subsequent oxidative wave passing throughout plant tissues, leading to triggering the defence networks in the hosts, on the one hand, and possible suppression of the growth and development of herbivorous insects, on the other hand [42]. In order to overcome the excessive accumulation of this highly reactive and cytotoxic ROS form, the superoxide anion radicals are converted in the dismutation reaction to molecular oxygen (O2) and less toxic hydrogen peroxide (H2O2) [6], [15]–[16], [39]. Furthermore, we revealed very slight changes in superoxide anion radicals content in non-infested maize seedlings of both tested cultivars with duration of experimental time, but the recorded differences were not statistically significant. It is probable that isolation of Z. mays seedlings with the cover gauze could cause a minor mechanical stress influencing negligible fluctuations in O2 •− amount.
Plants have developed a number of defence mechanisms that are involved with protecting the cells from the detrimental impact of exaggerated ROS formation in response to a variety of abiotic and biotic stresses [43]–[46]. Until now, it has been identified at least 42 genes encoding diverse isozymes of glutathione transferase in maize [47]. In recent years, an important role of cytosolic GSTs in the alleviation of oxidative stress in plant tissues has been increasingly described [48]–[52]. The GSTs predominantly occur as homo- or heterodimers, with subunits of 23–30 kDa [53]. It should be underlined that among diverse groups of GST isozymes, only Tau and Phi classes are plant specific [50]. The performed molecular studies revealed that the cereal aphid infestations led to significant increases in the relative expression of analysed gst genes (gst1, gst18, gst23 and gst24) in the seedling leaves of both Z. mays genotypes, exhibiting distinct susceptibility levels to the insect colonization. Time-course analysis revealed that the target genes encoding the relevant GST isoenzymes (GSTF1, GST18, GST23 and GST24) were maximally upregulated at different aphid exposure periods (gst23 at 8 hpi; gst1, gst18, and gst24 genes at 24 hpi). Interestingly, the bird cherry-oat aphid infestation caused more considerable increments in the amounts of all tested gst transcripts in the maize plants compared to grain aphid feeding. Additionally, relatively resistant Ambrozja plants that were attacked by the cereal aphids were characterized with a higher stimulation of the transcriptional activity of the gst genes in relation to the susceptible Tasty Sweet plants. There have been limited reports published evidencing aphid-stimulated transcriptional reprogramming in the attacked host plants [54]–[58]. Microarray experiments performed by Kuśnierczyk and co-workers revealed that feeding of M. persicae or Brevicoryne brassicae L. for 72 h led to significant alternations in the transcriptional activity of 13 gst genes in 22–30-day-old plants of three tested Arabidopsis thaliana ecotypes (Landsberg erecta/Ler/, Cape Verde Islands/Cvi/, and Wassilewskija/Ws/) [54]. The aphid colonization (8–12 insects per leaf) resulted in the overexpression of most analysed gst genes in the plants representing the tested ecotypes when compared to the non-stressed control. The opposite tendency was identified in the expression patterns of GSTU18 and GSTU20 transcripts (0.23–1.60-fold and 0.26–1.89-fold down regulation, respectively) in the investigated ecotypes in relation to the insect-free plants. Additionally, these authors demonstrated upregulation of the glutathione-conjugate transporter (MRP4) in the aphid-injured Cape Verde Islands and Wassilewskija plants. Another infestation experiments conducted by Kuśnierczyk et al. revealed that 21–25-day-old A. thaliana plants (Landsberg erecta/Ler/ecotype) infested with B. brassicae (4 aphids per leaf) were characterized with an early enhancement (at 6 hpi) of the amount of ATGST6, ATGST7 and ATGST10 transcripts relative to the control. Furthermore, prolonged aphid colonization (48 hpi) led to the strong upregulation of four glutathione transferase (ATGSTU3, ATGSTU10, ATGSTU11, ATGSTL1) genes, as well as increases in the transcript amounts of two glutathione S-conjugate transporters (MRP3 and MRP4) [55]. Similarly, Moran et al. elucidated that infestation of A. thaliana with M. persicae aphids for 72 h resulted in 2.9-fold- and 4.8-fold elevations in the expression of gst1 and gst11 genes, respectively, compared to the non-treated control [56]. Stotz et al. also ascertained that the diamondback moth (Plutella xylostella L.), feeding on the rosette leaves of wild-type A. thaliana plants, influenced profound increments in the expression of gst2 and gst6 genes compared to the insect-free control [57]. Likewise, Bandopadhyay and co-workers evidenced that the transcriptional activity of genes encoding various glutathione transferase isoforms may vary significantly depending on the duration of the aphid exposure period [58]. According to the cited authors, Rorippa indica L. plants infested with mustard aphids (Lipaphis erysimi/L./Kalt.) responded with a 2.5-fold elevation in the transcriptional activity of the AT1G78370 gene (glutathione transferase AtTAU20) at 12 hpi, but a dissimilar trend occurred when the insect colonization was extended to 48 hpi. Furthermore, it should be noted that other biotic stressors, such as pathogenic fungi or microorganisms are able to trigger notable modifications in the expression patterns of several gst genes within the hosts. For example, it has been elucidated that maize plants infected with Ustilago maydis possessed an increased transcriptional activity of seven transferase glutathione genes (gst15, gst18, gst20, gst24, gst25, gst30, and gst36) after 12 h post-fungal inoculation [59]. The upregulation levels ranged from a 3.1-fold increase of the gst18 gene expression to a 108-fold increment in the gst30 transcript abundance compared to non-treated plants. Microarray data achieved by Luo et al. revealed that the expression of transferase glutathione genes in maize kernels of aflatoxin-resistant (Eyl25) and aflatoxin-susceptible (Eyl31) lines differentially responded 72 h after inoculation with Aspergillus flavus [60]. Some authors have suggested that the induction of gst genes is involved in limiting the adverse effects of oxidative stress within plant tissues, including the reduction of cell death events occurring as a result of the hypersensitive reactions [17], [59]–[60].
In the present study, R. padi infestation contributed to a substantially greater upregulation of the analysed gst genes and to the increases in the O2 •− generation in seedlings of both Ambrozja and Tasty Sweet genotypes in comparison to grain aphid feeding. Oligophagous bird cherry-oat aphids alternate the host plants between members of the Prunus genus (winter hosts) and a broad set of Poaceae species (summer hosts), whereas the life cycle of monophagous S. avenae is associated with numerous grasses and cereals [10]–[11], [61]. Greater diversity of plant systems colonized with R. padi indicates a higher adaptation of this hemipteran species to the chemical composition of the hosts. Conceivable sources of distinct biochemical and molecular effects in aphid-infested maize plants may be caused by differences in the insect salivary compounds and specific routes of stylet insertion throughout the plant tissues. It has been reported that salivary secretions of the bird cherry-oat aphid contain a wide spectrum of biocatalysts, which are responsible for the hydrolysis of structural macromolecules in the primary and secondary cell walls, and plasmalemma [62]–[63]. Furthermore, microscopic observations conducted by some researchers have documented additional profound injuries within the mesophyll cells of both winter and summer hosts, whereas S. avenae infestation resulted in a much lower range of ultrastructural damages, and they displayed a typical intercellular mode of mouthparts passage within the winter wheat Sakva plants [64]–[65]. According to Urbańska et al., the bird cherry-oat aphid has evolved an adaptive enzymatic mechanism that allows detoxification of harmful cyanogenic constituents present in the leaves of primary hosts [66]. Łukasik et al. provided valuable findings, indicating that R. padi feeding caused greater depletion in the content of ascorbate and greater stimulation of ascorbate peroxidase activity in the triticale seedlings compared to S. avenae aphids [16]. Similarly, Sytykiewicz revealed that bird cherry-oat aphid infestation of maize plants evoked a more significant decrease in the total antioxidant capacity towards the DPPH (1,1-diphenyl-2-picrylhydrazyl) radical in relation to grain aphid colonization [6]. It may be assumed that a decreased efficacy of DPPH radical scavenging activity in aphid-infested maize plants might be associated with a continuous pressure of biotic stressing factor (aphid colonization) that triggered the oxidative stress in the host systems. It is particularly evident when massive and/or prolonged aphid infestation occurred. It is likely that the pool of available antioxidants under stressful conditions significantly depressed the total antioxidative capacity of extracts derived from the infested maize seedlings when compared to the control. On the other hand, lower contents of ascorbate and glutathione were evidenced, as well as higher levels of ascorbate peroxidase and glutathione transferase activities in tissues of the bird cherry-oat aphid, in comparison with S. avenae individuals, which proves that there are significant differences in the functioning of the antioxidative machinery within these cereal aphids [12], [67].
This report provides new insight into the molecular basis of highly complex antioxidative responses of the model maize plants colonized with cereal aphids. It was demonstrated, there is differential regulation of four gst genes, encoding various isoforms of glutathione transferase in the insect-challenged seedling leaves of Z. mays, representing high and low susceptibility to the aphid colonization. The obtained results revealed insect-triggering oxidative stress and the crucial role of glutathione transferases in constituting complex defence reactions in the attacked host systems. In order to gain a better understanding of the elicitation of the plant defence reactions which occur at the early stages of aphid infestation in maize plants, the extended molecular analyses comprising transcriptome-wide screening of other aphid-regulated genes, as well as identification of low molecular and regulatory RNA molecules (e.g. miRNA) and assessing their gene expression profiles should be performed.
Supporting Information
The set of Z. mays glutathione transferase genes analysed with the application of TaqMan Gene Expression Assays #. # TaqMan Gene Expression Assays used in the performed experiments were developed and supplied by Life Technologies (Poland).
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was financially supported by the National Science Centre (NCS, Poland) under the grant no. N N310 733940. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bender RS, Haegele JW, Ruffo ML, Below FE (2013) Nutrient uptake, partitioning, and remobilization in modern, transgenic insect-protected maize hybrids. Agron J 105: 161–170. [Google Scholar]
- 2. Dukowic-Schulze S, Harris A, Li J, Sundararajan A, Mudge, et al (2014) Comparative transcriptomics of early meiosis in Arabidopsis and maize. J Genet Genomics 41: 139–152. [DOI] [PubMed] [Google Scholar]
- 3. Zhao Y, Cai M, Zhang X, Li Y, Zhang J, et al. (2014) Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS One 9: e94126 10.1371/journal.pone.0094126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bosak EJ, Seidl-Adams IH, Zhu J, Tumlinson JH (2013) Maize developmental stage affects indirect and direct defense expression. Environ Entomol 42: 1309–1321. [DOI] [PubMed] [Google Scholar]
- 5.Seidl-Adams I, Richter A, Boomer K, Yoshinaga N, Degenhardt J, et al. (2014) Emission of herbivore elicitor-induced sesquiterpenes is regulated by stomatal aperture in maize (Zea mays) seedlings. Plant Cell Environ (in press). doi: 10.1111/pce.12347. Article first published online: 2014 May 13. [DOI] [PubMed]
- 6. Sytykiewicz H (2014) Differential expression of superoxide dismutase genes in aphid-stressed maize (Zea mays L.) seedlings. PLoS One 9: e94847 10.1371/journal.pone.009484715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Czapla A, Kurczak P, Kiełkiewicz M (2011) Elementy bionomii mszycy różano-trawowej (Metopolophium dirhodum Walker) na wybranych odmianach kukurydzy. Prog Plant Prot 51: 787–793 (In Polish) [Google Scholar]
- 8. Strażyński P (2008) Aphid fauna (Hemiptera, Aphidoidea) on maize crops in Wielkopolska – species composition and increase in number. Aphids and Other Homopterous Insects 14: 123–128. [Google Scholar]
- 9. Sprawka I, Goławska S, Czerniewicz P, Sytykiewicz H (2011) Insecticidal action of phytohemagglutinin (PHA) against the grain aphid, Sitobion avenae . Pestic Biochem Phys 100: 64–69. [Google Scholar]
- 10. Halarewicz A, Gabrys B (2012) Probing behavior of bird cherry-oat aphid Rhopalosiphum padi (L.) on native bird cherry Prunus padus L. and alien invasive black cherry Prunus serotina Erhr. in Europe and the role of cyanogenic glycosides. Arthropod-Plant Inte 6: 497–505. [Google Scholar]
- 11. Zielińska L, Trzmiel K, Jeżewska M (2012) Ultrastructural changes in maize leaf cells infected with maize dwarf mosaic virus and sugarcane mosaic virus. Acta Biol Cracov Bot 54: 97–104. [Google Scholar]
- 12. Sempruch C, Horbowicz M, Kosson R, Leszczyński B (2012) Biochemical interactions between triticale (Triticosecale; Poaceae) amines and bird cherry-oat aphid (Rhopalosiphum padi; Aphididae). Biochem Syst Ecol 40: 162–168. [Google Scholar]
- 13. Rodriguez PA, Stam R, Warbroek T, Bos JI (2014) Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol Plant Microbe Interact 27: 30–39. [DOI] [PubMed] [Google Scholar]
- 14. Pitino M, Hogenhout SA (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant Microbe Interact 26: 130–139. [DOI] [PubMed] [Google Scholar]
- 15. Mai VC, Bednarski W, Borowiak-Sobkowiak B, Wilkaniec B, Samardakiewicz S, et al. (2013) Oxidative stress in pea seedling leaves in response to Acyrthosiphon pisum infestation. Phytochemistry 93: 49–62. [DOI] [PubMed] [Google Scholar]
- 16. Łukasik I, Goławska S, Wójcicka A (2012) Effect of cereal aphid infestation on ascorbate content and ascorbate peroxidase activity in triticale. Pol J Environ Stud 21: 1937–1941. [Google Scholar]
- 17. Gong H, Jiao Y, Hu WW, Pua EC (2005) Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro . Plant Mol Biol 57: 53–66. [DOI] [PubMed] [Google Scholar]
- 18. Dixon DP, Hawkins T, Hussey PJ, Edwards R (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60: 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wisser RJ, Kolkman JM, Patzoldt ME, Holland JB, Yu J, et al. (2011) Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc Natl Acad Sci USA 108: 7339–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cummins I, Wortley DJ, Sabbadin F, He Z, Coxon CR, et al. (2013) Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc Natl Acad Sci USA 110: 5812–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lan T, Yang ZL, Yang X, Liu YJ, Wang XR, et al. (2009) Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 21: 3749–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dixon DP, Cole DJ, Edwards R (2000) Characterisation of a zeta class glutathione transferase from Arabidopsis thaliana with a putative role in tyrosine catabolism. Arch Biochem Biophys 384: 407–412. [DOI] [PubMed] [Google Scholar]
- 23. Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532. [DOI] [PubMed] [Google Scholar]
- 24. Kumar M, Yadav V, Tuteja N, Johri AK (2009) Antioxidant enzyme activities in maize plants colonized with Piriformospora indica . Microbiology 155: 780–790. [DOI] [PubMed] [Google Scholar]
- 25. Dean JD, Goodwin PH, Hsiang T (2005) Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J Exp Bot 56: 1525–1533. [DOI] [PubMed] [Google Scholar]
- 26. Yu T, Li YS, Chen XF, Hu J, Chang X, et al. (2003) Transgenic tobacco plants overexpressing cotton glutathione S-transferase (GST) show enhanced resistance to methyl viologen. J Plant Physiol 160: 1305–1311. [DOI] [PubMed] [Google Scholar]
- 27. Chronopoulou EG, Labrou NE (2009) Glutathione transferases: Emerging multidisciplinary tools in red and green biotechnology. Recent Pat Biotechnol 3: 211–223. [DOI] [PubMed] [Google Scholar]
- 28. Kampranis SC, Damianova R, Atallah M, Toby G, Kondi G, et al. (2000) A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. J Biol Chem 275: 29207–29216. [DOI] [PubMed] [Google Scholar]
- 29. Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, et al. (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of day length-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J 52: 640–657. [DOI] [PubMed] [Google Scholar]
- 30. Jain M, Ghanashyam C, Bhattacharjee A (2010) Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics 11: 1471–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dall'Asta P, Rossi GB, Arisi ACM (2013) Abscisic acid-induced antioxidant system in leaves of low flavonoid content maize. IV Simposio Brasileiro de Genetica Molecular de Planta (SBGMP), Bento Gonçalves, Brasil, 8-12 April. pp. 26.
- 32. Sytykiewicz H (2011) Expression patterns of glutathione transferase gene (Gst1) in maize seedlings under juglone-induced oxidative stress. Int J Mol Sci 12: 7982–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adejumo TO, Hettwer U, Nutz S, Karlovsky P (2009) Real-time PCR and agar plating method to predict Fusarium verticillioides and fumonisin B content in Nigerian maize. J Plant Protect Res 49: 399–404. [Google Scholar]
- 34. Chaitanya KSK, Naithani SC (1994) Role of superoxide, lipid peroxidation and superoxide dismutase in membrane perturbation during loss of viability in seeds of Shorea robusta Gaertn.f. New Phytol 126: 623–627. [Google Scholar]
- 35. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 36. Pieńkosz A, Leszczyński B, Warzecha R (2005) Podatność kukurydzy na mszyce zbożowe. Prog Plant Prot 45: 989–992 (In Polish) [Google Scholar]
- 37. Krawczyk A, Miętkiewski R, Hurej M (2006) Owadobójcze grzyby porażające mszyce żerujące na kukurydzy. Prog Plant Prot 46: 378–381 (In Polish) [Google Scholar]
- 38. Bereś PK, Pruszyński G (2008) Ochrona kukurydzy przed szkodnikami w produkcji integrowanej. Acta Sci Pol ser Agricultura 7: 19–32 (In Polish) [Google Scholar]
- 39. Morkunas I, Mai VC, Gabryś B (2011) Phytohormonal signaling in plant responses to aphid feeding. Acta Physiol Plant 33: 2057–2073. [Google Scholar]
- 40. Will T, van Bel AJ (2008) Induction as well as suppression: How aphid saliva may exert opposite effects on plant defense. Plant Signal Behav 3: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Will T, Furch AC, Zimmermann MR (2013) How phloem-feeding insects face the challenge of phloem-located defenses. Front Plant Sci 4: 336 10.3389/fpls.2013.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. War AR, Paulraj MG, Ahmad T, Buhroo AA, Hussain B, et al. (2012) Mechanisms of plant defense against insect herbivores. Plant Signal Behav 7: 1306–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Santamaria ME, Martínez M, Cambra I, Grbic V, Diaz I (2013) Understanding plant defence responses against herbivore attacks: an essential first step towards the development of sustainable resistance against pests. Transgenic Res 22: 697–708. [DOI] [PubMed] [Google Scholar]
- 44. Morkunas I, Formela M, Marczak L, Stobiecki M, Bednarski W (2013) The mobilization of defence mechanisms in the early stages of pea seed germination against Ascochyta pisi . Protoplasma 250: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prince DC, Drurey C, Zipfel C, Hogenhout SA (2014) The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis . Plant Physiol 164: 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Basantani M, Srivastava A, Sen S (2011) Elevated antioxidant response and induction of tau-class glutathione S-transferase after glyphosate treatment in Vigna radiata (L.) Wilczek. Plant Physiol Biochem 99: 111–117. [Google Scholar]
- 47. McGonigle B, Keeler SJ, Lau SMC, Koeppe MK, O'Keefe DP (2000) A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol 124: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moons A (2005) Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam Horm 72: 155–202. [DOI] [PubMed] [Google Scholar]
- 49. Wang Y, Tang Y, Zhang M, Cai F, Qin J, et al. (2012) Molecular cloning and functional characterization of a glutathione S-transferase involved in both anthocyanin and proanthocyanidin accumulation in Camelina sativa (Brassicaceae). Genet Mol Res 11: 4711–4719. [DOI] [PubMed] [Google Scholar]
- 50. Kitamura S, Akita Y, Ishizaka H, Narumi I, Tanaka A (2012) Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J Plant Physiol 169: 636–642. [DOI] [PubMed] [Google Scholar]
- 51. Abedini R, Zare S (2013) Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J Plant Physiol 170: 1277–1284. [DOI] [PubMed] [Google Scholar]
- 52. Liu YJ, Han XM, Ren LL, Yang HL, Zeng QY (2013) Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol 161: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lo Piero AR, Mercurio V, Puglisi I, Petrone G (2010) Different roles of functional residues in the hydrophobic binding site of two sweet orange tau glutathione S-transferases. FEBS J 277: 255–262. [DOI] [PubMed] [Google Scholar]
- 54. Kuśnierczyk A, Winge P, Midelfart H, Armbruster WS, Rossiter JT, et al. (2007) Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae . J Exp Bot 58: 2537–2552. [DOI] [PubMed] [Google Scholar]
- 55. Kuśnierczyk A, Winge P, Jørstad TS, Troczyńska J, Rossiter JT, et al. (2008) Towards global understanding of plant defence against aphids-timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ 31: 1097–1115. [DOI] [PubMed] [Google Scholar]
- 56. Moran PJ, Cheng Y, Cassell JL, Thompson GA (2002) Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch Insect Biochem Physiol 51: 182–203. [DOI] [PubMed] [Google Scholar]
- 57. Stotz HU, Pittendrigh BR, Kroymann J, Weniger K, Fritsche J, et al. (2000) Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol 124: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bandopadhyay L, Basu D, Sikdar SR (2013) Identification of genes involved in wild crucifer Rorippa indica resistance response on mustard aphid Lipaphis erysimi challenge. PLoS One 8: e73632 10.1371/journal.pone.0073632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, et al. (2008) Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis . Plant J 56: 181–195. [DOI] [PubMed] [Google Scholar]
- 60. Luo M, Brown RL, Chen ZY, Menkir A, Yu J, et al. (2011) Transcriptional profiles uncover Aspergillus flavus-induced resistance in maize kernels. Toxins 3: 766–786 10.3390/toxins3070766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Czerniewicz P, Leszczyński B, Chrzanowski G, Sempruch C, Sytykiewicz H (2011) Effects of host plant phenolics on spring migration of bird cherry-oat aphid (Rhopalosiphum padi L.). Allelopathy J 27: 309–316. [Google Scholar]
- 62. Rao SA, Carolan JC, Wilkinson TL (2013) Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS One 8: e57413 10.1371/journal.pone.0057413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Urbańska A, Niraz S (1990) Anatomiczne i biochemiczne aspekty żerowania mszyc zbożowych. Zesz Probl Post Nauk Roln 392: 201–213 (In Polish) [Google Scholar]
- 64.Urbańska A (2010) Histochemical analysis of aphid saliva in plant tissue. EJPAU ser Biology 13: #26. Available: www.ejpau.media.pl/volume13/issue4/art-26.html. Accessed 2014 Oct 13.
- 65.Sytykiewicz H, Leszczyński B (2008) Monitoring of anatomical changes within the bird cherry shoots evoked by Rhopalosiphum padi L. (Hemiptera, Aphididae). XXIII International Congress of Entomology, Durban, Republic of South Africa, 6–12 July, abstract no. 811.
- 66.Urbańska A, Leszczyński B, Matok H, Dixon AFG (2002) Cyanide detoxifying enzymes of bird cherry oat aphid. EJPAU ser Biology 5: #01. Available: www.ejpau.media.pl/volume5/issue2/biology/art-01.html. Accessed 2014 Oct 13.
- 67. Łukasik I (2006) Effect of o-dihydroxyphenols on antioxidant defence mechanisms of cereal aphids associated with glutathione. Pesticides 3–4: 67–73. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The set of Z. mays glutathione transferase genes analysed with the application of TaqMan Gene Expression Assays #. # TaqMan Gene Expression Assays used in the performed experiments were developed and supplied by Life Technologies (Poland).
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.