Abstract
The cellular immune response is the most important mediator of allograft rejection and is a major barrier to transplant tolerance. Delineation of the depth and breadth of the alloreactive T cell repertoire and subsequent application of the technology to the clinic may improve patient outcomes. As a first step toward this, we have used MLR and high-throughput sequencing to characterize the alloreactive T cell repertoire in healthy adults at baseline and 3 months later. Our results demonstrate that thousands of T cell clones proliferate in MLR, and that the alloreactive repertoire is dominated by relatively high-abundance T cell clones. This clonal make up is consistently reproducible across replicates and across a span of three months. These results indicate that our technology is sensitive and that the alloreactive TCR repertoire is broad and stable over time. We anticipate that application of this approach to track donor-reactive clones may positively impact clinical management of transplant patients.
Introduction
Cellular immune response is the most important mediator of transplant rejection and a major barrier to transplant tolerance [1]–[3]. It is largely mediated by memory T cell populations specific for allo-peptides presented either on allo-MHC (direct antigen presentation) or on self-MHC (indirect antigen presentation) [3]–[5]. Positive selection in the thymus requiring immature T cells to have some binding affinity for self-HLA means that a significant proportion of mature T cells also have off-target specificity for allo-HLA alleles. Negative selection removes T cells specific for self-peptides presented on self-HLA, buts leaves T cells specific for self-peptides presented on allo-HLA [6]–[12]. The production of the alloreactive T cell repertoire is further complicated by molecular mimicry. Thus, in one well-studied example a public T cell response specific to EBV in the context of HLA-B*08:01 has been shown to exhibit cross-reactivity with a self-peptide presented by HLA-B*44:02 [13]–[16]. These cross-reactive T cells have been observed in HLA-B*08:01/HLA-B*44:02 mismatched lung allografts, suggesting direct clinical relevance for this mode of T cell alloreactivity [17]. Even in individuals with no history of allo-HLA sensitization, viral exposure or vaccine administration can create HLA cross-reactive memory T cells [18]–[22].
Many studies have identified public and private alloreactive T cell clones that can be primed by a variety of immunogenic events. However, while public T cell clones may play an important role in specific exposures they represent a very small proportion of the entire T cell repertoire; investigating private T cell specificities allows for a much broader view of the alloreactive T cell repertoire but private T cell responses must be measured anew in each subject.
It is our hypothesis that the alloreactive T cell repertoire can be studied by performing mixed lymphocyte reaction cultures [23], [24], followed by molecular analysis of clonotypes thus generated. The availability of high-throughput sequencing of rearranged T cell receptor genes, which act as unique molecular tags for each clonal population, now allows for unprecedented depth and accuracy in the characterization of T cell repertoires. Here, we employ this high-throughput TCR sequencing to test our hypothesis by thoroughly interrogating the alloreactive T cell repertoire between three pairs of healthy adult subjects as well as the persistence of alloreactive T cell clones across biological replicates and across time.
Methods
Subjects
Human peripheral blood samples were obtained from laboratory volunteers under a protocol following written informed consent approved and supervised by a Northwestern University Institutional Review Board. These healthy volunteers were HLA-typed by the Northwestern HLA laboratory using molecular methods (reverse sequence specific oligonucleotide probe hybridization).
Mixed Lymphocyte Reaction (MLR) Culture and Alloreactive Responding Cell Isolation
Peripheral blood mononuclear cells (PBMC) were isolated using Ficoll-Hypaque. The responder cells were labeled with CFSE and the stimulator cells labeled with PKH26 as described previously [25], [26]. The responders and stimulators were matched for 1 HLA- DR antigen to mimic the minimum requirement for some clinical transplants [27]. The PKH26 labeled stimulator cells were also irradiated at 3000 rads. The responder and stimulator cells were cultured in bulk in 15% normal AB serum containing RPMI 1640 culture medium (NAB-CM) at 1×106/ml each. After 7 days these were harvested and the proliferating responders were then sorted on FACSAria (BD, San Jose, CA) by gating on the CFSE dim or negative cells after gating out both CFSE high non-proliferating and the very few PKH26+ stimulator cells that still survived.
In parallel, flow cytometric analysis of the above MLR cultures was performed to determine which subsets of responder cells proliferated in response to allostimulation, using fluorochrome conjugated monoclonal antibodies. The data were acquired on an FC500 flow cytometer (Beckman-Coulter) and analyzed for cell subsets by gating on the CFSE dim or negative cells after gating out both CFSE high non-proliferating and the very few PKH26+ stimulator cells [25], [26]. Additionally, standard 7-day 3H-thymidine incorporation assays were also performed to monitor the strength of the MLR responses as described previously [25], [26].
High-Throughput TCRβ Sequencing
Genomic DNA was extracted from cell samples using Qiagen DNeasy Blood extraction Kit (Qiagen, Gaithersburg, MD, USA). We sequenced the CDR3 region of rearranged TCRβ genes; the TCRβ CDR3 region was defined according to the IMGT collaboration [28]. TCRβ CDR3 regions were amplified and sequenced using previously-described protocols [29], [30]. Briefly, a multiplexed PCR method was employed using a mixture of 60 forward primers specific to TCR Vβ gene segments and 13 reverse primers specific to TCR Jβ gene segments. Reads of 87 bp were obtained using the Illumina HiSeq System. Raw HiSeq sequence data were preprocessed to remove errors in the primary sequence of each read, and to compress the data. A nearest neighbor algorithm was used to collapse the data into unique sequences by merging closely related sequences, to remove both PCR and sequencing errors.
PCR Template Abundance Estimation
To estimate the average read coverage per input template in our PCR and sequencing approach, we employed a set of approximately 850 unique types of synthetic TCR analog, comprising each combination of Vβ and Jβ gene segments [31]. These molecules were included in each PCR reaction at very low concentration so that only some types of synthetic template were observed. Using the known concentration of the synthetic template pool, we simulated the relationship between the number of observed unique synthetic molecules and the total number of synthetic molecules added to the reaction (this is very nearly one-to-one at the low concentrations we employed). These molecules then allowed us to calculate for each PCR reaction the mean number of sequencing reads obtained per molecule of PCR template, and thus to estimate the number of T cells in the input material bearing each unique TCR rearrangement.
Results and Discussion
Isolation of the Alloreactive T Cell Repertoire
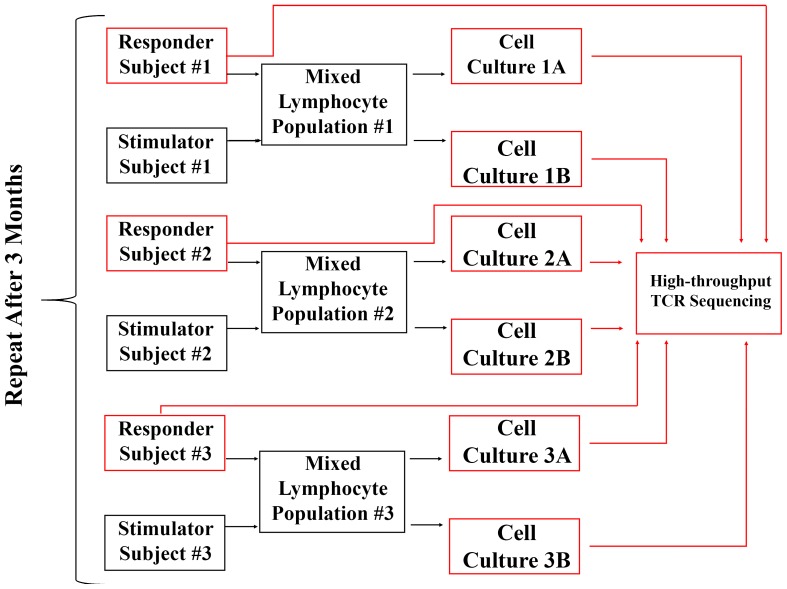
In order to study the breadth, clonal structure and dynamics of the alloreactive T cell repertoire, we performed a one-way mixed lymphocyte culture using CFSE-labeled responder cells and PKH26-labeled stimulator cells on each of three pairs of healthy adult subjects [25], [26], with cell culture performed in duplicate. Three months after the first experiment, we repeated this cell culture protocol for the same three pairs of subjects. In total, we generated 18 samples of T cells, comprising six samples from each pair of subjects: uncultured total PBMC and purified proliferating T cells from duplicate MLR, at baseline and after three months (Figure 1 summarizes experimental design).
Figure 1. Experimental design.
We assayed three pairs of healthy adult subjects using mixed lymphocyte reaction cultures. For each pair, lymphocytes from a responder subject were mixed with inactivated lymphocytes from a stimulator subject and cultured in duplicate. Uncultured freshly isolated PBMC from the responder as well as proliferating T cell populations from the duplicate cultures were subjected to high-throughput sequencing: we sequenced nine samples in total across the three pairs of subjects. Three months later, the experiments were repeated to generate nine more samples for high-throughput TCRβ sequencing.
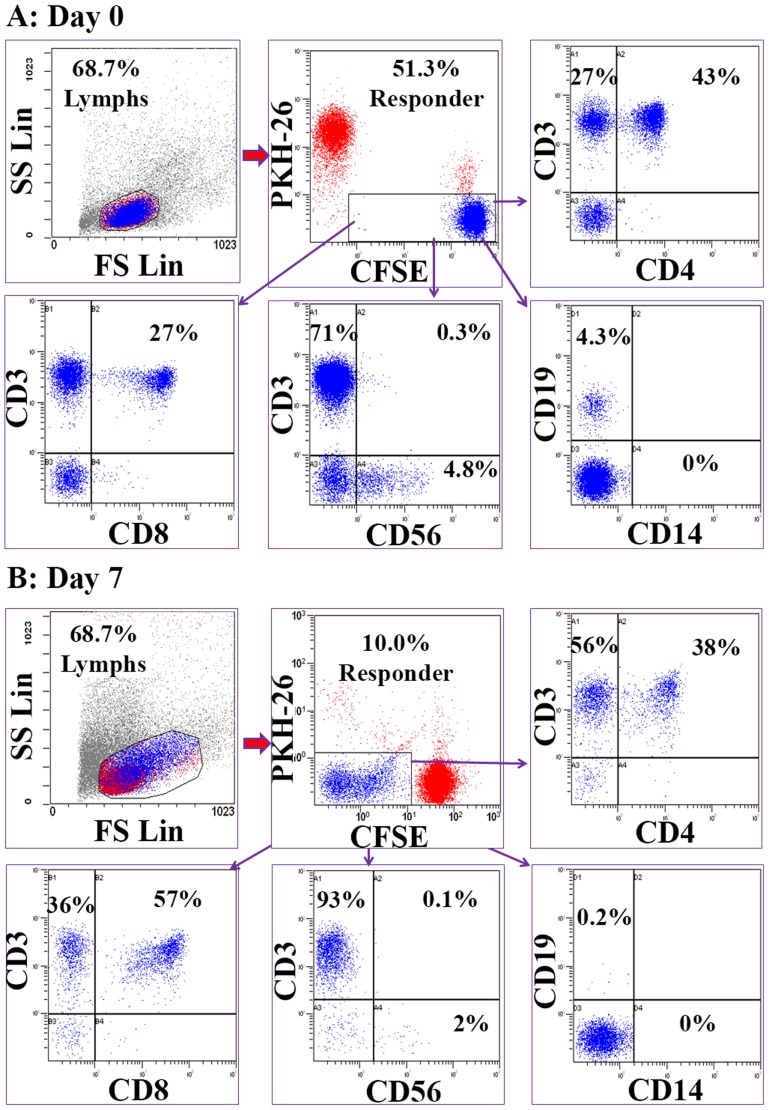
For each MLR reaction, after 7 days the proliferating responders were sorted by gating on the CFSE dim or negative cells after gating out both CFSE high non-proliferating and the very few PKH26+ stimulator cells that still survived (Figure 2A). The proliferating cells consisted of 40.3±4.7% CD3+CD4+ and 57.2±5.1% CD3+CD8+ T cells as well as minor subset of CD56+ NK cells (Figure 2B). Each population of uncultured PBMC or proliferating T cells was subjected to amplification and high-throughput sequencing of the CDR3 region of TCRβ, which somatically rearranges during T cell maturation and acts as a unique molecular tag for each clonal population of T cells. Sequencing results are presented in Table 1.
Figure 2. Alloreactive Cellular Subset Profile Generated in MLR.
Bulk MLRs were prepared as described in Materials and Methods. The cellular makeup of responder cell populations were delineated at the onset and after 7 days in culture using fluorochrome coupled monoclonal antibodies. The cells were analyzed first by gating on lymphocytes and then after gating either on total CFSE positive responder cells (A: Day 0) or on CFSE diluted proliferating responder cells (B: Day 7).
Table 1. Summary of TCRβ sequencing results.
| Sample | T cells assayed (estimated) a | Unique TCRβ sequences | Sequencing reads b |
| Fresh PBMC sample #1, 0 months | 4,336,812 | 750,211 | 51,160,577 |
| Fresh PBMC sample #2, 0 months | 4,774,312 | 1,375,340 | 46,370,325 |
| Fresh PBMC sample #3, 0 months | 4,016,260 | 991,848 | 33,633,101 |
| Fresh PBMC sample #1, 3 months | 713,990 | 264,159 | 17,437,692 |
| Fresh PBMC sample #2, 3 months | 1,847,987 | 1,046,492 | 23,507,950 |
| Fresh PBMC sample #3, 3 months | 2,197,064 | 1,061,154 | 18,766,880 |
| Proliferated MLR responder #1A, 0 months | 1,885,973 | 33,677 | 23,366,016 |
| Proliferated MLR responder #1B, 0 months | 1,997,723 | 33,387 | 26,098,554 |
| Proliferated MLR responder #2A, 0 months | 1,575,201 | 79,174 | 24,704,053 |
| Proliferated MLR responder #2B, 0 months | 1,527,643 | 68,505 | 13,832,785 |
| Proliferated MLR responder #3A, 0 months | 3,372,150 | 58,382 | 37,022,643 |
| Proliferated MLR responder #3B, 0 months | 3,190,902 | 53,316 | 23,126,368 |
| Proliferated MLR responder #1A, 3 months | 640,366 | 57,778 | 12,741,642 |
| Proliferated MLR responder #1B, 3 months | 587,681 | 53,260 | 9,806,707 |
| Proliferated MLR responder #2A, 3 months | 1,022,417 | 68,565 | 10,736,335 |
| Proliferated MLR responder #2B, 3 months | 522,273 | 53,337 | 10,679,864 |
| Proliferated MLR responder #3A, 3 months | 685,126 | 64,615 | 9,788,942 |
| Proliferated MLR responder #3B, 3 months | 760,990 | 67,586 | 10,999,866 |
| 35,654,870 | 6,180,786 | 403,780,300 |
see Methods.
the total number of 87-bp sequencing reads generated.
Size of the Alloreactive T Cell Repertoire
To determine the number of T cell clonal lineages involved in the alloreactive T cell response, we analyzed the number of unique CDR3 sequences observed in the proliferated T cell samples in comparison to uncultured bulk T cells from the same subjects. We defined alloreactive T cell clones as those observed in at least 10 cells in the proliferated sample and unobserved in the uncultured T cell sample, or T cells whose frequency in the proliferated sample was at least ten-fold higher than in the uncultured T cell sample. We defined two sets of alloreactive T cell clones: low-abundance alloreactive clones (below the threshold of detection in the subject's baseline T cell repertoire) and high-abundance alloreactive clones (present at measurable frequency in the subject's baseline T cell repertoire). On average, we observed 14,000 alloreactive T cell clones in each experiment; 84% of alloreactive T cell clones were low-abundance before proliferation, but in total low-abundance clones made up 40% and high-abundance clones made up 60% of the alloreactive T cell repertoire when weighting by post-proliferation clonal abundance (Table 2). While the number of proliferated low-abundance clones varied considerably, variation in the number of high-abundance (thus, presumably antigen-experienced) T cell clones between subjects was much smaller, at about 2,000 clones in each of the six experiments. These data indicate that thousands of different clonal populations of T cells comprise the alloreactive T cell repertoire.
Table 2. Size of the alloreactive T cell repertoire.
| Mean (N = 6) | SD | % of proliferated T cells | |
| Number of alloreactive clones | 13750 | 6823 | 100% |
| Low-abundance pre-culture a | 11610 | 6494 | 40.0% |
| High-abundance pre-culture b | 2140 | 539 | 60.0% |
unobserved in pre-culture sample and ≥10 T cells after MLR.
present in pre-culture sample and ≥10× enriched after MLR.
Reproducibility of the Alloreactive T Cell Repertoire
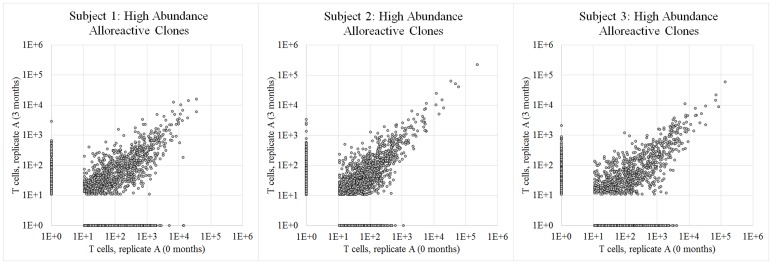
To assay the consistency of the alloreactive T cell repertoire, we examined the persistence of each T cell clone. After defining high-abundance and low-abundance alloreactive T cells, we compared the set of alloreactive T cell clones generated in duplicate cell culture experiments (Figure 3). In each subject, essentially all clones that were highly expanded in proliferated cell culture assorted to the high-abundance subset (i.e., were present at appreciable frequency in the peripheral T cell repertoire to begin with). Reproducibility between duplicate cell culture experiments was high among this set of abundant and highly alloreactive T cell clones (average r2 among three subjects = 0.96), indicating that when presented with identical stimuli these clonal populations of T cells responded in a very reproducible manner.
Figure 3. T cell clonal frequency among biological replicates of mixed lymphocyte culture.
Above are six scatter plots showing the number of T cells bearing each unique CDR3 sequence in replicate mixed lymphocyte culture experiments performed on three pairs of healthy adult subjects. Each column corresponds to one pair of subjects; the top row of plots show T cell clones that were previously observed in a pre-MLR sample of peripheral T cells (high-abundance), and the bottom row of plots show T cell clones unobserved in a pre-MLR sample of peripheral T cells (low-abundance). Each point represents a unique T cell clone, and points are plotted at (# of observed T cells +1), so that clones unobserved in one sample are plotted on the axes.
Since our replicate cell culture experiments did not address the stability of the alloreactive T cell repertoire over time, we repeated the T cell isolation and duplicate MLR experiments with the same three pairs of subjects three months after our initial experiment. Specifically, we hypothesized that high-abundance alloreactive clones, which we presume to represent memory T cells due to their frequency in the peripheral T cell repertoire, should be stable over time and thus should remain in the alloreactive T cell compartment. Figure 4 presents the high-abundance T cell repertoire after three months in each pair of subjects. Many T cell clones identified as part of the high-abundance alloreactive T cell repertoire at baseline were observed in the high-abundance alloreactive T cell repertoire three months later, at similar clonal frequencies (Figure 4; average r2 = 0.78). To quantify similarity between sets of T cells, we calculated a TCR overlap metric (the proportion of T cells belonging to clones found in both samples) [29]. Table 3 presents the TCR overlap between duplicate cell culture experiments and between experiments spaced three months apart. While duplicate cell culture experiments generated more concordant sets of alloreactive T cell clones than experiments from different time-points, overlap between different time-points was nonetheless quite high (mean overlap = 0.97 for duplicate experiments vs. 0.87 across time-points). We hypothesize that the lower overlap over time might be due to the emergence of naïve T cell clones of exceptional size which would not be expected to persist in the periphery and/or the noise in the estimation of absolute cellular abundance could have caused a subset of low-abundance clones to be erroneously classified as high-abundance in our experiment [32]–[34].
Figure 4. T cell clonal frequency among temporal replicates of mixed lymphocyte culture.
Above are three scatter plots showing the number of T cells bearing each unique CDR3 sequence in replicate mixed lymphocyte culture experiments performed three months apart on each of three pairs of healthy adult subjects. Considering only T cell clones previously observed in a pre-MLR T cell sample from each time-point and enriched at least ten-fold after mixed lymphocyte culture, each point represents a unique T cell clone and points are plotted at (# of observed T cells +1) so that clones unobserved in one sample are plotted on the axes.
Table 3. TCR overlap between biological & temporal replicate mixed lymphocyte culture experiments.
| Biological replicates (N = 2) a | Temporal replicates (N = 4) b | |
| High-abundance pre-culture c | ||
| Subject 1 | 0.96 | 0.78 |
| Subject 2 | 0.98 | 0.93 |
| Subject 3 | 0.98 | 0.89 |
| Average | 0.97 | 0.87 |
| Low-abundance pre-culture d | ||
| Subject 1 | 0.54 | 0.15 |
| Subject 2 | 0.43 | 0.06 |
| Subject 3 | 0.67 | 0.08 |
| Average | 0.55 | 0.10 |
MLR Cultured in duplicate.
MLR performed at three months apart.
Present in pre-culture sample and ≥10× enriched after MLR.
Unobserved in pre-culture sample and ≥10 T cells after MLR.
The low-abundance alloreactive T cell clones, however, showed lower reproducibility between duplicate cell culture experiments (Table 3, bottom) and appeared to be considerably more transient. Comparisons between biological duplicates were much more concordant than comparisons between time-points (mean overlap = 0.55 for duplicate experiments vs. 0.10 across time-points). Several hypotheses may explain why T cell clones were not reproducibly found in the low-abundant alloreactive T cell compartment; first, the lower overlap between biological replicates is mostly due to sample error (most unique T cell lineages are at very low abundance, and we cannot reliably find a T cell clone in two biological replicates unless it is present in at least several cells); second, the even lower reproducibility after three months can be attributed to a preponderance of newly emerged naïve T cell clones among this subset; lastly, these clones may represent memory T cell populations that did not persist at detectable levels in the periphery over the intervening time [32]–[34].
Taken together, our TCR repertoire analysis is highly sensitive and reproducible. Further, our results indicated that a majority of the alloreactivity observed between three pairs of healthy adults was attributable to a set of several thousand T cell clones, present at reasonably high frequency in the peripheral T cell repertoire, whose alloreactive potential remained stable over at least several months. While we cannot conclusively demonstrate using TCR sequencing that the proliferating clones identified are specifically alloreactive, our screening algorithm (requiring a T cell clone to represent a 10× higher proportion of the proliferated than the fresh sample) should ensure that only a minimal number of nonspecifically-proliferating clones are identified. Likewise, we do not anticipate that our method will reliably identify all T cell clones which will react to and infiltrate an allograft. However, we expect neither tracking of some irrelevant T cell clones nor failure to track some genuinely alloreactive clones should compromise clinical utility so long as a sufficient number of truly alloreactive clones are also identified and tracked, providing a means of quantitating the host cellular immune response to the allograft.
We anticipate that application of our approach to transplantation could have a positive impact in the clinical management of patients. This is to be achieved by performing donor-specific MLR at transplant to pre-define the donor-reactive T cell repertoire, and then tracking their presence, abundance and dynamics in recipient primary tissues (e.g. peripheral blood, allograft biopsies, urine) during the post-transplant period. Such an approach will make this technology utilizable in both living donor and deceased donor transplants. We speculate that foreknowledge of the alloreactive T cell repertoire could thus be combined with post-transplant immune profiling in the recipient peripheral blood for non-invasive monitoring of cellular allograft rejection. Conversely, absence of the donor reactive clones from the post-transplant repertoire would indicate immune tolerance.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All TCR profiling data reported on in this manuscript are available from the Dryad data repository (http://www.datadryad.org; doi:10.5061/dryad.3f60c).
Funding Statement
The authors have no support or funding to report.
References
- 1. Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, et al. (2003) Heterologous immunity provides a potent barrier to transplantation tolerance. J Clin Invest 111: 1887–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brook MO, Wood KJ, Jones ND (2006) The impact of memory T cells on rejection and the induction of tolerance. Transplantation 82: 1–9. [DOI] [PubMed] [Google Scholar]
- 3. D'Orsogna LJ, Roelen DL, Doxiadis, II, Claas FH (2012) TCR cross-reactivity and allorecognition: new insights into the immunogenetics of allorecognition. Immunogenetics 64: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burrows SR, Khanna R, Silins SL, Moss DJ (1999) The influence of antiviral T-cell responses on the alloreactive repertoire. Immunol Today 20: 203–207. [DOI] [PubMed] [Google Scholar]
- 5. Welsh RM, Selin LK (2002) No one is naive: the significance of heterologous T-cell immunity. Nat Rev Immunol 2: 417–426. [DOI] [PubMed] [Google Scholar]
- 6. Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, et al. (2009) T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 31: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, et al. (2007) How a single T cell receptor recognizes both self and foreign MHC. Cell 129: 135–146. [DOI] [PubMed] [Google Scholar]
- 8. Ely LK, Burrows SR, Purcell AW, Rossjohn J, McCluskey J (2008) T-cells behaving badly: structural insights into alloreactivity and autoimmunity. Curr Opin Immunol 20: 575–580. [DOI] [PubMed] [Google Scholar]
- 9. Griesemer AD, Sorenson EC, Hardy MA (2010) The role of the thymus in tolerance. Transplantation 90: 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macedo C, Orkis EA, Popescu I, Elinoff BD, Zeevi A, et al. (2009) Contribution of naive and memory T-cell populations to the human alloimmune response. Am J Transplant 9: 2057–2066. [DOI] [PubMed] [Google Scholar]
- 11. Marrack P, Kappler J (1988) T cells can distinguish between allogeneic major histocompatibility complex products on different cell types. Nature 332: 840–843. [DOI] [PubMed] [Google Scholar]
- 12. Schild H, Rotzschke O, Kalbacher H, Rammensee HG (1990) Limit of T cell tolerance to self proteins by peptide presentation. Science 247: 1587–1589. [DOI] [PubMed] [Google Scholar]
- 13. Argaet VP, Schmidt CW, Burrows SR, Silins SL, Kurilla MG, et al. (1994) Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J Exp Med 180: 2335–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burrows SR, Khanna R, Burrows JM, Moss DJ (1994) An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTL) cross-reactive with a single Epstein-Barr virus CTL epitope: implications for graft-versus-host disease. J Exp Med 179: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burrows SR, Silins SL, Moss DJ, Khanna R, Misko IS, et al. (1995) T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J Exp Med 182: 1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gras S, Burrows SR, Kjer-Nielsen L, Clements CS, Liu YC, et al. (2009) The shaping of T cell receptor recognition by self-tolerance. Immunity 30: 193–203. [DOI] [PubMed] [Google Scholar]
- 17. Mifsud NA, Nguyen TH, Tait BD, Kotsimbos TC (2010) Quantitative and functional diversity of cross-reactive EBV-specific CD8+ T cells in a longitudinal study cohort of lung transplant recipients. Transplantation 90: 1439–1449. [DOI] [PubMed] [Google Scholar]
- 18. Amir AL, D'Orsogna LJ, Roelen DL, van Loenen MM, Hagedoorn RS, et al. (2010) Allo-HLA reactivity of virus-specific memory T cells is common. Blood 115: 3146–3157. [DOI] [PubMed] [Google Scholar]
- 19. Danziger-Isakov L, Cherkassky L, Siegel H, McManamon M, Kramer K, et al. (2010) Effects of influenza immunization on humoral and cellular alloreactivity in humans. Transplantation 89: 838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Orsogna LJ, Roelen DL, Doxiadis II, Claas FH (2011) Screening of viral specific T-cell lines for HLA alloreactivity prior to adoptive immunotherapy may prevent GvHD. Transpl Immunol 24: 141. [DOI] [PubMed] [Google Scholar]
- 21. D'Orsogna LJ, van Besouw NM, van der Meer-Prins EM, van der Pol P, Franke-van Dijk M, et al. (2011) Vaccine-induced allo-HLA-reactive memory T cells in a kidney transplantation candidate. Transplantation 91: 645–651. [DOI] [PubMed] [Google Scholar]
- 22. Wang T, Chen L, Ahmed E, Ma L, Yin D, et al. (2008) Prevention of allograft tolerance by bacterial infection with Listeria monocytogenes. J Immunol 180: 5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bain B, Lowenstein L (1964) Genetic Studies On The Mixed Leukocyte Reaction. Science 145: 1315–1316. [DOI] [PubMed] [Google Scholar]
- 24. Bain B, Vas MR, Lowenstein L (1964) The Development Of Large Immature Mononuclear Cells In Mixed Leukocyte Cultures. Blood 23: 108–116. [PubMed] [Google Scholar]
- 25. Levitsky J, Leventhal JR, Miller J, Huang X, Chen L, et al. (2012) Favorable effects of alemtuzumab on allospecific regulatory T-cell generation. Human Immunology 73: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levitsky J, Miller J, Huang X, Chandrasekaran D, Chen L, et al. (2013) Inhibitory Effects of Belatacept on Allospecific Regulatory T-Cell Generation in Humans. Transplantation 96: 689–696 610.1097/TP.1090b1013e31829f31607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mathew JM, Garcia-Morales RO, Carreno M, Jin Y, Fuller L, et al. (2003) Immune responses and their regulation by donor bone marrow cells in clinical organ transplantation. Transplant Immunology 11: 307–321. [DOI] [PubMed] [Google Scholar]
- 28. Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP (2004) IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics 20 Suppl 1: i379–385. [DOI] [PubMed] [Google Scholar]
- 29. Emerson RO, Sherwood AM, Rieder MJ, Guenthoer J, Williamson DW, et al. (2013) High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathol 231: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, et al. (2009) Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 114: 4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, et al. (2013) Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 4: 2680. [DOI] [PubMed] [Google Scholar]
- 32. Jenkins MK, Chu HH, McLachlan JB, Moon JJ (2010) On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol 28: 275–294. [DOI] [PubMed] [Google Scholar]
- 33. Jenkins MK, Moon JJ (2012) The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol 188: 4135–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Surh CD, Sprent J (2008) Homeostasis of naive and memory T cells. Immunity 29: 848–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All TCR profiling data reported on in this manuscript are available from the Dryad data repository (http://www.datadryad.org; doi:10.5061/dryad.3f60c).