Abstract
We have performed 50 independent molecular dynamics (MD) simulations to determine the effect of pseudophosphorylation mutants on the structural dynamics of smooth muscle myosin (SMM) regulatory light chain (RLC). We previously showed that the N-terminal phosphorylation domain of RLC simultaneously populates two structural states in equilibrium, closed and open, and that phosphorylation at S19 induces a modest shift toward the open state, which is sufficient to activate smooth muscle. However, it remains unknown why pseudophosphorylation mutants poorly mimic phosphorylation-induced activation of SMM. We performed MD simulations of unphosphorylated, phosphorylated, and three pseudophosphorylatedRLC mutants: S19E, T18D/S19D and T18E/S19E. We found that the S19E mutation does not shift the equilibrium toward the open state, indicating that simple charge replacement at position S19 does not mimic the activating effect of phosphorylation, providing a structural explanation for previously published functional data. In contrast, mutants T18D/S19D and T18E/S19E shift the equilibrium toward the open structure and partially activate in vitro motility, further supporting the model that an increase in the mol fraction of the open state is coupled to SMM motility. Structural analyses of the doubly-charged pseudophosphorylation mutants suggest that alterations in an interdomain salt bridge between residues R4 and D100 results in impaired signal transmission from RLC to the catalytic domain of SMM, which explains the low ATPase activity of these mutants. Our results demonstrate that phosphorylation produces a unique structural balance in the RLC. These observations have important implications for our understanding of the structural aspects of activation and force potentiation in smooth and striated muscle.
Keywords: Smooth muscle myosin, striated muscle, regulatory light chain, phosphorylation, pseudophosphorylation, molecular dynamics simulations
Introduction
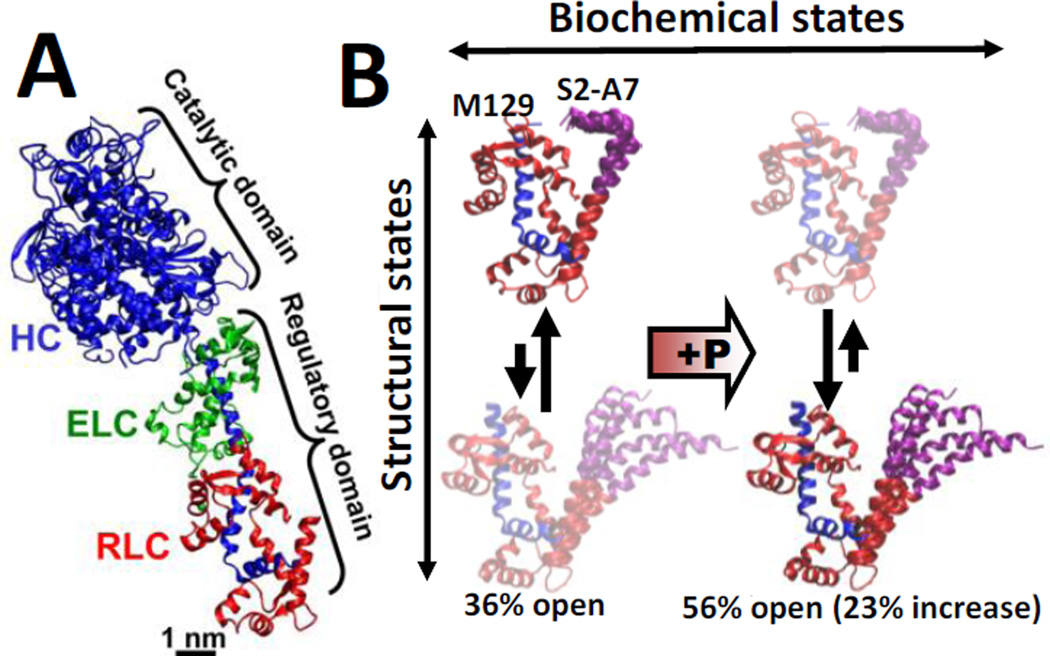
Smooth muscle myosin (SMM) is a member of the myosin super family of motor proteins, which use chemical energy from ATP hydrolysis to perform mechanical work on act in. Activation of smooth muscle requires phosphorylation of the regulatory light chain (RLC), which is distant from the myosin active site on the catalytic domain(Fig. 1A). Unphosphorylated SMM is auto-inhibited by interactions between the two catalytic domains1, 2 that are relieved by RLC phosphorylation3, 4, through a mechanism that is proposed to involve interactions among the heavy chain, the essential light chain (ELC), and the RLC5, 6.
Fig. 1.
(A) Crystal structure of myosin S1, with heavy chain (HC, blue), essential light chain (ELC, green), and regulatory light chain (RLC, red). (B) Structural model of phosphorylation-induced structural changes within the RLC in solution, based on FRET experiments and MD simulations (Ref. 10). Left (unphosphorylated RLC): the closed (top) and open (bottom) states are in equilibrium, with the more helical open state of PD (purple) being more populated. Right: Upon phosphorylation, the structure shifts toward the open state, activating smooth muscle.
Our previous EPR experiments7 and molecular dynamics (MD) simulations8, 9 showed that upon phosphorylation of S19, the 25-residue phosphorylation domain (PD) of RLC undergoes a disorder-to-order transition accompanied by an increase in helical structure. Complementary FRET and MD simulations showed that the biochemical and structural states of the RLC are loosely coupled: the PD of the RLC simultaneously populates two structural states, closed and open, in both unphosphorylated and phosphorylated biochemical states10. This complementary approach revealed that unphosphorylated RLC preferably populates a closed conformation, and that phosphorylation shifts the conformational equilibrium toward the open state, increasing its mol fraction by 23%10. Based on these observations, we proposed a model in which a modest change in the conformational equilibrium of RLC is sufficient and necessary to activate smooth muscle(Fig. 1B).
These complementary studies suggest that incorporation of negative charge into the phosphorylation sites of the RLC should be sufficient increase the mol fraction of the open structural state, resulting in SMM activation. However, experiments have shown that simple charge replacement or deletion of PD causes partial or no activation11, 12, suggesting that full activation requires specific structural changes within PD. In particular, functional assays have shown that pseudophosphorylation mutants have minimal or no effect on SMM activation12. For example, mutant S19E was studied to determine the effect of simple charge replacement in the PD; functional assays performed on this mutant showed that S19E does not activate ATPase activity or motility of SMM12. Biochemical experiments also showed that pseudophosphorylation mutants T18D/S19D and T18E/S19Epoorlymimic phosphorylation-inducedATPase activation, although they retain in vitro motility12. Only in the case of T18E/S19Ewasthere both a slight activation of ATPase activity and a moderate activation of motility12.
These observations suggest that a structural shift of theRLC toward the open state is required for smooth muscle activation, and that pseudo-phosphorylation mutants do not induce this structural shift. To test this hypothesis, we have performed 50 independent 0.2-µs MD simulations to determine the effect of three pseudophosphorylation mutants(S19E, T18D/S19D and T18E/S19E) on the structural dynamics of the RLC.
Methods
Preparation of the systems
The structure of the RLC bound to the IQ motif of myosin heavy chain was obtained by homology modeling using the procedure described previously10.Unphosphorylated, phosphorylated (at position S19) and pseudophosphorylationmutants S19E, T18D/S19D and T18E/S19E were modeled using the PSFGEN utility of NAMD 2.913. Phosphate was assigned a charge of −2, based on pKa = 6.5 for phosphoserine and pH > 7 in smooth muscle14, and previous time-resolved FRET experiments10. We adjusted the side-chain ionization states of the RLC to pH = 7.0 with PROPKA15. The system was embedded in TIP3P water boxes with a minimum distance of 2 nm between the protein and the edges of the periodic box. Na+ and Cl− ions were added to neutralize charge and produce an ion concentration of approximately 150 mM, as in experiments. The final systems contained ~100,000 atoms. Topologies and parameters were used according to the CHARMM22 force field with CMAP correction16, 17.
Molecular dynamics protocol
MD simulations were performed by using the program NAMD 2.913. Periodic boundary conditions18, particle mesh Ewald19, 20, a nonbonded cutoff of 9 Å and a 2 fs time step were used. The NPT ensemble was maintained with a Langevin thermostat (310K) and a Langevin piston barostat (1 atm). The system was first subjected to energy minimization for 1000 steps, followed by a warming up period for 200 ps. This procedure was followed by equilibration for 0.01 µs with backbone atoms harmonically restrained using a force constant of 100 kcal mol−1 nm−2. To improve conformational sampling, we performed ten independent MD simulations for each system, with randomly selected initial atomic velocities21. Ten independent simulations of unphosphorylated, phosphorylated and pseudophosphorylation mutants were continued for 0.2 µs, for a total of 50 MD simulations and acumulative simulation time of 10µs.
Analysis and visualization
VMD22 was used for analysis, visualization, and rendering of the structures. STRIDE23 was used to analyze the evolution of the secondary structure of the PD in unphosphorylated, phosphorylated and four pseudophosphorylation mutants. STRIDE recognizes secondary structural elements in proteins from their atomic coordinates. It utilizes both hydrogen-bond energy and main chain dihedral angles to define the secondary structure pattern, relying on database-derived recognition parameters with the crystallographers’ secondary structure definitions as a standard-of-truth.
We calculated Cα-Cα distances between residues S2, K3 and A7 of PD and M129 of the C-terminal lobe of the RLCcore (Fig. 1B). The distances for each set of ten independent MD simulations were then combined into a single histogram. Finally, each histogram was fitted to one-Gaussian and two-Gaussian distance distributions ρ(R). We found that the best fits, indicated by a lower χ2 (Table S1, Supplementary material), were consistently obtained for the two-Gaussian model:
Each fit yielded six independent parameters of ρ(R): centers for open (j=1) and closed (j=2) structural states, R1 and R2, widths FWHM1 and FWHM2, and mol fractions X1 and X2.
Results and discussion
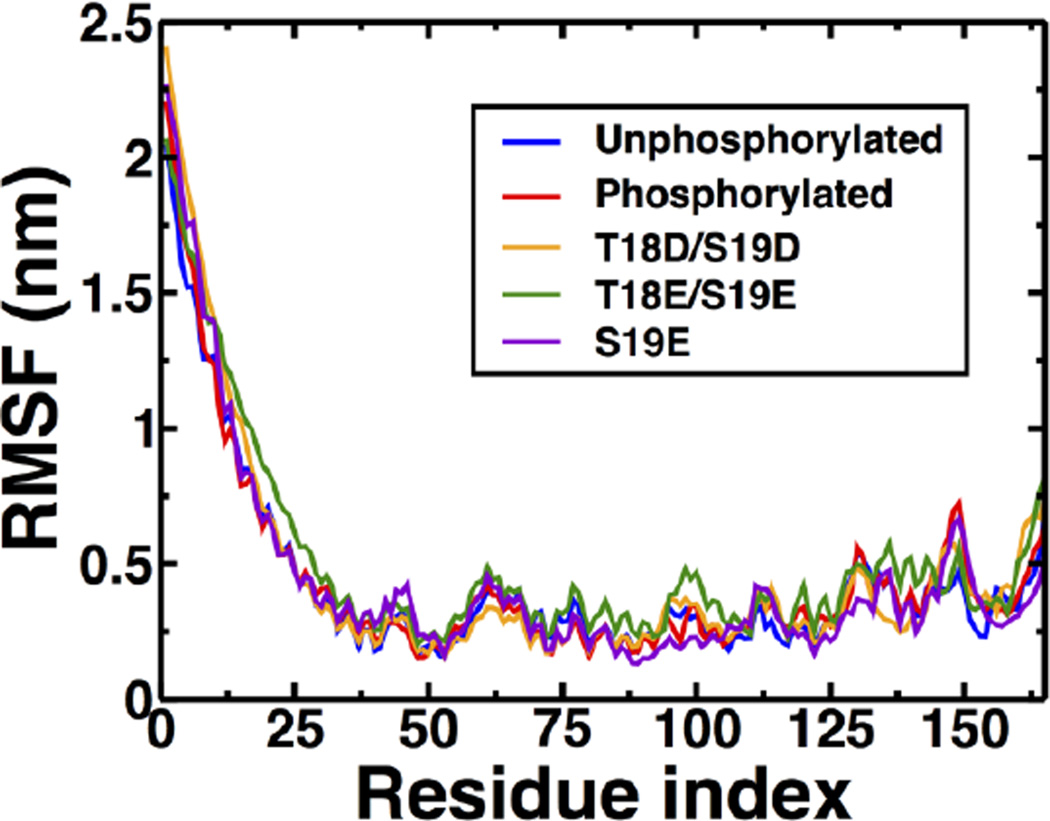
We previously showed that phosphorylation of PD at S19 does not induce noticeable global structural changes on the RLC; instead, phosphorylation induces structural changes only around residue K11 of the PD10. To determine whether pseudophosphorylation mutants alter the structural stability of the RLC, PD, or both, we calculated the Cαroot-mean square fluctuations (RMSF) averaged over all individual MD simulations (Fig. 2). Analysis of the RMSF plots revealed negligible differences between unphosphosphorylated, phosphorylated and pseudophosphorylated RLC. For example, variable residue mobility is observed in the N- and C-termini of the RLC. Despite these small differences, the RMSF patterns are similar for phosphorylated and pseudophosphorylated RLC, indicating that phosphorylation and pseudophosphorylation mutants do not alter the global structural stability of the RLC. Furthermore, RMSF plots indicate that the 24-residue PD is very flexible in phosphorylated and unphosphorylated RLC (i.e., maximum RMSF values of 2.4 nm), in agreement with a previous study10.This suggests that the effects of pseudophosphorylation mutants on the structural dynamics of the RLC are localized to the PD.
Fig. 2.
Root-mean-square fluctuations (RMSF) of the RLC. RMSF values were calculated about the average position of Cα atoms. Each plot represents the average RMSF of ten independent MD trajectories.
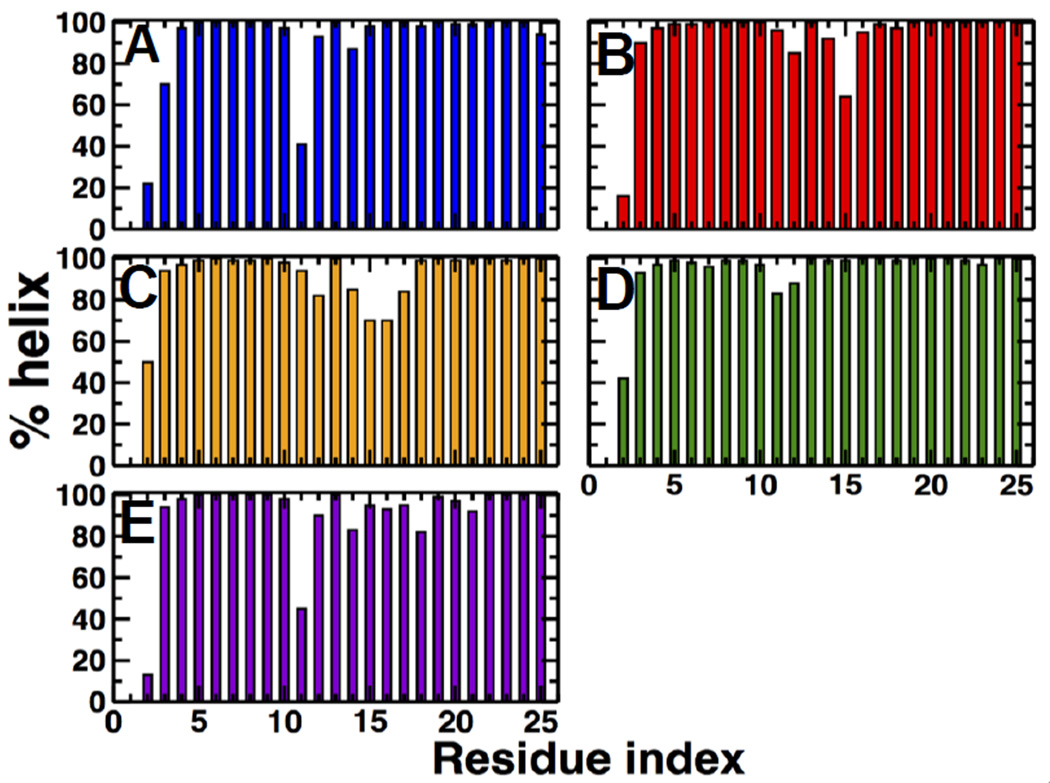
Because RMSF plots do not directly indicate secondary structure, we calculated the percentage of helical content for each individual residue in PD(Fig. 3).In agreement with previous MD simulations, we found that helical periodicity of unphosphorylated PD is disrupted at residue K11, and phosphorylation increases the helical content of this residue by ~60%8–10.Analysis of mutant S19E revealed a secondary structure pattern was similar to that of unphosphorylated RLC, indicating that this mutant does not induce a disorder-to-order transition of PD. On the other hand, mutants T18D/S19D and T18E/S19E restore the helical periodicity of PD at position K11 in a similar manner as phosphorylation. These observations indicate that, except for S19E, pseudophosphorylation mimics the local disorder-to-order transition of PD.These observations also indicate that, if present, the structural effect of pseudophosphorylation is localized around residue K11, which suggests that the inability of pseudophosphorylation to optimally activate SMM does not result from global structural alterations in the RLC.
Fig. 3. Percentage of time that each residue of the PD spends an in α-helical conformation.
(A) unphosphorylated, (B) phosphorylated, (C) T18D/S19D,(D) T18E/S19E, and (E) S19E.
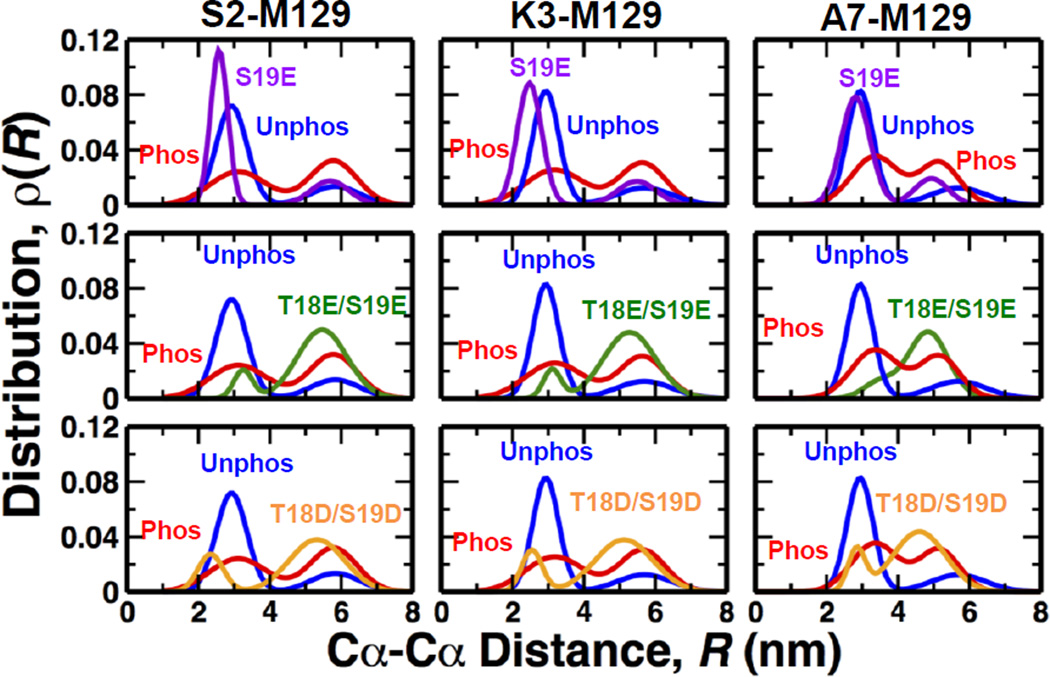
We calculated the distributions of distances from residues in PD(S2, K3 and A7) to M129 in the C-terminal lobe of theRLC (Fig. 4), because these are the sites previously labeled in FRET experiments10. As shown in Table S1 (Supporting Material), a two-Gaussian distribution best fits the data in all cases, indicating two clearly resolved structural states: with a short distance assigned to the closed state and a longer distance assigned to the open state. Table 1 shows the mol fraction in the open state, compared with previous experimental results. Phosphorylation shifts the distribution by 23% toward the open state, in excellent agreement with previous time-resolved FRET experiments10. These findings indicate that although MD simulations were not performed on the entire regulatory domain and were performed in the sub microsecond time scale, the MD simulations capture the structural dynamics of RLC observed in experiments10. These observations are also consistent with a previous study showing that 5–10 independent MD simulations are sufficient to capture the structural properties of a protein observed in experiments21. In contrast, pseudophosphorylation gave results substantially different from that of phosphorylation: S19E showed no shift at all toward the open state, while the other mutants showed shifts much greater than that of phosphorylation (Fig. 4, Table 1). Thus simple charge replacement does not explain the structural effects of phosphorylation, and this is consistent with the observation that the functional effects are also not equivalent (Table 1).
Fig. 4. Comparison of the distance distributions of unphosphorylated (blue) and phosphorylated (red, pS19) RLC with those of pseudophosphorylation mutants.
Each distance distribution, determined for each Cα-Cα distance between S2, K3, A7 and M129, is the best 2-Gaussian fit to the average of histograms from 10 trajectories.
Table 1.
Summary of structural and functional data on RLC
| RLC species | Mol fraction in open state1 |
Increase of mol fraction in open state2 |
Relative motility3 | Relative actin- activated ATPase activity3 |
|---|---|---|---|---|
| Unphosphorylated | 0.25 (0.33)1 | − | 0.00 ± .02 | 0.02 ± .01 |
| Phosphorylated | 0.48 (0.56)1 | 0.23 (0.23)1 | 1.00 | 1.00 |
| S19E | 0.25 | 0 | 0.00 ± .02 | 0.00 ± .01 |
| T18E/S19E | 0.88 | 0.63 | 0.45 ± .03 | 0.16 ± .01 |
| T18D/S19D | 0.79 | 0.54 | 0.59 ± .02 | 0.0 ± .01 |
Functional experiments have shown that that, except for mutant S19E, pseudophosphorylation partially restores SMM motility in vitro12. Our simulations showed that only mutant S19E is unable to induce a shift toward the open state (Table 1); this evidence suggests a relationship between an increase in the mol fraction and motility. Indeed, we found a relationship between the mol fraction of the open state of the RLC and in vitro motility of SMM. This linear relationship indicates that an increase in mol fraction of the open state and motility are coupled. These observations suggest that increasing the mol fraction of the open state is sufficient to partially activate motility of SMM.
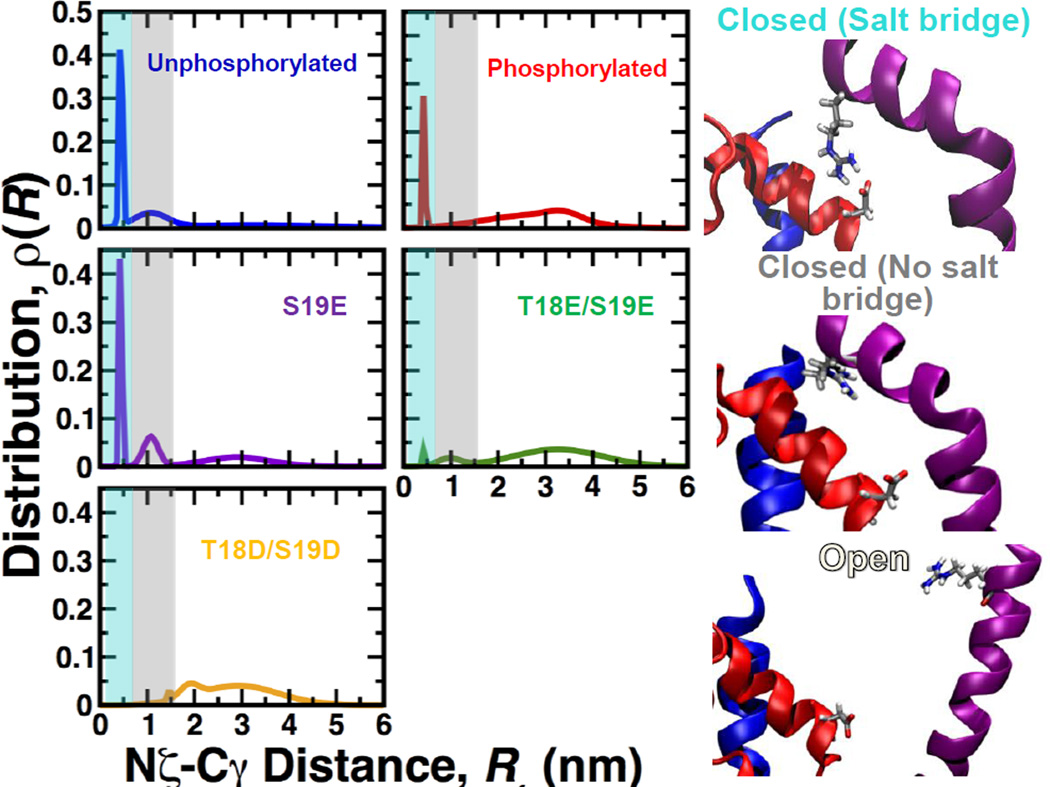
We found that the inability of mutant S19E to shift the structure of the RLC toward the open state correlate with in vitro assays showing that this mutant does not activate ATPase activity12.MutantsT18D/S19D and T18E/S19E substantially increase the mol fraction of the open structural state, sometimes even more effectively than phosphorylation. However, functional assays showed that these mutants poorly mimic phosphorylation-induced act in-activated ATPase activity of SMM. If T18D/S19D and T18E/S19E can effectively shift the equilibrium toward the open state and increase motility, why are they unable to optimally regulate ATPase activation? We have previously shown that an interdomain salt bridge between R4 (PD) and D100 (C-lobe) plays a role in the stabilization the closed state in both unphosphorylated and phosphorylated RLC10. The presence of this PD-to-C-lobe salt bridge in both phosphorylated and unphosphorylated RLC indicates that the R4-D100 salt bridge serves as an anchor to properly position PD onto the C-terminal lobe in the closed state. To evaluate the presence of this salt bridge, we calculated the distance distribution between atoms Nζ of R4 and Cγ of D100 (Fig. 5). We found that unphosphorylated RLC features three peaks, which correspond to the closed state stabilized by salt bridge R4-D100 (closed(SB), Rmean=0.4 nm), a closed state not stabilized by a salt bridge (closed(noSB), Rmean=1.1 nm) and an open state (Open, Rmean=3.3 nm) (Fig. 5, right). In phosphorylated RLC, the peak representing closed(SB) is still present, but the closed(noSB)state is no longer present. The distance distribution of S19E is very similar to that of unphosphorylated RLC, explaining the absence of ATPase activity12. Among the mutants that increase the fraction of the open state, only T18E/S19Eretains features of both unphosphorylated and phosphorylated RLC: two small peaks corresponding to closed(SB) and closed(noSB),and broad open states, characteristic of dynamic tertiary structure (Fig. 5). This pattern is completely disrupted in mutant T18D/S19D.
Fig. 5. Distance distribution between atoms Nζ of R4 and Cγ of D100.
Distance distributions are the average of histograms from the 10 trajectories combined for the five RLC species. Distance distributions were constructed by fitting distance histograms to a 3-Gaussian model. Right: Typical structural details. Closed(SB) is the closed state stabilized by a salt bridge between R4 and D100. Closed(no SB) is the closed state that is not stabilized by this salt bridge. Open is the open state, which also lacks the salt bridge. PD, C-terminal lobe and HC are colored in purple, red and blue, respectively. The shaded areas in the distribution plots indicate the presence of the closed(SB) (cyan), closed(noSB) (grey) and open (white) states.
These results suggest that upon phosphorylation, only the closed state stabilized by the salt bridge R4-D100 and the open states are present in equilibrium. The presence of a closed(noSB)population in unphosphorylated RLC, and its absence in phosphorylated RLC, indicates that the closed(noSB)conformation is probably responsible for inhibition of ATPase activity. We found that the state closed(SB) is present in both phosphorylated and unphosphorylated RLC;the proximity of the C-lobe of the RLC to the IQ and ELC(Fig. 1A) suggests that the salt bridge R4-D100 is necessary for the correct placement of the highly mobile PD onto the C-terminal lobe of the RLC during the open-to-closed transition. Because salt bridge R4-D100 is still partially present in phosphorylated RLC, it is likely that the alignment of PD and C-terminal lobe facilitated by this salt bridge is required for signal transmission from the RLCtoward the catalytic domain via the ELC. It is possible that these differences are linked to the unique physicochemical properties of phosphoserine. For instance, it has been shown that salt bridge interactions are stronger for phosphoserine than for acidic residues24, 25. In addition, phosphoserine has a significantly higher probability than glutamate to form hydrogen bonds with arginein and lysine25. Stronger electrostatic interactions in the presence of phosphoserine might also impose additional structural restraints necessary for the correct placement of the PD domain onto the C-lobe (i.e., via pS19-R16 salt bridges9). We propose that the absence of the salt bridge R4-D100 in mutant T18D/T19D results in an impaired signal transmission from the RLC to the catalytic domain of SMM through the ELC5, which explains the low ATPase activity of these mutants12. Conversely, partial activation of ATPase activity of T18E/S19E probably results from a shift toward the open state, the presence of a small fraction of the closed(SB)state, and the presence of inhibitory closed(noSB) state (Fig. 5).
In conclusion, we found that simple charge replacement at position S19 is not sufficient to induce the structural changes necessary for activation of both ATPase activity and motility of SMM. We also found that pseudophosphorylation mutants that induce a structural shift toward the open state can also partially or fully mimic the motility observed with phosphorylated RLC. This finding suggests that an increase in the mol fraction of the open state and motility in SMM are coupled. Structural analyses of pseudophosphorylation mutants T18D/S19D and T18E/S19E suggest that alterations of an interdomain salt bridge between residues R4 and D100 result in impaired signal transmission from the RLC to the catalytic domain of SMM, which explains the low of ATPase activity of these mutants. Our results demonstrate that phosphorylation produces a unique conformational balance in the RLC, which plays a key role in smooth muscle activation. Our findings regarding the structural landscape of the RLC that is vital for activation reach beyond smooth muscle to different muscle types and species, including the residues we have investigated, where the RLC sequence remains quite conserved. RLCs are very powerful governors of motile effects, underscored by the striking capability of smooth muscle RLC to regulate and modulate force generation characteristic of smooth muscle, even when it is bound to the IQ motif of striated myosin12. Taken together, these findings are not only relevant to smooth muscle, but also to other muscle types. For instance, mono- and biphosphorylation of RLC in tarantula thick filaments from striated muscle are important for activation (i.e., destabilization of the super-relaxed state26) and force potentiation, respectively. These observations have important implications for our understanding of the structural aspects of activation and force potentiation in smooth and striated muscle.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matt Mauseth for insightful discussions. This work was supported by grants to L.M.E-F. from the American Heart Association (10POST4350076) and to D.D.T from NIH (GM27906). This project made extensive use of the outstanding high-performance computing resources at the University of Minnesota Supercomputing Institute.
REFERENCES
- 1.Liu J, Wendt T, Taylor D, Taylor K. J Mol Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- 2.Wendt T, Taylor D, Trybus KM, Taylor K. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellison PA, Sellers JR, Cremo CR. J Biol Chem. 2000;275:15142–15151. doi: 10.1074/jbc.275.20.15142. [DOI] [PubMed] [Google Scholar]
- 4.Sellers JR. J Biol Chem. 1985;260:15815–15819. [PubMed] [Google Scholar]
- 5.Ni S, Hong F, Haldeman BD, Baker JE, Facemyer KC, Cremo CR. J Biol Chem. 2012;287:22068–22079. doi: 10.1074/jbc.M112.343491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor KA, Feig M, Brooks CL, 3rd, Fagnant PM, Lowey S, Trybus KM. Journal of structural biology. 2014;185:375–382. doi: 10.1016/j.jsb.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson WD, Blakely SE, Nesmelov YE, Thomas DD. Proc Natl Acad Sci U S A. 2005;102:4000–4005. doi: 10.1073/pnas.0401664102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinoza-Fonseca LM, Kast D, Thomas DD. Biophys J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinoza-Fonseca LM, Kast D, Thomas DD. J Am Chem Soc. 2008;130:12208–12209. doi: 10.1021/ja803143g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Proc Natl Acad Sci U S A. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikebe M, Ikebe R, Kamisoyama H, Reardon S, Schwonek JP, Sanders CR, 2nd, Matsuura M. The Journal of biological chemistry. 1994;269:28173–28180. [PubMed] [Google Scholar]
- 12.Sweeney HL, Yang Z, Zhi G, Stull JT, Trybus KM. Proc Natl Acad Sci U S A. 1994;91:1490–1494. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aickin CC. J Physiol. 1994;479(Pt 2):331–340. doi: 10.1113/jphysiol.1994.sp020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Robertson AD, Jensen JH. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 16.MacKerell J, Bashford ADD, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher I, Roux WEB, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 17.MacKerell AD, Jr, Feig M, Brooks CL., 3rd J Am Chem Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 18.Weber W, Hünenberger PH, McCammon JA. J Phys Chem B. 2000;104:3668–3675. [Google Scholar]
- 19.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 20.Essmann U, Perera L, Berkowitz ML. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 21.Day R, Daggett V. Proc Natl Acad Sci U S A. 2005;102:13445–13450. doi: 10.1073/pnas.0501773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.Frishman D, Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 24.Mandell DJ, Chorny I, Groban ES, Wong SE, Levine E, Rapp CS, Jacobson MP. J Am Chem Soc. 2007;129:820–827. doi: 10.1021/ja063019w. [DOI] [PubMed] [Google Scholar]
- 25.Rapp C, Klerman H, Levine E, McClendon CL. PLoS One. 2013;8:e57804. doi: 10.1371/journal.pone.0057804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Proc Natl Acad Sci U S A. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.