Abstract
Pseudomonas aeruginosa ParA belongs to a large subfamily of Walker-type ATPases acting as partitioning proteins in bacteria. ParA has the ability to both self-associate and interact with its partner ParB. Analysis of the deletion mutants defined the part of the protein involved in dimerization and interactions with ParB. Here, a set of ParA alanine substitution mutants in the region between E67 and L85 was created and analysed in vivo and in vitro. All mutants impaired in dimerization (substitutions at positions M74, H79, Y82 and L84) were also defective in interactions with ParB, suggesting that ParA–ParB interactions depend on the ability of ParA to dimerize. Mutants with alanine substitutions at positions E67, C68, L70, E72, F76, Q83 and L85 were not impaired in dimerization, but were defective in interactions with ParB. The dimerization interface partly overlapped the pseudo-hairpin, involved in interactions with ParB. ParA mutant derivatives tested in vitro showed no defects in ATPase activity. Two parA alleles (parA84, whose product can neither self-interact nor interact with ParB, and parA67, whose product is impaired in interactions with ParB, but not in dimerization) were introduced into the P. aeruginosa chromosome by homologous gene exchange. Both mutants showed defective separation of ParB foci, but to different extents. Only PAO1161 parA84 was visibly impaired in terms of chromosome segregation, growth rate and motility, similar to a parA-null mutant.
Introduction
The faithful segregation of low-copy-number plasmids depends on the plasmid-specific partition complex of two Par proteins and a centromere-like sequence parS. Despite the variety of plasmid partitioning systems, there are common features: component A, an NTPase (Walker-type ATPase, actin-type ATPase, tubulin-like GTPase), forms the dynamic scaffold for plasmid movement to the progeny cells (Ah-Seng et al., 2013; Vecchiarelli et al., 2010), whereas component B, a DNA-binding protein, recognizes and binds parS, forming the segregating unit – the segrosome (Barillà & Hayes, 2003; Gerdes et al., 2010; Pratto et al., 2008). Interactions between these two proteins stimulate NTP hydrolysis and lead the movement of segrosomes over the nucleoid (Gerdes et al., 2010).
The homologues of class I plasmidic Par proteins, encoded on bacterial chromosomes in close proximity to the origin of replication, form their own subgroups (Gerdes et al., 2000; Livny et al., 2007; Ringgaard et al., 2011; Yamaichi & Niki, 2000). Chromosomal ParA proteins belong to the subgroup of P-loop ATPases with a deviant Walker A motif (Koonin, 1993; Motallebi-Veshareh et al., 1990) without N-terminal DNA-binding domains (Fig. 1). Their partners, DNA-binding proteins with an HTH motif in the central part, belong to the highly conserved family of ParB proteins. Chromosomally encoded ParA and ParB homologues can mimic their plasmid counterparts in stabilizing otherwise unstable plasmids (Bartosik et al., 2004; Lin & Grossman, 1998; Yamaichi & Niki, 2000). The importance of chromosomal Par systems in condensation and segregation of oriC domains of newly replicated chromosomes prior to cell division has been confirmed (Bartosik et al., 2009; Figge et al., 2003; Fogel & Waldor, 2006; Gruber & Errington, 2009; Lasocki et al., 2007; Ogura et al., 2003; Ptacin et al., 2010; Schofield et al., 2010; Sullivan et al., 2009; Umbarger et al., 2011).
Fig. 1.
Comparison of the best-studied chromosomal ParA family members. Amino acids similar in at least four proteins are marked by a black background, those similar in three are marked by a dark grey background and homologous residues in two proteins are marked by a light grey background. Conserved ATP-binding Walker A, A′ and B (III) motifs (Koonin, 1993) and motif C (IV) characteristic of ParA-like proteins are indicated. The P. aeruginosa ParA region defined by deletion mapping is shown by a thin line above the sequence and the region analysed by alanine scanning is shown by a dark grey box. Pa, Pseudomonas aeruginosa; Pp, Pseudomonas putida; Cc, Caulobacter crescentus; Bs, Bacillus subtilis; Tt, Thermus thermophilus.
Apart from the main function in the separation of oriC domains during chromosome segregation, ParA–ParB proteins participate in the control of replication initiation, cell division, growth and motility (Mierzejewska & Jagura-Burdzy, 2012). These additional roles are species-dependent and seem to depend on their ability to interact with other proteins (Bowman et al., 2008; Gruber & Errington, 2009; Ptacin et al., 2010; Ringgaard et al., 2011; Sullivan et al., 2009; Yamaichi et al., 2012).
The object of our investigations was the par system of Pseudomonas aeruginosa consisting of ParA, ParB and 10 potential parS sites, most of them localized in close proximity to the oriC region (Bartosik et al., 2004). ParB creates one to four regularly distributed foci in P. aeruginosa cells, which co-localize with the nucleoid and undergo dynamic changes (Bartosik et al., 2009). When in excess, ParB is able to silence the expression of genes placed on the test plasmid near parS. Overproduction of ParA and ParB in P. aeruginosa causes growth inhibition and defects in chromosome segregation (Bartosik et al., 2004; Lasocki et al., 2007). The parA and parB genes are not essential for P. aeruginosa viability, but the chromosomal par mutants show defects in growth, chromosome partitioning and motility. Both proteins interact with each other, forming complexes that are protected from proteolytic digestion (Bartosik et al., 2009; Lasocki et al., 2007). Molecular characterization of P. aeruginosa ParB revealed its domain structure (Bartosik, et al., 2004, 2009). The central part of ParB is involved in DNA-binding activity not only through the HTH motif, but also through an additional DNA-binding interface (Kusiak et al., 2011). The C terminus of ParB plays an essential role in self-association (primary dimerization domain) with the vital role of hydrophobic residues at the C terminus of the protein (Bartosik et al., 2004; Mierzejewska et al., 2012), whereas the N-terminal part of ParB is involved in oligomerization of the protein (Kusiak et al., 2011).
Previous analysis of P. aeruginosa ParA revealed that this protein has the ability to both self-associate and interact with ParB (Bartosik et al., 2004). This study was aimed at the dissection of ParA from P. aeruginosa and identification of the dimerization interface as well as the domains involved in interactions with the ParB partner.
Methods
Bacterial and yeast strains, and growth conditions.
The Escherichia coli strains used were: DH5α [F− (ϕ80dlacZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17(rk−mk+) supE44 relA1 deoR Δ(lacZYA–argF)U196], BTH101 [F− cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1] (Karimova et al., 1998), BL21 [F− ompT hsdSB (rB−mB−) gal dcm (λ DE3)] (Novagen) and S17-1 [recA pro hsdR RP4-2-Tc : : Mu-Km : : Tn7] (Simon et al., 1986). P. aeruginosa PAO1161 (leu− r−m+) was kindly provided by B. M. Holloway (Monash University, Australia). PAO1161 RifR derivative was used as a recipient strain in conjugation. Saccharomyces cerevisiae strain L40 [MATa trp1 leu2 his3 ade2 LYS : : lexA–HIS3 URA3 : : lexA–lacZ] was provided by Clontech.
Bacteria were grown in L-broth (Kahn et al., 1979) at 37 °C. Some experiments were performed in M9 minimal medium with glucose (Sambrook et al., 1989) supplemented with leucine (132 mM) for propagation of PAO1161 derivative strains. L-agar (L-broth with 1.5 % w/v agar) was supplemented with antibiotics at appropriate concentrations. For E. coli strains, benzyl penicillin sodium salt (150 µg ml−1 in liquid media and 300 µg ml−1 in agar plates), kanamycin sulfate (50 µg ml−1) or chloramphenicol (10 µg ml−1) were added. For P. aeruginosa strains, carbenicillin (300 µg ml−1) and rifampicin (300 µg ml−1) were applied. L-agar used for blue/white screening contained 0.1 mM IPTG and 40 µg X-Gal ml−1. MacConkey agar was supplemented with 1 % maltose, antibiotics and 0.1 mM IPTG.
Plasmid DNA isolation, analysis, DNA amplification and manipulation.
Plasmid DNA was isolated and manipulated by standard procedures (Sambrook et al., 1989). Standard PCRs (Mullis et al., 1986) were performed with the appropriate pairs of primers listed in Table S1 (available in the online Supplementary Material). E. coli competent cell preparation and DNA transformation were performed according to standard protocols (Sambrook et al., 1989). The fidelity of PCR-derived clones was checked by DNA sequencing (DNA Sequencing and Oligonucleotide Synthesis Laboratory, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland). Plasmids used in this study are listed in Tables 1 and 2. Construction of modified parA alleles is described in detail in the Supplementary Materials and Methods. Requests for strains and plasmids constructed in this work should be addressed to the corresponding author.
Table 1. Plasmids used in this work.
| Plasmid | Relevant features | Reference/source |
| pAKE600 | oriMB1, oriTRK2, ApR, sacB | El-Sayed et al. (2001) |
| pAMB9.37 | pBBRMCS-1 expression vector, lacIq tacp | Ludwiczak et al. (2013) |
| pBBR1MCS-1 | IncA/C broad-host-range cloning vector, lacZα–MCS, mob, T7p, T3p, CmR | Kovach et al. (1994) |
| pBGS18 | oriMB1, KmR, cloning vector | Spratt et al. (1986) |
| pBTM116 | oriMB1, 2μ, ApR, trp1, shuttle vector lexABD | Clontech |
| pET28a(+) | oriMB1, KmR, T7p, lacO, His6-tag, T7 tag, expression vector | Novagen |
| pET28mod | oriMB1, KmR, T7p, lacO, His6-tag, modified to remove T7 tag | Lukaszewicz et al. (2002) |
| pGAD424 | oriMB1, 2μ, ApR, leu2, shuttle vector gal4AD | Clontech |
| pKLB1.4 | pGAD424 with gal4AD–parA translational fusion | Bartosik et al. (2004) |
| pKLB1.6 | pBTM116 with lexADB–parA translational fusion | Bartosik et al. (2004) |
| pKLB2.4 | pGAD424 with gal4AD–parB translational fusion | Bartosik et al. (2004) |
| pKLB2.6 | pBTM116 with lexADB–parB translational fusion | Bartosik et al. (2004) |
| pKLB28 | pET28mod with T7p–parB transcriptional fusion | Bartosik et al. (2004) |
| pKLB60.1 | pAKE600 derivative lacking BamHI site | Lasocki et al. (2007) |
| pKLB60.2 | pAKE600 derivative with parA | Lasocki et al. (2007) |
| pKLB8.1 | pET28mod with T7p–parA transcriptional fusion | Lasocki et al. (2007) |
| pKNT25 | orip15, KmR, lacp–MCS–cyaT25 | Karimova et al. (1998) |
| pKT25 | orip15, KmR, lacp–cyaT25–MCS | Karimova et al. (1998) |
| pKT25-zip | pKT25 with lacp–cyaT25–GCN4 leucine zipper fragment | Karimova et al. (1998) |
| pLKB2 | pKT25 with modified MCS | L. Kusiak* |
| pLKB220 | pLKB2 with translationally fused cyaT25–parA | L. Kusiak |
| pLKB233 | pLKB2 with translationally fused cyaT25–parB | L. Kusiak |
| pLKB4 | pUT18C derivative with modified MCS | L. Kusiak |
| pMKB5.1 | pLKB4 with cyaT18–parA translational fusion | M. Kusiak* |
| pMKB5.2 | pLKB4 with cyaT18–parB translational fusion | M. Kusiak |
| pMKB5.3 | pUT18 with parA–cyaT18 translational fusion | M. Kusiak |
| pMKB5.4 | pUT18 with parB–cyaT18 translational fusion | M. Kusiak |
| pMKB6.1 | pKNT25 with parA–cyaT25 translational fusion | M. Kusiak |
| pMKB6.2 | pKNT25 with parB–cyaT25 translational fusion | M. Kusiak |
| pUC18 | oriMB1, ApR, cloning vector | Yanisch-Perron et al. (1985) |
| pUT18 | oriColE1, ApR, lacp–MCS–cyaT18 | Karimova et al. (1998) |
| pUT18C | oriColE1, ApR, lacp–cyaT18–MCS | Karimova et al. (1998) |
| pUT18C-zip | pUT18C with lacp–cyaT18–GCN4 leucine zipper fragment | Karimova et al. (1998) |
MCS, multiple cloning site.
Institute of Biochemistry and Biophysics, Polish Academy of Sciences.
Table 2. Plasmids constructed during this work.
| Plasmid | Relevant features |
| pAKE600 derivatives | |
| pKGB6.67 | parA67 inserted as EcoRI/SalI fragment |
| pKLB60.4 | PCR-amplified parA1–47 with the use of primers 1 and 6, inserted as EcoRI/HindIII fragment |
| pKLB60.5 | parAΔ48–59, PCR-amplified fragment (primers 8 and 2) encoding parA60–262 cloned as HindIII/SalI fragment into pKLB60.4 |
| pKLB60.6 | PCR-amplified parA1–76 with the use of primers 1 and 7, inserted as EcoRI/HindIII fragment |
| pKLB60.7 | parAΔ77–85, PCR-amplified fragment (primers 9 and 2) encoding parA86–262 cloned as HindIII/SalI fragment into pKLB60.6 |
| pKLB60.8* | parA84 inserted as EcoRI/SalI fragment |
| pUC18 derivatives | |
| pGMB11* | parA74 inserted as EcoRI/SalI fragment |
| pGMB12* | parA75 inserted as EcoRI/SalI fragment |
| pGMB13* | parA76 inserted as EcoRI/SalI fragment |
| pGMB14* | parA78 inserted as EcoRI/SalI fragment |
| pGMB15* | parA79 inserted as EcoRI/SalI fragment |
| pBGS18 derivatives | |
| pGMB33* | parA inserted as EcoRI/SalI fragment with internal SacI site without amino acid change |
| Bacterial two-hybrid vectors | |
| pKGB4 | pUT18 derivative with modified MCS to facilitate ORF cloning as EcoRI/KpnI or EcoRI/SacI fragments in-frame with the N terminus of CyaT18 |
| pKGB4.14 | pKGB4 with parA67–cyaT18 translational fusion |
| pKGB4.15 | pKGB4 with parA68–cyaT18 translational fusion |
| pKGB4.16 | pKGB4 with parA70–cyaT18 translational fusion |
| pKGB4.17 | pKGB4 with parA72–cyaT18 translational fusion |
| pKGB4.18 | pKGB4 with parA74–cyaT18 translational fusion |
| pKGB4.19 | pKGB4 with parA75–cyaT18 translational fusion |
| pKGB4.20 | pKGB4 with parA76–cyaT18 translational fusion |
| pKGB4.21 | pKGB4 with parA78–cyaT18 translational fusion |
| pKGB4.22 | pKGB4 with parA79–cyaT18 translational fusion |
| pKGB4.24 | pKGB4 with parA82–cyaT18 translational fusion |
| pKGB4.25 | pKGB4 with parA83–cyaT18 translational fusion |
| pKGB4.26 | pKGB4 with parA84a†–cyaT18 translational fusion |
| pKGB4.27 | pKGB4 with parA84–cyaT18 translational fusion |
| pKGB4.28 | pKGB4 with parA85–cyaT18 translational fusion |
| pKGB5 | pKNT25 derivative with modified MCS to facilitate ORF cloning as EcoRI/KpnI or EcoRI/SacI fragments in-frame with the N terminus of CyaT25 |
| pKGB5.14 | pKGB5 with parA67–cyaT25 translational fusion |
| pKGB5.15 | pKGB5 with parA68–cyaT25 translational fusion |
| pKGB5.16 | pKGB5 with parA70–cyaT25 translational fusion |
| pKGB5.17 | pKGB5 with parA72–cyaT25 translational fusion |
| pKGB5.18 | pKGB5 with parA74–cyaT25 translational fusion |
| pKGB5.19 | pKGB5 with parA75–cyaT25 translational fusion |
| pKGB5.20 | pKGB5 with parA76–cyaT25 translational fusion |
| pKGB5.21 | pKGB5 with parA78–cyaT25 translational fusion |
| pKGB5.22 | pKGB5 with parA79–cyaT25 translational fusion |
| pKGB5.24 | pKGB5 with parA82–cyaT25 translational fusion |
| pKGB5.25 | pKGB5 with parA83–cyaT25 translational fusion |
| pKGB5.26 | pKGB5 with parA84a–cyaT25 translational fusion |
| pKGB5.27 | pKGB5 with parA84–cyaT25 translational fusion |
| pKGB5.28 | pKGB5 with parA85–cyaT25 translational fusion |
| pBBRMCS-1 derivative | |
| pABB1.0 | pBBRMCS-1 with modified EcoRI restriction site within the CmR cassette (PCR site-directed mutagenesis with primers 26 and 27) |
| pABB1.2 | pAMB9.37 with tacp–parB transcriptional fusion |
| pABB84 | pAMB9.37 with tacp–parA84a transcriptional fusion |
| pET28a (+) or pET28mod derivatives | |
| pABB8.0 | parA fragment PCR-amplified with primers 3 and 4 and inserted as NcoI/XhoI |
| pABB8.67 | parA67 PCR-amplified and inserted as above |
| pABB8.78 | parA78 PCR-amplified and inserted as above |
| pABB8.83 | parA83 PCR-amplified and inserted as above |
| pABB8.84 | parA84a PCR-amplified and inserted as above |
| pKLB8.3 | parA40–262 inserted as EcoRI/SalI fragment |
| pKLB8.4 | parA1–40 inserted as EcoRI/SalI fragment |
| pKLB8.5 | parA1–151 inserted as EcoRI/SalI fragment |
| pKLB8.6 | parA152–262 inserted as EcoRI/SalI fragment |
| pKLB8.7 | parA40–151 inserted as EcoRI/SalI fragment |
| pKLB8.8 | parA1–85 inserted as EcoRI/SalI fragment |
| pKLB8.9 | parA86–151 inserted as EcoRI/SalI fragment |
| pKLB8.10 | parAΔ48–59 inserted as EcoRI/SalI fragment |
| pKLB8.11 | parAΔ77–85 inserted as EcoRI/SalI fragment |
| Yeast two-hybrid vectors | |
| pABB482* | pGMB57 with parA82 inserted as EcoRI/SalI fragment |
| pABB483* | pGMB57 with parA83 inserted as EcoRI/SalI fragment |
| pABB484* | pGMB57 with parA84a inserted as EcoRI/SalI fragment |
| pABB485* | pGMB57 with parA85 inserted as EcoRI/SalI fragment |
| pGMB21* | pGAD424 with parA67 inserted as EcoRI/SalI fragment |
| pGMB22* | pGAD424 with parA68 inserted as EcoRI/SalI fragment |
| pGMB24* | pGAD424 with parA70 inserted as EcoRI/SalI fragment |
| pGMB25* | pGAD424 with parA72 inserted as EcoRI/SalI fragment |
| pGMB57 | pGAD424 with gal4AD–parA40–262 fragment with internal SacI site inserted as BamHI/SalI fragment |
| pKLB4.5 | pGAD424 with gal4AD–parA1–151 inserted as EcoRI/SalI fragment |
| pKLB4.8 | pKLB4.5 with gal4AD–parA1–85 with SalI oligo with stop codon inserted in NaeI site |
| pKLB4.9 | pKLB4.5 with gal4AD–parA86–151 with EcoRI oligo with ATG codon inserted in NaeI site, EcoRI fragment deleted |
| pKLB6.3 | pBTM116 with lexADB–parA40–262 inserted as BamHI/SalI fragment |
| pKLB6.4 | pBTM116 with lexADB–parA1–40 inserted as EcoRI/SalI fragment |
| pKLB6.5 | pBTM116 with lexADB–parA1–151 inserted as EcoRI/SalI fragment |
| pKLB6.6 | pBTM116 with lexADB–parA152–262 inserted as EcoRI/SalI fragment |
| pKLB6.7 | pBTM116 with lexADB–parA40–151 inserted as EcoRI/SalI fragment |
| pKLB6.8 | pBTM116 with lexADB–parA1–85 inserted as EcoRI/SalI fragment |
| pKLB6.9 | pBTM116 with lexADB–parA86–151 inserted as EcoRI/SalI fragment |
| pKLB6.10 | pBTM116 with lexADB–parAΔ48–59 inserted as EcoRI/SalI fragment |
| pKLB6.11 | pBTM116 with lexADB–parAΔ77–85 inserted as EcoRI/SalI fragment |
Construction of these plasmids is described in detail in Supplementary Materials and Methods.
parA84a encodes ParAL84A, whereas parA84 encodes ParAL84K.
Yeast two-hybrid (YTH) system.
The parA alleles were cloned in two vectors, pGAD424 and pBTM116, to form translational fusions with C termini of GAL4 and LexA, respectively. S. cerevisiae strain L40 was transformed with pGAD424 and pBTM116 parA mutant derivatives in various combinations with adequate plasmids encoding either WT ParA or WT ParB. Yeast strain transformation and LacZ activity assay were performed as described previously (Bartosik et al., 2004). Colonies of double transformants were transferred on nitrocellulose filters, immersed in liquid nitrogen to lyse and treated with X-Gal solution as a substrate.
Bacterial two-hybrid system.
A bacterial adenylate cyclase two-hybrid (BACTH) system was used to analyse in vivo interactions between ParA single-amino-acid substitution derivatives and WT ParA or ParB (Karimova et al., 1998, 2000). The multiple cloning site (MCS) sequences in the original BACTH system vectors were modified according to requirements to construct two pairs of vectors: pLKB2 and pLKB4 or pKGB4 and pKGB5 (Table 2).
For protein–protein interactions in the BACTH assay, pairs of appropriate vectors were used to co-transform E. coli BTH101 cyaA− cells. The co-transformants were selected onto MacConkey agar supplemented with 1 % maltose, 0.1 mM IPTG, kanamycin and penicillin, and grown at 30 °C for 48 h. The β-galactosidase activity assays were performed according to Miller (1972).
His6-tagged proteins purification.
The E. coli BL21(DE3) strain was transformed with pET28 derivatives encoding ParA variants His6-tagged at C ends. Overproduction of ParA-His6 or its mutant forms was carried out overnight in M9 medium with glucose supplemented with kanamycin (50 µg ml−1) and 0.1 mM IPTG. Tris buffer (10 mM Tris/HCl, pH 8, 1 M NaCl, 0.1 mM EDTA and 5 % glycerol) was used during the purification procedure on Protino Ni-TED 1000 columns (Macherey-Nagel). Purified proteins were stored in small portions at −80 °C prior to further analysis.
His6-ParB purification was carried out as described previously (Bartosik et al., 2004); however, Tris buffer (as for ParA) was used instead of phosphate buffer.
ATPase activity assay.
ATPase activity of purified ParA-His6 and its mutant derivatives was determined using the Malachite Green Phosphate Assay kit (BioAssay Systems). The purified protein (100 pmol) was incubated in 50 mM Tris/HCl, pH 8, 150 mM NaCl, 10 mM MgCl2, 5 % glycerol, 1 mM ATP and 2 mg BSA ml−1 in a final volume of 50 µl. The amount of released phosphate was assayed at 30 min intervals (up to 2 h) and calculated according to the phosphate standard curve prepared under the same conditions. At least 10 measurements in three independent experiments were performed for each ParA derivative using a 96-well microplate (Greiner Bio-One) in a Bio Tek plate reader at 630 nm.
Co-immunoprecipitation after in vivo protein cross-linking with formaldehyde.
E. coli BL21(DE3) strain was transformed with pET28 derivatives carrying different parA alleles and pABB1.2 carrying a tacp–parB fusion. Formaldehyde was added to a final concentration of 1 % to the exponentially growing cultures induced for 4 h by 0.5 mM IPTG. The cells were incubated for 30 min at room temperature, pelleted and washed twice in 10 ml PBS buffer (15 mM KCl, 150 mM NaCl and 10 mM NaPi, pH 7.4). The pellet was resuspended in 100 µl lysis buffer (10 mM Tris/HCl, pH 8, 20 % sucrose and 40 mM EDTA) with 1 mg lysozyme ml−1 and incubated on ice for 30 min. Then 100 µl 2× IP buffer (1.5 M Tris/HCl, pH 7, 300 mM NaCl and 0.2 % Triton X-100) with 1 mM PMSF was added. After 10 min on ice, the cell suspensions were sonicated and cleared by centrifugation. An aliquot of 100 µl anti-ParB antibodies was added to the soluble fraction and incubated overnight on an orbital shaker at 4 °C. Then 50 µl Protein A–Sepharose (Amersham Biosciences) was added to each sample and treated according to the protocol. Western blotting with anti-His6-tag antibodies (1 : 3000 dilution; Pierce) was carried out after protein separation by SDS-PAGE and transfer onto a nitrocellulose membrane.
Introduction of mutant alleles into the PAO1161 backbone by homologous recombination.
The parA mutant alleles were cloned as EcoRI/SalI fragments into pKLB60.1, a derivative of the suicide vector pAKE600 (El-Sayed et al., 2001). The constructed plasmids were transformed into E. coli S17-1 and then mobilized into P. aeruginosa PAO1161 RifR. The allele exchange procedure was performed as described previously (Lasocki et al., 2007). The allele exchange was verified by sequencing of PCR fragments amplified on the mutant chromosomal DNA as the template.
Fluorescence microscopy.
DAPI staining and immunofluorescence microscopy were carried out as described previously (Bartosik et al., 2004; Bignell et al., 1999). Cells were analysed using a Nikon Eclipse EC 800 microscope. The images were collected and analysed in Lucia software, and prepared for publication using Adobe Photoshop CS4.
Motility assays.
Motility assays were performed as described previously (Rashid & Kornberg, 2000). All sets of plates were standardized by using the same medium volume.
Modelling.
A structural model of the monomeric ParA of P. aeruginosa was obtained using sybyl-x 2.0 (Tripos) on the basis of ParA of P. aeruginosa and Soj of Thermus thermophilus alignment as well as the Soj crystal structure (Protein Data Bank ID: 1WCV; Leonard et al., 2005). To check if introduced amino acid substitutions significantly influenced ParA structure, the models of WT ParA and its derivatives were subjected to energy minimization using the AmberFF99 force field as implemented in sybyl-x 2.0.
Results
In vivo deletion mapping of the ParA domains involved in dimerization and interactions with ParB
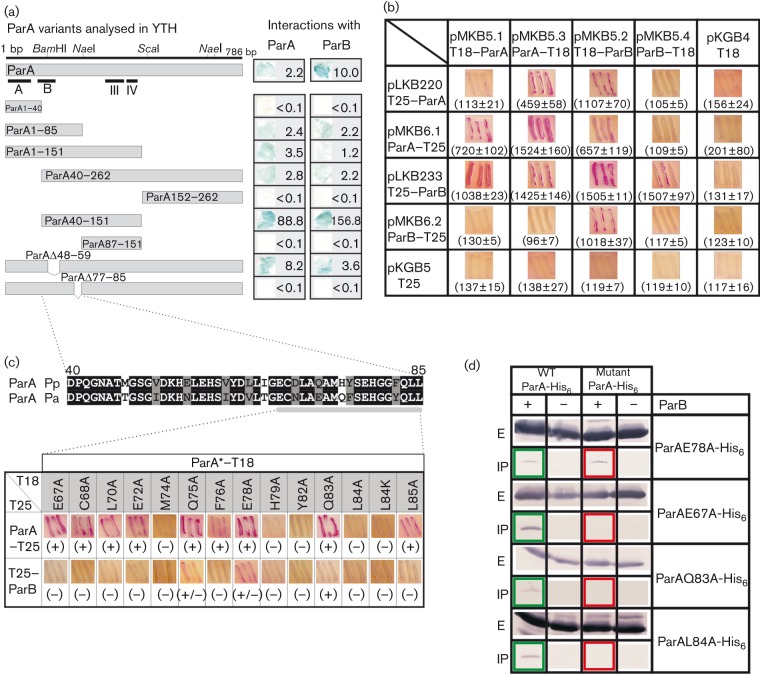
It has been shown previously using the YTH system that ParA of P. aeruginosa dimerizes and is able to interact with ParB (Bartosik et al., 2004). To establish which part of ParA is responsible for dimerization and interactions with ParB, a set of parA deletion mutants (Fig. 2a) was constructed (Supplementary Materials and Methods) and tested in the YTH system. The parA derivatives were used as bait or prey by translational linking with the activation domain of GAL4 or the DNA-binding domain of LexA (pGAD424 and pBTM116, respectively), and tested for interactions with hybrid parA and parB cloned into complementary vectors. The S. cerevisiae L40 strain was transformed with the appropriate pairs of plasmids. The expression of the lexA–lacZ fusion, activated by interactions between hybrid proteins, was monitored in double transformants by plate tests and β-galactosidase activity assays in the liquid cultures. Regardless of ParA derivatives being the bait or prey, the results were the same so only the results for one combination of hybrid proteins are demonstrated in Fig. 2(a) as scanned nitrocellulose filters, the corresponding enzyme activities assayed in the liquid cultures are shown.
Fig. 2.
Protein–protein interactions of ParA mutants. (a) ParA deletion mutant analysis in the YTH system. S. cerevisiae strain L40 was transformed with the appropriate pairs of pGAD424 and pBTM116 derivatives carrying parA, parB and different parA deletion alleles. The interactions between hybrid proteins were visualized by the plate test and β-galactosidase activity assays in liquid cultures. The mean values of LacZ activities from at least three independent experiments are shown. (b) BACTH system analysis of P. aeruginosa ParA and ParB interactions. E. coli BTH101 cyaA− was transformed with the pairs of BACTH vectors. As the control, double transformants of the plasmids encoding hybrid proteins and empty vectors were included. Data in parentheses represent the mean±sd β-galactosidase values from at least three experiments. (c) BACTH analysis of ParA substitution derivatives. (Upper) Comparison of ParA region D40–L85 from P. aeruginosa (Pa) and P. putida (Pp), with identical (black background) and similar (grey background) residues indicated. The ParA region analysed by alanine scanning is enlarged. (Lower) Summary of BACTH results between mutated ParAs linked to CyaAT18 and ParA–CyaAT25 or CyaAT25–ParB. ParA* represents ParA derivatives with amino acid substitutions. (+), Interactions detected; (+/−), weak interactions; (–), no interactions. (d) Co-immunoprecipitation of ParB with ParA derivatives. The extracts of strains producing His6-tagged ParA derivatives with/without WT ParB (‘+’ and ‘–’, respectively) were treated with anti-ParB antibodies. The ParA derivatives in immunoprecipitated pellets (‘IP’ panels) were detected with anti-His6 antibodies. ‘E’ panels show ParA protein levels in the initial extracts. In each set of experiments WT ParA was co-immunoprecipitated with ParB as the control.
Both truncated proteins, ParA40–262 and ParA1–151, self-associated in the YTH system, indicating that neither the N-terminal 39 aa nor the C-terminal 111 aa were important for ParA dimerization in vivo (Fig. 2a). Interestingly, when both the N and C terminus were removed (ParA40–151), the dimerization ability (measured by β-galactosidase activity) was significantly stronger than that detected for the single deletions. ParA1–85 was also able to self-associate in the YTH system and, as ParA1–40 and ParA87–151 could not, it was concluded that the short region between D40 and L85 played an important role in ParA self-association.
The same truncated forms of ParA that gave positive results in the dimerization test demonstrated interactions with ParB (Fig. 2a). ParA40–151 interacted with ParB significantly more strongly than any other modified form of ParA (similarly to the enhanced effect of association with ParA). This suggested that the 46 aa segment located between D40 and L85 was involved in both ParA dimerization and ParA–ParB interactions, and the removal of the N and C termini unmasked the interaction domain of ParA.
The ParA alignment (Fig. 1) showed that the D40–L85 segment encompassed two highly conserved blocks and two short sequences quite diverse among various bacterial strains, but very similar in two Pseudomonas species, P. aeruginosa and Pseudomonas putida (Fig. 2c). As such regions of variability between homologous proteins from different species may reflect important species specificity of the partitioning components, two short internal deletion alleles encoding ParAΔ48–59 and ParAΔ77–85, lacking the amino acid residues from the Pseudomonas ‘specific’ regions, were constructed (Supplementary Materials and Methods). The YTH assay revealed that the deletion of 12 aa in ParAΔ48–59 did not eliminate self-association and interactions with ParB, whereas the ParAΔ77–85 derivative was not able to dimerize or interact with ParB (Fig. 2a).
The low level of hybrid protein production in the yeast strains did not allow their detection by Western blotting, so the possibility that lack of interactions resulted from the instability of some derivatives in the yeast strains could not be excluded.
Fine mapping of dimerization and ParB interaction determinants in ParA
As the YTH analysis indicated quite weak ParA self-association and ParA–ParB interactions, it was decided to shift to the BACTH system (Karimova et al., 1998, 2000) to potentially increase the sensitivity of detection of interactions between Par proteins and possibly monitor the stability of hybrid proteins.
The parA and parB alleles were cloned into two pairs of BACTH vectors, derivatives of pairs pUT18C/pKT25 and pUT18/pKNT25, facilitating the linkage of Par proteins with two adenylate cyclase (CyaA) fragments (T18 and T25) of Bordetella pertussis, either at the N or C-terminus of tested proteins (Tables 1 and 2). Reconstruction of adenylate cyclase activity due to the interactions between proteins fused to the CyaA fragments led to the production of cAMP, which bound to the activator CAP and turned on the expression of sugar catabolism genes (e.g. lac or mal operon). The plate test for maltose fermentation and β-galactosidase assays in the liquid cultures of double transformants of E. coli BTH101 cyaA− strain demonstrated that the strongest ParB–ParB interactions occurred between ParB linked to the CyaA fragment by its N terminus (CyaA–ParB fusions), whereas the strongest dimerization of ParA was detected when ParA was linked to the CyaA fragment by its C terminus (ParA–CyaA fusions) (Fig. 2b).
Interactions between ParA and ParB were undetectable when ParB–CyaA was tested, suggesting that the C terminus of ParB is important for both homo- and heterologous interactions. The CyaA–ParB fusions demonstrated strong interactions with ParA regardless of which ParA end was linked to the CyaA fragment.
The YTH analysis indicated that region 59–84 in ParA is important for dimerization and interactions with ParB. Part of this region (60–64) is highly conserved among various chromosomal ParA homologues (Fig. 1). We decided to modify residues that were variable in ParAs from different classes, but similar in the Pseudomonas genus.
Thirteen parA point mutant alleles with alanine substitutions in the region E67–L85 (Fig. 2c) were constructed (Supplementary Materials and Methods). The previously constructed allele parAL84K was also included in the analysis. All mutant alleles were cloned into two BACTH system vectors, pKGB4 and pKGB5, facilitating their translational fusion via C-termini to two CyaA fragments, T18 and T25, respectively. Western analysis of extracts from DH5α cells carrying pKGB4 derivatives showed no differences in the stability of the hybrid proteins (Fig. S1). The BTH101 cyaA− strain was transformed with new constructs in pairs with vectors carrying parA–cyaA and cyaA–parB fusions to check the ability of mutant derivatives to dimerize and interact with ParB. The data shown in Fig. 2(c) confirmed that the E67–L85 region was very important for protein–protein interactions and divided the ParA substitution mutants into three categories. The first category consisted of two variants (ParAQ75A and E78A) that could dimerize similar to WT ParA and had slightly decreased ability to interact with ParB. The second category encompassed five mutants (ParAM74A, H79A, Y82A, L84A and L84K) that were defective in dimerization and interactions with ParB. The third category included seven mutants (ParAE67A, C68A, L70A, E72A, F76A, Q83A and L85A) that were capable of self-interactions, but did not interact with ParB, suggesting that the interface involved in ParA dimerization is not identical to the interface involved in interactions with ParB. As none of the ParA mutants defective in dimerization interacted with ParB, it was concluded that ParA dimerization was vital for association with ParB.
Verification of ParA–ParB interactions
Four parA alleles were chosen for further analysis: parAE67A and parAQ83A encoded ParAs that could dimerize, but did not interact with ParB, parAE78A encoded a product that was only slightly impaired in its ability to interact with ParB, and parAL84A encoded a product that showed neither dimerization nor interactions with ParB.
An immunoprecipitation assay was used to check the interactions between Par proteins. E. coli BL21(DE3) cells were transformed with pET28 derivatives overproducing the four mutant ParAs His6-tagged at their C-termini and with a pBBR1-MCS1 derivative carrying a tacp–parB transcriptional fusion (pABB1.2). In each set of immunoprecipitation experiments, strain BL21(DE3) (pABB8.0) (pABB1.2), overproducing WT ParA-His6 and WT ParB, was included as the positive control. After protein overproduction, the formaldehyde cross-linked complexes were immunoprecipitated with polyclonal anti-ParB antibodies, and anti-His6-tag antibodies were used to detect the presence of ParA in the initial extracts and immunoprecipitation pellets (Fig. 2d). The results showed that ParB formed complexes only with WT ParA and ParAE78A. ParAE67A, ParAQ83A and ParAL84A were not co-precipitated with ParB, confirming that the introduced amino acid substitutions significantly impaired the interactions between partners.
ATPase activity of WT ParA and its mutant derivatives.
Previous studies on various ParA homologues demonstrated their weak ATPase activities (Batt et al., 2009; Barillà & Hayes, 2003; Bouet & Funnell, 1999; Bouet et al., 2007; Leipe et al., 2002; Leonard et al., 2005; Lutkenhaus & Sundaramoorthy, 2003; Pratto et al., 2008).
The parA gene was cloned under the control of T7p into pET28a(+), expressed in E. coli BL21(DE3) and C-terminally His6-tagged ParA (ParA-His6) was purified (Fig. S2). The ATPase activity of ParA-His6 was assayed by the spectrophotometric detection of released inorganic phosphate (Pi). ParA-His6 demonstrated ATPase activity (Fig. 3) of ~1.2 pmol Pi released min−1 (pmol ParA)−1.
Fig. 3.
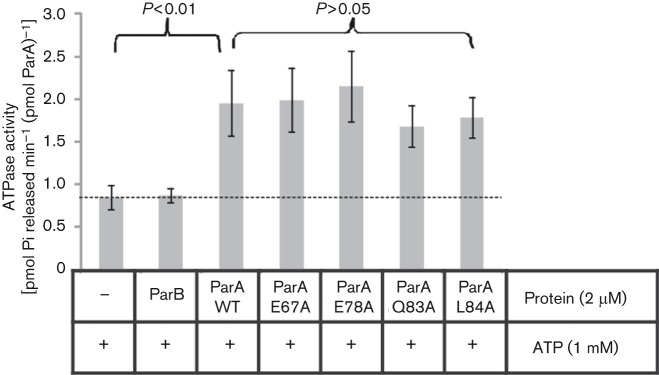
ATPase activities of ParA mutant derivatives. The C-terminally His6-tagged purified ParA derivatives (2 µM) were incubated with ATP and released inorganic phosphate (Pi) was detected spectrophotometrically. The control reactions (no protein added or His6-ParB alone) were included. Data represent the mean±sd values from at least 10 experiments. Statistical analysis revealed significant differences between WT ParA and two control samples (P<0.01; t-test), but not between different ParA variants (P>0.05; t-test).
Four ParA variants (tested by immunoprecipitation) were purified by affinity chromatography after overproduction in E. coli BL21(DE3) transformed with pET28a(+) derivatives carrying these parA alleles. All analysed ParA derivatives, among them dimerization-deficient ParAL84A, demonstrated ATPase activities similar to WT ParA (Fig. 3). This suggested that the amino acid substitutions did not drastically change the protein folding responsible for enzymic activity (see also Fig. 5b and Discussion).
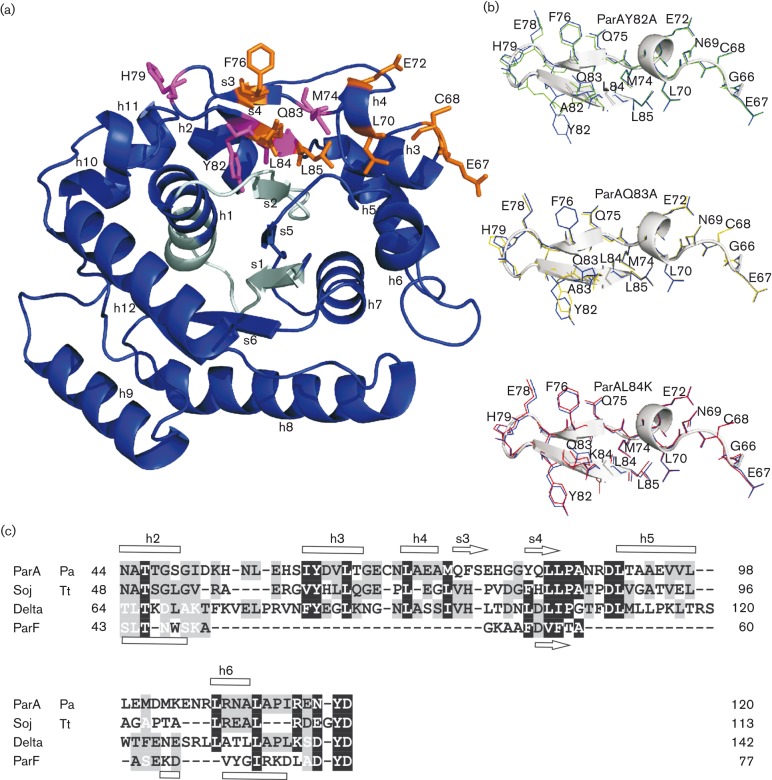
Fig. 5.
Model of the P. aeruginosa ParA monomer. (a) Model structure of ParA shown as a ribbon representation. The region of Walker motifs is highlighted in light blue. Amino acid residues important for interactions with ParB are shown as orange sticks; those playing roles in dimerization and interactions with ParB are shown as magenta sticks. Secondary structure elements marked according to the structure of Soj from T. thermophilus (Leonard et al., 2005). (b) Overlaid structures of pseudo-hairpins in the WT and the most ‘distorted’ mutant derivatives of ParA. Structures forced by Y82A, Q83A and L84K substitutions are shown in green, yellow and red, respectively, whereas WT residues are shown in blue. (c) Alignment of P. aeruginosa (Pa) ParA fragment (44–120 aa) with Soj [T. thermophilus (Tt)], Delta (pSM19035) and ParF (TP228). The alignment was prepared using cobalt (Papadopoulos & Agarwala, 2007), HHpred (Söding et al., 2005) and fatcat (Ye & Godzik, 2003) servers, and then corrected manually. Secondary structures are indicated according to Protein Data Bank entries 1WCV and 4DZZ for Soj and ParF, respectively. The P. aeruginosa ParA interactive pseudo-hairpin loop encompassed amino acid residues between G66 and L85. h, Helix; s, sheet.
Introduction of parA67 and parA84 mutations into the chromosome of PAO1161
The alleles parA84 and parA67 were chosen to be introduced into the P. aeruginosa PAO1161 genome as representatives of two different classes of parA mutants. ParAL84K was unable to dimerize and interact with ParB, whereas ParAE67A was defective in interactions with ParB, but capable of self-association. The mutant alleles were cloned into the suicide vector pAKE600 (El-Sayed et al., 2001) and then introduced into the PAO1161 chromosome via homologous recombination (allele exchange).
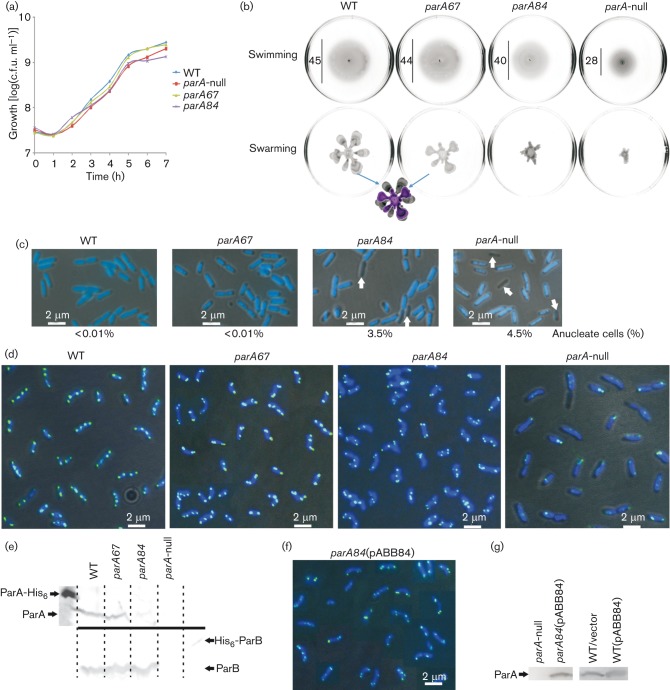
Growth experiments performed in L-broth (Fig. 4a) at 37 °C showed a slower growth rate of the PAO1161 parA84 mutant strain (mean division time 45 min) as compared with the parental PAO1161 strain (mean division time 36 min), the phenotype that was observed previously for the parA-null mutant (Lasocki et al., 2007). The mutant PAO1161 parA67 showed ~10 % reduction in growth rate (mean division time 39 min).
Fig. 4.
Effect of parA84 and parA67 mutations on growth, motility, nucleoid segregation and ParB localization in P. aeruginosa. Data for the PAO1161 WT strain and PAO1161 parA-null mutant are included for comparison. (a) Growth of WT, parA-null, parA67 and parA84 strains (L-broth, 37 °C) presented as log(c.f.u. ml−1). (b) Swimming and swarming assays. Representative images are shown. The diameters of swimming zones are indicated (in mm). The swarming zones of WT PAO1161 and parA67 are overlaid for comparison. (c) Anucleate cell formation. Representative images of cells from the exponential growth phase (L-broth, 37 °C) are shown. Anucleate cells are indicated by arrows. (d) Subcellular localization of ParB. Representative images of cells from the exponential growth phase (L-broth, 37 °C) are shown. The dark background in the merged micrographs is a phase-contrast image, the dark blue is the DAPI-stained chromosome and the green/light blue is the FITC-stained ParB. (e) Par protein levels in the mutant strains. ParA (top, anti-ParA antibodies) and ParB (bottom, anti-ParB antibodies) detected in 109 cells from the exponentially growing cultures of analysed strains by Western blotting. ParA-His6 (200 ng) and His6-ParB (30 ng) were used as the respective controls. (f) Fluorescence detection of ParB in the merodiploid strain PAO1161 parA84 (pABB84). Description as in (d). (g) ParA levels in the merodiploid strains. Western blotting with anti-ParA antibodies was used to visualize ParA proteins in extracts from 109 cells. Bar, 2 μm.
The PAO1161 parA84 and parA67 mutants were also tested for motility properties (Fig. 4b). The PAO1161 parA84 mutant showed defects in swarming and swimming, although not as strong as the parA-null mutant used as the control strain (Lasocki et al., 2007). The PAO1161 parA67 mutant was not impaired in swimming and only slightly defective in swarming as compared with the WT strain.
Cells collected from the exponential phase of culture growth were stained with DAPI, and the presence of anucleate cells was monitored in the newly constructed parA mutants and control strains, PAO1161 and PAO1161 parA-null mutant. Visible defects in chromosome segregation were observed for the PAO1161 parA84 mutant strain (Fig. 4c); a significant fraction (>3.5 %) of chromosome-less cells or cells with partially segregated chromosomes appeared. A similar proportion of chromosome-less cells was observed for the PAO1161 parA-null, whereas parental PAO1161 and mutant PAO1161 parA67 strains produced <0.01 % visibly defective cells.
As both ParA derivatives encoded by the mutated alleles were impaired in interactions with ParB, it was important to determine the effect of such modifications on ParB distribution in P. aeruginosa cells. Immunofluorescence microscopy using anti-ParB antibodies and FITC-conjugated secondary antibodies was used to observe cells from exponential-phase cultures grown on rich medium at 37 °C.
The majority of the actively dividing cells of PAO1161 contained two to four ParB foci distributed symmetrically along the long cell axis, marking the positions of oriC domains of the P. aeruginosa chromosome (Bartosik et al., 2009), whereas the signals for ParB in the parA-null mutant were much weaker, irregularly distributed and often in pairs (Fig. 4d). The PAO1161 parA84 strain showed perturbations in ParB localization that did not conform to the WT pattern of distribution (Fig. 4d). ParB signals were visibly paired at the poles in 90 % of the cells as compared with 10 % of cells with paired foci in the WT strain. In the case of PAO1161 parA67, a similar pattern of non-separated ParB foci was observed although not as frequent as for parA84 mutant (~40 % of the cells). This indicated that the mutant strains producing ParA impaired in its ability to interact with ParB were defective in proper separation of the oriC domains. The stronger mutant phenotype was observed for the ParA derivative impaired not only in interactions with ParB, but also in self-interactions (PAO1161 parA84).
The extracts from 109 cells from exponentially growing cultures of WT PAO1161, PAO1161 parA84 and PAO1161 parA67 mutants were analysed by Western blotting with anti-ParA antibodies to determine the level of production of ParA variants. Whereas ParAE67A was produced in quantities comparable with the WT ParA in PAO1161 (estimated 400 molecules per cell), the amount of ParAL84K seemed to be at least fivefold lower (Fig. 4e). Both parA mutant strains produced similar levels of ParB. To exclude the possibility that the observed mutant phenotypes resulted from insufficient quantities of ParAL84K, the parA84 allele was cloned into the medium-copy broad-host-range expression vector under tacp control to give pABB84. The amount of ParA (with substituted leucine at position 84) produced in the transformant PAO1161 parA84 (pABB84) was approximately twofold higher than WT ParA in PAO1161 as demonstrated by Western blotting (Fig. 4g). Immunofluorescence experiments with PAO1161 parA84 (pABB84) cells confirmed that disturbed ParB foci localization/separation was not associated with the ParA level, but resulted from the change in amino acid sequence (Fig. 4f). Additionally, the presence of pABB84 in PAO1161 parA84 did not reverse other defects, i.e. slower growth rate, motility defects and anucleate cells production (data not shown).
Discussion
Many studies on Par homologues (plasmid and chromosomally encoded) have demonstrated a role for a dynamic ParA scaffold and the importance of ParA–ParB interactions in bacterial DNA segregation (reviewed by Gerdes et al., 2010; Banigan et al., 2011; Hwang et al., 2013; Lim et al., 2014; Scholefield et al., 2011; Vecchiarelli et al., 2013). In the case of chromosomally encoded Par proteins, their roles in regulation of other cellular processes in a species-specific manner were also demonstrated (Kadoya et al., 2011; Murray & Errington, 2008; Scholefield et al., 2011; Thanbichler & Shapiro, 2006; Yamaichi et al., 2012). Whilst the ParB homologues have been dissected with respect to DNA binding, and dimerization/oligomerization domains, and in many cases the domains of interactions with the ParA homologues identified (Ah-Seng et al., 2009; Barillà et al., 2007; Bartosik et al., 2004; Figge et al., 2003; Kim & Shim, 1999; Leonard et al., 2004; Lukaszewicz et al., 2002; Scholefield et al., 2011; Surtees & Funnell, 1999), less is known about regions of ParA homologues involved in reciprocal interactions with the cognate partners (Jakimowicz et al., 2007; Leonard et al., 2005; Ravin et al., 2003; Scholefield et al., 2011).
Here, deletion analysis of P. aeruginosa parA using the YTH system roughly assigned the homo- and hetero-oligomerization to the N-terminal part of the protein. Alanine scanning of charged and hydrophobic residues in the ParA region E67–L85 and analysis of ParA variants in the BACTH system clearly demonstrated the essentiality of this region for both ParA self-interactions and interactions with ParB. Residues M74, H79, Y82 and L84 were shown to mediate ParA dimerization, and mutations at these positions were defective in interactions with ParB, indicating that only dimeric forms of ParA may associate with ParB. ParA variants with alanine substitutions at positions E67, C68, L70, E72, F76, Q83 and L85 were still capable of dimerization in the BACTH system, but impaired in interactions with ParB, thus defining a patch likely specific for hetero-oligomerization.
The 3D model of P. aeruginosa ParA (Fig. 5a) built on the basis of crystallographic data for the representative of the chromosomal ParA subfamily, Soj of T. thermophilus (Leonard et al., 2005), demonstrated that the region defined experimentally as important for dimerization and interactions with ParB encompasses two β-sheets folded into a pseudo-hairpin between helix 3 and 5 (Fig. 5c). The residues important for interactions with ParB are located at the external part of the loop, consistent with the idea that this part of the protein may be involved in partner binding. Comparison of predicted 3D structures of WT and mutant ParAs did not indicate any important conformational changes, with root-mean-square deviations for the backbone atoms of regions 1–65 and 86–254 being ≤0.1 Å. Only three mutant proteins, ParAY82A, ParAQ83A and ParAL84K, showed slight distortions in the region G66–L85 of the pseudo-hairpin structure as presented in Fig. 5(b). However, the introduced modifications did not affect loop folding, and were strictly limited to the substituted and adjacent residues. Two proteins, ParAQ83A and ParAL84A, were tested for ATPase activity, and proved capable of ATP hydrolysis at a comparable rate to the WT ParA (Fig. 3). This strongly suggested no major changes in the structures of mutant proteins.
The ATPase activity of P. aeruginosa ParA is in the range of 60–70 mol Pi released h−1 (mol protein)−1, much higher than observed for other proteins of this subfamily, e.g. 0.3−1.8 mol Pi released h−1 (mol protein)−1 (Ah-Seng et al., 2009; Batt et al., 2009; Leonard et al., 2005; Schofield et al., 2010). The ATPase activity of the dimerization-deficient ParAL84K suggests that impairment of self-interactions does not affect its enzymic function. Further studies are required to define the role of ATP binding in the ParA dimerization process.
Apart from the 3D structure of Soj from T. thermophilus, crystallographic data for three plasmid representatives of partitioning Walker-type ATPases are available: ParA of P1 plasmid prophage, Delta of pSM19035 and ParF of pTP228 (Dunham et al., 2009; Pratto et al., 2008; Schumacher et al., 2012). Only Delta protein of pSM19035 (Pratto et al., 2008) contains a pseudo-hairpin structure similar to the two chromosomal homologues (Fig. 5c). We reported previously that a Delta variant with alanine substitutions of two hydrophobic residues corresponding to L84 and L85 in ParA was impaired in dimerization and interactions with the Omega partner (Dmowski & Jagura-Burdzy, 2011). This suggests that some members of the ParA family of deviant Walker-type ATPases may rely on the same interface of interactions with their partners.
Screening of the database with the amino acid sequence from I60 to N110 of P. aeruginosa ParA encompassing the region we have identified as important for ParA–ParB interactions demonstrated that the Pseudomonas clade is separated from other species (the closest homologues are encoded by betaproteobacteria). The alignment of these regions originated from predicted ParAs encoded by the 35 Pseudomonas genomes clearly discriminated between different Pseudomonas species (Fig. S3), suggesting that observed subtle changes may determine the extent/lack of cross-reactivity between Par systems.
The PAO1161 parA84 mutant (encoding a ParA impaired in dimerization and interactions with ParB) showed defects in growth rate, nucleoid segregation, ParB localization, and swarming and swimming motility comparable to the parA-null mutant (Fig. 4). Immunofluorescence microscopy revealed that ParB can form one to four foci in cells of PAO1161 parA84; however, these are mislocalized in the majority of cells, positioned asymmetrically, close to the cell poles and paired (Fig. 4d). The PAO1161 parA84 mutant produces anucleate cells 300-fold more frequently than the WT strain. Despite such an increase in production of anucleate cells, the vast majority of the cells can survive and segregate their chromosomes, although with elongated division times. To distinguish between the role of ParA alone and in complex with ParB, we also constructed the PAO1161 parA67 mutant encoding a ParA that can dimerize, but is impaired in interactions with ParB in the tests applied. Phenotypic characterization of the mutant demonstrated only mild changes when compared with the WT strain. No defects in the growth rate and no increase in the production of anucleate cells were observed for this mutant, but the noticeable impairment of swarming ability as well as evident coupling of ParB foci in the polar regions of the cells clearly indicated the role of ParA–ParB interactions in these processes. Whereas it is well established that ParA–ParB interactions are crucial for their function in the separation of ori domains, the unique role of both Par proteins in swimming and swarming of P. aeruginosa cells is not fully understood (Bartosik et al., 2009; Lasocki et al., 2007). In our search for ParB partners we have identified a protein affecting expression of ‘motility’ operons (manuscript in preparation). It is feasible that ParB interactions with this partner are influenced by association with ParA. The stronger mutant phenotype of PAO1161 parAL84 than PAO1161 parAE67 suggests that the dimerization/polymerization of ParA may be more important than ParA–ParB interactions in the biology of P. aeruginosa. However, we cannot exclude the possibility that the weak phenotype of the ParAE67A mutant protein is because in live cells it is still capable of limited interactions with ParB.
Our transcriptomic studies (Bartosik et al., 2014) on parA and parB mutants of P. aeruginosa indicated major changes in gene expression patterns in both mutant strains. Despite the large overlap between the ParA and ParB regulon in P. aeruginosa, there are also clusters of genes regulated by ParA and ParB separately, which fits with the data presented here. Further studies are therefore required to explain the mechanisms that Par proteins use to regulate specific cell functions.
Acknowledgements
This work was funded by Wellcome Trust Collaborative Research Initiative grants 056022/Z/98/Z and 067068/Z/02/Z, and in part by MNiSW grant 2913/B/PO1/2008/34. We gratefully thank Dr Gouzel Karimova (Unité de Biochimie Cellulaire, Institut Pasteur, France) for providing vectors and strains for BACTH analysis. We thank Dr Magdalena Kusiak (Institue of Biochemistry and Biophysics, Polish Academy of Sciences) and Lukasz Kusiak (Institute of Biochemistry and Biophysics, Polish Academy of Sciences) for the construction of part of the BACTH system vector derivatives. We thank the Laboratory of Confocal and Fluorescence Microscopy of the Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland for the opportunity to conduct the microscopic observations.
Abbreviations:
- BACTH
bacterial adenylate cyclase two-hybrid
- MCS
multiple cloning site
- YTH
yeast two-hybrid
Footnotes
Supplementary material is available with the online version of this paper.
References
- Ah-Seng Y., Lopez F., Pasta F., Lane D., Bouet J. Y. (2009). Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. J Biol Chem 284, 30067–30075. 10.1074/jbc.M109.044800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Seng Y., Rech J., Lane D., Bouet J. Y. (2013). Defining the role of ATP hydrolysis in mitotic segregation of bacterial plasmids. PLoS Genet 9, e1003956. 10.1371/journal.pgen.1003956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan E. J., Gelbart M. A., Gitai Z., Wingreen N. S., Liu A. J. (2011). Filament depolymerization can explain chromosome pulling during bacterial mitosis. PLOS Comput Biol 7, e1002145. 10.1371/journal.pcbi.1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillà D., Hayes F. (2003). Architecture of the ParF·ParG protein complex involved in prokaryotic DNA segregation. Mol Microbiol 49, 487–499. 10.1046/j.1365-2958.2003.03564.x [DOI] [PubMed] [Google Scholar]
- Barillà D., Carmelo E., Hayes F. (2007). The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc Natl Acad Sci U S A 104, 1811–1816. 10.1073/pnas.0607216104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A. A., Lasocki K., Mierzejewska J., Thomas C. M., Jagura-Burdzy G. (2004). ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J Bacteriol 186, 6983–6998. 10.1128/JB.186.20.6983-6998.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A. A., Mierzejewska J., Thomas C. M., Jagura-Burdzy G. (2009). ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology 155, 1080–1092. 10.1099/mic.0.024661-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A. A., Glabski K., Jecz P., Mikulska S., Fogtman A., Koblowska M., Jagura-Burdzy G. (2014). Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS ONE 9, e87276. 10.1371/journal.pone.0087276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt S. M., Bingle L. E. H., Dafforn T. R., Thomas C. M. (2009). Bacterial genome partitioning: N-terminal domain of IncC protein encoded by broad-host-range plasmid RK2 modulates oligomerisation and DNA binding. J Mol Biol 385, 1361–1374. 10.1016/j.jmb.2008.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell C. R., Haines A. S., Khare D., Thomas C. M. (1999). Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol Microbiol 34, 205–216. 10.1046/j.1365-2958.1999.01565.x [DOI] [PubMed] [Google Scholar]
- Bouet J.-Y., Funnell B. E. (1999). P1 ParA interacts with the P1 partition complex at parS and an ATP–ADP switch controls ParA activities. EMBO J 18, 1415–1424. 10.1093/emboj/18.5.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J.-Y., Ah-Seng Y., Benmeradi N., Lane D. (2007). Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol Microbiol 63, 468–481. 10.1111/j.1365-2958.2006.05537.x [DOI] [PubMed] [Google Scholar]
- Bowman G. R., Comolli L. R., Zhu J., Eckart M., Koenig M., Downing K. H., Moerner W. E., Earnest T., Shapiro L. (2008). A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134, 945–955. 10.1016/j.cell.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmowski M., Jagura-Burdzy G. (2011). Mapping of the interactions between partition proteins Delta and Omega of plasmid pSM19035 from Streptococcus pyogenes. Microbiology 157, 1009–1020. 10.1099/mic.0.045369-0 [DOI] [PubMed] [Google Scholar]
- Dunham T. D., Xu W., Funnell B. E., Schumacher M. A. (2009). Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J 28, 1792–1802. 10.1038/emboj.2009.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A. K., Hothersall J., Thomas C. M. (2001). Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586. Microbiology 147, 2127–2139. [DOI] [PubMed] [Google Scholar]
- Figge R. M., Easter J., Gober J. W. (2003). Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol Microbiol 47, 1225–1237. 10.1046/j.1365-2958.2003.03367.x [DOI] [PubMed] [Google Scholar]
- Fogel M. A., Waldor M. K. (2006). A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20, 3269–3282. 10.1101/gad.1496506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Møller-Jensen J., Jensen R. B. (2000). Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol 37, 455–466. 10.1046/j.1365-2958.2000.01975.x [DOI] [PubMed] [Google Scholar]
- Gerdes K., Howard M., Szardenings F. (2010). Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942. 10.1016/j.cell.2010.05.033 [DOI] [PubMed] [Google Scholar]
- Gruber S., Errington J. (2009). Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696. 10.1016/j.cell.2009.02.035 [DOI] [PubMed] [Google Scholar]
- Hwang L. C., Vecchiarelli A. G., Han Y. W., Mizuuchi M., Harada Y., Funnell B. E., Mizuuchi K. (2013). ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J 32, 1238–1249. 10.1038/emboj.2013.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., Zydek P., Kois A., Zakrzewska-Czerwińska J., Chater K. F. (2007). Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol 65, 625–641. 10.1111/j.1365-2958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- Kadoya R., Baek J. H., Sarker A., Chattoraj D. K. (2011). Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J Bacteriol 193, 1504–1514. 10.1128/JB.01067-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C. M., Figurski D., Meyer R., Remaut E., Helinski D. R. (1979). Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol 68, 268–280. 10.1016/0076-6879(79)68019-9 [DOI] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95, 5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Ullmann A., Ladant D. (2000). A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol 328, 59–73. 10.1016/S0076-6879(00)28390-0 [DOI] [PubMed] [Google Scholar]
- Kim S.-K., Shim J. (1999). Interaction between F plasmid partition proteins SopA and SopB. Biochem Biophys Res Commun 263, 113–117. 10.1006/bbrc.1999.1317 [DOI] [PubMed] [Google Scholar]
- Koonin E. V. (1993). A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res 21, 2541–2547. 10.1093/nar/21.11.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E., Phillips R. W., Elzer P. H., Roop R. M., II, Peterson K. M. (1994). pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802. [PubMed] [Google Scholar]
- Kusiak M., Gapczynska A., Plochocka D., Thomas C. M., Jagura-Burdzy G. (2011). Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J Bacteriol 193, 3342–3355. 10.1128/JB.00328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasocki K., Bartosik A. A., Mierzejewska J., Thomas C. M., Jagura-Burdzy G. (2007). Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J Bacteriol 189, 5762–5772. 10.1128/JB.00371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317, 41–72. 10.1006/jmbi.2001.5378 [DOI] [PubMed] [Google Scholar]
- Leonard T. A., Butler P. J., Löwe J. (2004). Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol Microbiol 53, 419–432. 10.1111/j.1365-2958.2004.04133.x [DOI] [PubMed] [Google Scholar]
- Leonard T. A., Butler P. J., Löwe J. (2005). Bacterial chromosome segregation: structure and DNA binding of the Soj dimer – a conserved biological switch. EMBO J 24, 270–282. 10.1038/sj.emboj.7600530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. C., Surovtsev I. V., Beltran B. G., Huang F., Bewersdorf J., Jacobs-Wagner C. (2014). Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife 3, e02758. 10.7554/eLife.02758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. C.-H., Grossman A. D. (1998). Identification and characterization of a bacterial chromosome partitioning site. Cell 92, 675–685. 10.1016/S0092-8674(00)81135-6 [DOI] [PubMed] [Google Scholar]
- Livny J., Yamaichi Y., Waldor M. K. (2007). Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189, 8693–8703. 10.1128/JB.01239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak M., Dolowy P., Markowska A., Szarlak J., Kulinska A., Jagura-Burdzy G. (2013). Global transcriptional regulator KorC coordinates expression of three backbone modules of the broad-host-range RA3 plasmid from IncU incompatibility group. Plasmid 70, 131–145. 10.1016/j.plasmid.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Lukaszewicz M., Kostelidou K., Bartosik A. A., Cooke G. D., Thomas C. M., Jagura-Burdzy G. (2002). Functional dissection of the ParB homologue (KorB) from IncP-1 plasmid RK2. Nucleic Acids Res 30, 1046–1055. 10.1093/nar/30.4.1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J., Sundaramoorthy M. (2003). MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol 48, 295–303. 10.1046/j.1365-2958.2003.03427.x [DOI] [PubMed] [Google Scholar]
- Mierzejewska J., Jagura-Burdzy G. (2012). Prokaryotic ParA–ParB–parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67, 1–14. 10.1016/j.plasmid.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Mierzejewska J., Bartosik A. A., Macioszek M., Płochocka D., Thomas C. M., Jagura-Burdzy G. (2012). Identification of C-terminal hydrophobic residues important for dimerization and all known functions of ParB of Pseudomonas aeruginosa. Microbiology 158, 1183–1195. 10.1099/mic.0.056234-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Motallebi-Veshareh M., Rouch D. A., Thomas C. M. (1990). A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol 4, 1455–1463. 10.1111/j.1365-2958.1990.tb02056.x [DOI] [PubMed] [Google Scholar]
- Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. (1986). Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51, 263–273. 10.1101/SQB.1986.051.01.032 [DOI] [PubMed] [Google Scholar]
- Murray H., Errington J. (2008). Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135, 74–84. 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- Ogura Y., Ogasawara N., Harry E. J., Moriya S. (2003). Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J Bacteriol 185, 6316–6324. 10.1128/JB.185.21.6316-6324.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos J. S., Agarwala R. (2007). cobalt: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Pratto F., Cicek A., Weihofen W. A., Lurz R., Saenger W., Alonso J. C. (2008). Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res 36, 3676–3689. 10.1093/nar/gkn170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin J. L., Lee S. F., Garner E. C., Toro E., Eckart M., Comolli L. R., Moerner W. E., Shapiro L. (2010). A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12, 791–798. 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M. H., Kornberg A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97, 4885–4890. 10.1073/pnas.060030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin N. V., Rech J., Lane D. (2003). Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J Mol Biol 329, 875–889. 10.1016/S0022-2836(03)00525-4 [DOI] [PubMed] [Google Scholar]
- Ringgaard S., Schirner K., Davis B. M., Waldor M. K. (2011). A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev 25, 1544–1555. 10.1101/gad.2061811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schofield W. B., Lim H. C., Jacobs-Wagner C. (2010). Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 29, 3068–3081. 10.1038/emboj.2010.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G., Whiting R., Errington J., Murray H. (2011). Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol 79, 1089–1100. 10.1111/j.1365-2958.2010.07507.x [DOI] [PubMed] [Google Scholar]
- Schumacher M. A., Ye Q., Barge M. T., Zampini M., Barillà D., Hayes F. (2012). Structural mechanism of ATP-induced polymerization of the partition factor ParF: implications for DNA segregation. J Biol Chem 287, 26146–26154. 10.1074/jbc.M112.373696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., O’Connell M., Labes M., Pühler A. (1986). Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol 118, 640–659. 10.1016/0076-6879(86)18106-7 [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A. N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33 (Web Server issue), W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Hedge P. J., te Heesen S., Edelman A., Broome-Smith J. K. (1986). Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41, 337–342. 10.1016/0378-1119(86)90117-4 [DOI] [PubMed] [Google Scholar]
- Sullivan N. L., Marquis K. A., Rudner D. Z. (2009). Recruitment of SMC by ParB–parS organizes the origin region and promotes efficient chromosome segregation. Cell 137, 697–707. 10.1016/j.cell.2009.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees J. A., Funnell B. E. (1999). P1 ParB domain structure includes two independent multimerization domains. J Bacteriol 181, 5898–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M., Shapiro L. (2006). MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126, 147–162. 10.1016/j.cell.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Umbarger M. A., Toro E., Wright M. A., Porreca G. J., Baù D., Hong S.-H., Fero M. J., Zhu L. J., Marti-Renom M. A. & other authors (2011). The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44, 252–264. 10.1016/j.molcel.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli A. G., Han Y. W., Tan X., Mizuuchi M., Ghirlando R., Biertümpfel C., Funnell B. E., Mizuuchi K. (2010). ATP control of dynamic P1 ParA–DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol 78, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli A. G., Hwang L. C., Mizuuchi K. (2013). Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci U S A 110, E1390–E1397. 10.1073/pnas.1302745110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y., Niki H. (2000). Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci U S A 97, 14656–14661. 10.1073/pnas.97.26.14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y., Bruckner R., Ringgaard S., Möll A., Cameron D. E., Briegel A., Jensen G. J., Davis B. M., Waldor M. K. (2012). A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26, 2348–2360. 10.1101/gad.199869.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- Ye Y., Godzik A. (2003). Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19 (Suppl 2), ii246–ii255. 10.1093/bioinformatics/btg1086 [DOI] [PubMed] [Google Scholar]