Abstract
Background
Essential tremor is a common movement disorder with a strong heritable component. Large families with inherited forms of essential tremor have undergone genetic analyses by different approaches. However, our knowledge of genetic variants unequivocally linked to essential tremor is remarkably limited. Several explanations have been put forth to explain this challenge, including the possibility of mutations in non-coding areas of the genome.
Methods
We encountered a family with highly penetrant, autosomal dominant tremor. We hypothesized that, if a single coding gene mutation was responsible for the phenotype, novel genetic tools would allow us to identify it. We employed single nucleotide polymorphism (SNP) arrays in 17 members of this family followed by next generation whole-exome sequencing in five affected subjects.
Results
We did not identify any copy number variant or mutation that segregated with the disease phenotype.
Discussion
This study emphasizes the remarkably challenging field of tremor genetics and indicates that future studies should perhaps shift to analysis of the non-coding genome.
Keywords: Essential tremor, genetics, dystonia
Introduction
Essential tremor (ET) is one of the most common movement disorders.1,2 A clinically and etiologically heterogeneous syndrome, both genetic and environmental causes have been implicated in the pathogenesis of ET.3–6 The identification of gene variants that cause ET would facilitate the development of animal models to study the neurobiology underlying this syndrome and for rational therapeutic design. However, ET genetics is a surprisingly challenging area of movement disorders research.6 Despite the high prevalence of ET families following a Mendelian pattern of inheritance, the number of causative genes conclusively linked to ET remains negligible. This contrasts with the multiple genes that are known to be implicated in the etiology of other movement disorders, such as Parkinson's disease (PD), despite a lower prevalence and infrequent monogenic pedigrees.7
Microsatellite-based linkage studies in single families identified several loci linked to ET and candidate genes,4,5,7 but were not successful in the identification of gene variants that unequivocally segregate with the disease. Recently, we have witnessed remarkable advances in the availability of tools used to identify disease-causing genes.8 Two examples are the use of genome-wide single nucleotide polymorphism (SNP) arrays and next-generation sequencing. These novel techniques significantly increased efficiency and reduced costs of human genetic studies. They have been applied to the study of ET. For instance, a genome-wide association study (GWAS) completed in a large Icelandic and North American population identified an association between a SNP close to the LINGO1 gene and ET.9 However, mutations in LINGO1 do not cause Mendelian forms of ET, and this SNP is located in a non-coding sequence. Therefore, how this variant is associated with ET remains unknown. Another GWAS in patients with ET identified SLC1A2 as a potential susceptibility gene for ET.10 More recently, whole-exome sequencing identified mutations in the FUS gene in patients with ET,11 though follow-up studies suggest that this is a very rare cause of ET. An important step in this direction would be to identify genetic variants that cause ET in families that follow an autosomal dominant pattern of inheritance.
In this study, we identified a family with autosomal dominant ET and hypothesized that combining the use of SNP arrays and next generation sequencing would allow us to identify causative genetic variants that segregate with the ET phenotype.
Methods
Subjects
Through the evaluation of an index case in our Movement Disorders Clinic, we identified a family with multiple members affected by ET. All family members were invited to participate in the study. After obtaining informed consent, participants underwent an interview and neurological examination by a neurologist with expertise on movement disorders, recording pertinent information on the presence of tremor, other medical problems, and treatments received. Specific maneuvers used to elicit the tremor included arm extension, pouring water, drinking water, finger to nose, and drawing a spiral. The Washington Heights–Inwood Genetic Study of Essential Tremor criteria for probable or definite ET were used. Information on the presence of tremor in family members not examined, and therefore not included in the genetic study, was obtained from participants to determine type of inheritance. To generate a pedigree, non-examined relatives were considered as affected if at least two other family members reported the unequivocal presence of tremor. This study was reviewed and approved by the University of Iowa Institutional Review Board.
Genetic studies
Blood samples were obtained from participating subjects and genomic DNA was isolated following standard procedures. Genome-wide genotyping was performed using Affymetrix GeneChip Human Mapping 250K Array in the DNA Core Facility at the University of Iowa following the manufacturer's protocols (Affymetrix, Santa Clara, CA). Genotypes were called using GeneChip DNA Analysis Software (GDAS version 3).
For linkage, nonparametric and parametric analyses were performed using ALLEGRO (version 2.0) through the graphical user interface EasyLinkage Plus. Non-informative SNPs were removed from analysis. The marker spacing was set to 0.8 cM, and we assumed equal marker allele frequencies. For the parametric analyses, we specified an autosomal dominant model with 0.005 disease allele frequency, 0.99 penetrance for both heterozygous and homozygous disease allele carriers, and 0.01 phenocopy rate. Simulations (1,000 replicates) were also performed using Simwalk (version 2.91) and the same genetic parameters. The .CEL files from the Affymetrix arrays were analyzed for copy number using the default parameters within the Hidden Markov Model (HMM) within the Partek Genomics Suite (Partek Inc, St. Louis, MO). Copy Number Variants were annotated for gene content and overlap with CNVs in the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home). Using an autosomal dominant model, we tested for CNVs that were common to all the affected individuals but not detected in unaffected family members.
For next-generation sequencing, the Agilent SureSelect Version 4 (51 Mb) exome enrichment kit was used prior to sequencing on an Illumina HiSeq 2000 instrument. One hundred nucleotide-long paired end reads were used to sequence each exome to an average coverage of approximately 50× before the removal of PCR duplicates. We followed the best practice variant detection guidelines for sequence analysis developed by the Broad Institute for the Genome Analysis Tool Kit (GATK) suite of software utilities.12 The overall process, which we have previously described in more detail,13 consists of three stages—alignment to the reference genome, identification of high-quality variations and genotypes, and annotation with biologically relevant information. We identified variants that were found at greater than 1% in the 1000 Genomes Project, dbSNP, and the Exome Variant Server. All variants were also compared to our own internal database of exome-sequencing results from patients without the same phenotype. Following these initial variant filtration steps, annotation of remaining variants was performed using SnpEff and SnpSift,14 the dbNSFP,15 and the Human Gene Mutation Database Professional Version (HGMD).16 Variation prioritization was accomplished by three methods: 1) heuristic filtering based on suspected disease inheritance, 2) likely variant functional consequence, and 3) a probabilistic search algorithm, VAAST.17 Various quality control metrics were used throughout the analysis process. Fastq files were evaluated using FastQC, mapping was evaluated using both Samtools and Qualimap and variant quality was assessed using parallel strategies of hard filtering and variant quality score recalibration. We assumed that any causative variant would be transmitted in a dominant manner and exhibit complete penetrance. VarSifter18 was used to perform the inheritance based, variant segregation analysis.
Results
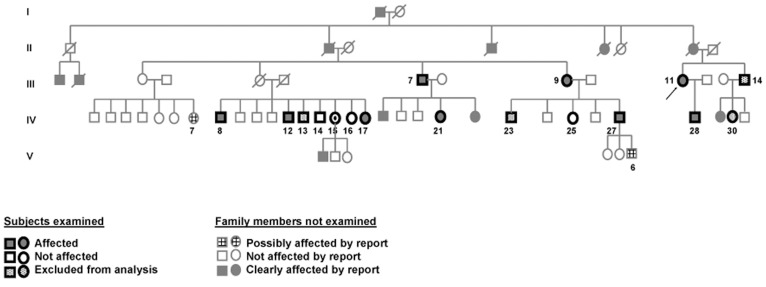
A total of 17 subjects participated in this study, undergoing an interview and examination followed by genetic analysis. Nine were determined to be affected by tremor, a woman assigned to be an obligate carrier (subject IV.15, who had a son with the typical head and hand tremor as described by all subjects) and three subjects were non-affected. Four subjects were excluded from the genetic analysis because the presence of ET was equivocal. One had late onset PD with a component of action tremor (subject III.14). Two reported occasional mild head tremor, but this was not observed by examination or noted by other family members (subjects IV.13 and IV.23). Another subject described the rare presence of mild action tremor in both upper extremities that was not detected by exam (subject IV.30). All participants provided information on the presence of tremor on non-participating family members. There was agreement in all but two subjects (subjects IV.7 and V.6). This information was used to generate a pedigree that indicated autosomal dominant inheritance shown by the presence of male to male transmission (Figure 1).
Figure 1. The Pedigree of the Reported Family Is Consistent with an Autosomal Dominant Inheritance Pattern.
The proband is indicated with an arrow. Deceased individuals are crossed by a slash. Note that many subjects of the V generation are not included in the pedigree as most were in the first two decades of life. Only the reportedly affected subjects from that generation and their siblings are shown.
All affected subjects but one met criteria for definite or probable ET according to the Washington Heights–Inwood Genetic Study of Essential Tremor criteria,19 and in more than half the head was prominently involved. The subject who did not meet criteria did not exhibit arm tremor, but isolated head tremor. We are confident that this phenotype represents the same tremor as other subjects, as the head tremor was significant and phenotypically comparable to other affected subjects. Another had very mild cervical dystonia without head tremor but with upper extremity action tremor (Table 1). However, the head tremor present in other subjects had the characteristics of head tremor of ET and not dystonic tremor.
Table 1. Clinical Characteristics of Tremor in Included Subjects.
| Subject | Current Age | Age Onset1 | Location | Severity (arm)2 |
|---|---|---|---|---|
| III.7 | 80 | 16 | Arms/head/voice | 3 |
| III.9 | 78 | 50 | Arms/head/voice | 3 |
| III.11 | 70 | 40 | Arms/head/voice | 3 |
| IV.8 | 51 | 35 | Arms3 | 2 |
| IV.12 | 56 | 20 | Arms/head | 2 |
| IV.17 | 42 | 38 | Head | 0 (only head) |
| IV.21 | 48 | 45 | Arms | 2 |
| IV.27 | 48 | 20 | Arms | 2 |
| IV.28 | 42 | 14 | Arms | 3 |
| Mean (±SD) | 57.2 (±14.9) | 30.9 (±13.5) |
Approximate age reported by the subjects.
Washington Heights–Inwood Genetic Study of Essential Tremor Severity Score.
Mild cervical dystonia.
The Affymetrix GeneChip array data were analyzed for CNVs. We compared CNVs detected in affected versus unaffected individuals. We did not detect any CNVs that segregated with the disease phenotype. We detected an average of 34 CNVs per individual and did not detect any variation in CNV burden between affected and unaffected individuals. There were three CNVs within 2p11.2, 8p11.23-p11.22 and 14q32.33, respectively that were detected in all affected subjects. But, upon further analysis they were also detected in some or all of the tested, unaffected family members and were later confirmed as common CNVs in the general population.
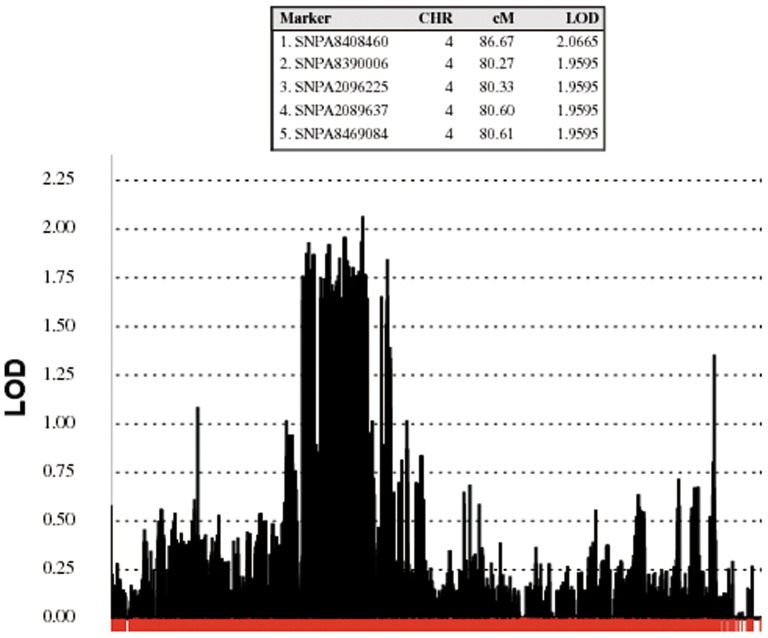
Next, we used the SNP data to complete a linkage analysis. We identified a two-point logarithm of odds score >2 in chromosome 4 (Figure 2), although this did not reach the LOD>3 required for evidence of linkage. To identify any potential mutations in this or other genomic loci, we then pursued next-generation, whole-exome sequencing in five affected subjects (III.7, III.9, III.11, IV.12, and IV.17). Surprisingly, this analysis also did not find any mutations shared by the five subjects. We specifically looked for variants in known ET genes. There were no rare (<1% Minor allele frequency), coding mutations within any of the five affected members in the genes LINGO1, SLC1A2, or FUS.
Figure 2. Linkage Analysis.
Illustration of the Only Genomic Region with a LOD Score >2 in the Linkage Analysis.
Discussion
In this study, we employed novel genetic technologies to study a family with autosomal dominant ET. However, we did not identify any CNV or mutation segregating with the disease. These results are consistent with previous studies that highlight the challenge of ET genetics. The absence of a shared mutation in the exonic sequence of five affected subjects indicates that either non-coding genes play an important role in the etiology of monogenic ET, or that ET follows a more complex inheritance pattern as previously suggested by others.20
The family described here has clinical features similar to other previously reported pedigrees, although with some interesting characteristics. It includes many subjects with typical ET affecting the upper extremities, but also several subjects with more prominent head tremor and a woman with comorbid mild cervical dystonia. The presence of prominent head tremor in many subjects and at a relatively young age is not very common. It is possible that this is a result of the interesting relationship between ET and dystonia. While these subjects exhibited the typical characteristics of ET-related head tremor, and not a dystonic head tremor, the potential contribution of a dystonic disorder cannot be entirely ruled out. Lou and Jankovic21 reported the presence of dystonia in a large proportion of patients with ET and the same group studied described families with inherited ET co-existing with dystonia.22 More recently, Hedera and colleagues23 reported 97 kindreds with autosomal dominant ET, and found that 28% of those pedigrees included subjects with dystonia. Consistent with previous studies illustrating the clinical heterogeneity of ET, these and other reports indicate that families with autosomal dominant ET that include subjects with dystonia represent a subtype of ET. Our family would belong to this subgroup of ET. Clustering large families into similar phenotypes, such as the ET-dystonia subtype, will likely aid in future genetic studies to reduce the noise caused by etiological heterogeneity.
It is possible that sequence variants in non-coding genes could underlie at least some forms of inherited ET. This would explain the difficulty in identifying causative genes in families with an apparent Mendelian mode of inheritance when focusing on the analysis of protein-coding genes. The role of non-coding RNAs in the pathogenesis of neurological diseases is just beginning to unfold.24,25 In the future, sequencing studies in families with ET should include protocols to isolate and sequence small RNAs. For instance, RNA-Seq of small RNAs extracted from blood samples or brains obtained from ET patients could serve to both sequence and quantify expression levels of hundreds of neurally expressed microRNAs. Furthermore, whole-genome, rather than whole-exome, sequencing could help identify mutations in non-coding regions. However, the increased cost and lower degree of sequence conservation in non-expressed regions will add some difficulty to this task. Other potential causes for the repeated lack of success in genetic studies in ET include complex modes of inheritance or the presence of phenocopies in this frequent syndrome. In fact, a fundamental problem with ET genetics resides in the definition of the phenotype and its highly variable penetration and expressivity. In the context of rare coding variants, it is plausible that we are not facing single highly penetrant coding variants that segregate with the phenotype, but a collection of rare variants with variable penetrance that result in a heterogeneous phenotype. Finally, the existing publication bias against “negative” studies might represent another obstacle, as studies of families with common phenotypical and genetic features performed by different groups probably remain unknown, preventing the design of collaborative studies with larger sample sizes.
In summary, the identification of genetic variants that cause ET in families with an apparent autosomal dominant mode of inheritance remains a challenge. Large collaborative studies, including multiple families with similar phenotypic subtypes and employing novel sequencing techniques that evaluate coding and non-coding sequences, should be pursued in the quest to identify genetic mutations that cause ET. In parallel, establishing collaborations to group and analyze linkage and sequencing data already obtained by different groups in different families, such as ours, could yield interesting data with no added costs. Either way, the field would benefit from the organization of an international network of investigators focused on the study of ET genetics.
Footnotes
Funding: None.
Financial Disclosures: Dr. Gonzalez-Alegre has received honoraria from Lundbeck for his participation in the Xenazine Advisory Board.
Conflict of Interests: The authors report no conflict of interest.
References
- 1.Louis ED. Essential tremor. Handb Clin Neurol. 2011;100:433–448. doi: 10.1016/B978-0-444-52014-2.00033-1. [DOI] [PubMed] [Google Scholar]
- 2.Louis ED. Clinical practice. Essential tremor. N Engl J Med. 2001;345:887–891. doi: 10.1056/NEJMcp010928. [DOI] [PubMed] [Google Scholar]
- 3.Louis ED. Environmental epidemiology of essential tremor. Neuroepidemiology. 2008;31:139–149. doi: 10.1159/000151523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasinska-Myga B, Wider C. Genetics of essential tremor. Parkinsonism Relat Disord. 2012;18((Suppl 1)):S138–139. doi: 10.1016/S1353-8020(11)70043-8. [DOI] [PubMed] [Google Scholar]
- 5.Deng H, Le W, Jankovic J. Genetics of essential tremor. Brain. 2007;130:1456–1464. doi: 10.1093/brain/awm018. [DOI] [PubMed] [Google Scholar]
- 6.Testa CM. Key issues in essential tremor genetics research: Where are we now and how can we move forward? Tremor Other Hyperkinet Mov. 2013;3 doi: 10.7916/D8Q23Z0Z. http://www.tremorjournal.org/article/view/105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wider C, Ross OA, Wszolek ZK. Genetics of Parkinson disease and essential tremor. Curr Opin Neurol. 2010;23:388–393. doi: 10.1097/WCO.0b013e32833b1f4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci. 2012;13:453–464. doi: 10.1038/nrn3271. [DOI] [PubMed] [Google Scholar]
- 9.Stefansson H, Steinberg S, Petursson H, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41:277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thier S, Lorenz D, Nothnagel M, et al. Polymorphisms in the glial glutamate transporter SLC1A2 are associated with essential tremor. Neurology. 2012;79:243–248. doi: 10.1212/WNL.0b013e31825fdeed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merner ND, Girard SL, Catoire H, et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet. 2012;91:313–319. doi: 10.1016/j.ajhg.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darbro BW, Mahajan VB, Gakhar L, et al. Mutations in extracellular matrix genes NID1 and LAMC1 cause autosomal dominant Dandy-Walker malformation and occipital cephaloceles. Hum Mutat. 2013;34:1075–1079. doi: 10.1002/humu.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mut. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson PD, Ball EV, Mort M, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mut. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 17.Yandell M, Huff C, Hu H, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: Visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benito-Leon J, Louis ED. Essential tremor: Emerging views of a common disorder. Nat Clin Pract Neurol. 2006;2:666–678. doi: 10.1038/ncpneuro0347. quiz 91. [DOI] [PubMed] [Google Scholar]
- 20.Ma S, Davis TL, Blair MA, et al. Familial essential tremor with apparent autosomal dominant inheritance: Should we also consider other inheritance modes? Mov Disord. 2006;21:1368–1374. doi: 10.1002/mds.20950. [DOI] [PubMed] [Google Scholar]
- 21.Lou JS, Jankovic J. Essential tremor: Clinical correlates in 350 patients. Neurology. 1991;41:234–238. doi: 10.1212/wnl.41.2_part_1.234. [DOI] [PubMed] [Google Scholar]
- 22.Jankovic J, Beach J, Pandolfo M, Patel PI. Familial essential tremor in 4 kindreds. Prospects for genetic mapping. Arch Neurol. 1997;54:289–294. doi: 10.1001/archneur.1997.00550150047015. [DOI] [PubMed] [Google Scholar]
- 23.Hedera P, Phibbs FT, Fang JY, Cooper MK, Charles PD, Davis TL. Clustering of dystonia in some pedigrees with autosomal dominant essential tremor suggests the existence of a distinct subtype of essential tremor. BMC Neurol. 2010;10:66. doi: 10.1186/1471-2377-10-66. Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niland CN, Merry CR, Khalil AM. Emerging roles for long non-coding RNAs in cancer and neurological disorders. Front Genet. 2012;3:25. doi: 10.3389/fgene.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]