Abstract
Epigenetic silencing through promoter hypermethylation is an important hallmark for the inactivation of tumor-related genes in carcinogenesis. Here we identified the ATP-binding cassette sub-family B member 4 (ABCB4) as a novel epigenetically silenced target gene. We investigated the epigenetic regulation of ABCB4 in 26 human lung, breast, skin, liver, head and neck cancer cells lines and in primary cancers by methylation and expression analysis. Hypermethylation of the ABCB4 CpG island promoter occurred in 16 out of 26 (62%) human cancer cell lines. Aberrant methylation of ABCB4 was also revealed in 39% of primary lung cancer and in 20% of head and neck cancer tissues. In 37% of primary lung cancer samples, ABCB4 expression was absent. For breast cancer a significant hypermethylation occurred in tumor tissues (41%) compared to matching normal samples (0%, p = 0.002). Silencing of ABCB4 was reversed by 5-aza-2'-deoxycytidine and zebularine treatments leading to its reexpression in cancer cells. Overexpression of ABCB4 significantly suppressed colony formation and proliferation of lung cancer cells. Hypermethylation of Abcb4 occurred also in murine cancer, but was not found in normal tissues. Our findings suggest that ABCB4 is a frequently silenced gene in different cancers and it may act tumor suppressivly in lung cancer.
Epigenetic mechanisms play an important role for initiating and maintaining memory effects on gene expression. In mammalians, the epigenetic regulation is essential for normal development by regulating gene imprinting, X-chromosome inactivation and transcriptional inactivation of repetitive genomic elements. Moreover, epigenetic inactivation of tumor suppressor genes is frequently observed during carcinogenesis1. Especially hypermethylation of promoters harboring a CpG island is a hallmark of gene silencing during malignant transformation. CpG islands are sequences greater than 500 bp of GC-rich and CpG-dense elements in the genome. About 70% of known genes harbor a CpG island within the promoter and the first exon. During tumorigenesis CpG islands of the promoter regions of tumor suppressive genes become hypermethylated and this aberrant methylation is accompanied by the formation of repressive chromatin and gene inactivation. The most frequently epigenetically inactivated tumor suppressor genes are the cyclin-dependent kinase inhibitor 2A (p16) and the Ras association domain family (RASSF) genes1,2,3.
The ABC (ATP-binding cassette) transporters are a large family of transmembrane proteins, with seven subfamilies that are designated A to G4. These subfamilies are also termed ABC1, MDR/TAP, MRP, ALD, OABP, GCN20 and White, respectively. ABCB4 (ATP-binding cassette, sub-family B, member 4) belongs to the MDR/TAP subfamily and the protein is also known as MDR2 or MDR3 (multi drug resistance). ABC proteins transport various molecules (e.g. xenobiotics, drugs, lipid and other metabolic products) across the plasma and intracellular membranes4. ABCB4 is such a transporter and a member of the p-glycoprotein family of membrane proteins and translocates phospholipids (e.g. phosphatidylcholine) from the inner to the outer membrane of the hepatocyte5. The exact function of ABCB4 has not been determined in detail, however it is not involved in drug resistance of ovarian carcinoma cells to cisplatin6.
The ABCB4 gene is localized on chromosome 7 at q21.12 (Fig. 1). Genetic alterations of ABCB4 are associated with progressive familial intrahepatic cholestasis type 3, low phospholipid associated cholelithiasis and also found in women with intrahepatic cholestasis of pregnancy7,8,9,10,11,12. The ABCB4 protein consists of 1273 aa and harbors two ABC transporter transmembrane regions and two ATP-binding cassette domains (Fig. 1A). In accordance with its function, ABCB4 is highly expressed in human liver, but lower mRNA levels were also found in other normal tissues13. The gene promoter of ABCB4 harbors a CpG island that is usually unmethylated in normal cells (Fig. 1A)14. The epigenetic regulation ABCB4 in cancer has not been analyzed in detail. In our study we report frequent hypermethylation of ABCB4 in human cancers. Interestingly, we also observed a growth suppressive function of ABCB4 in lung cancer cells.
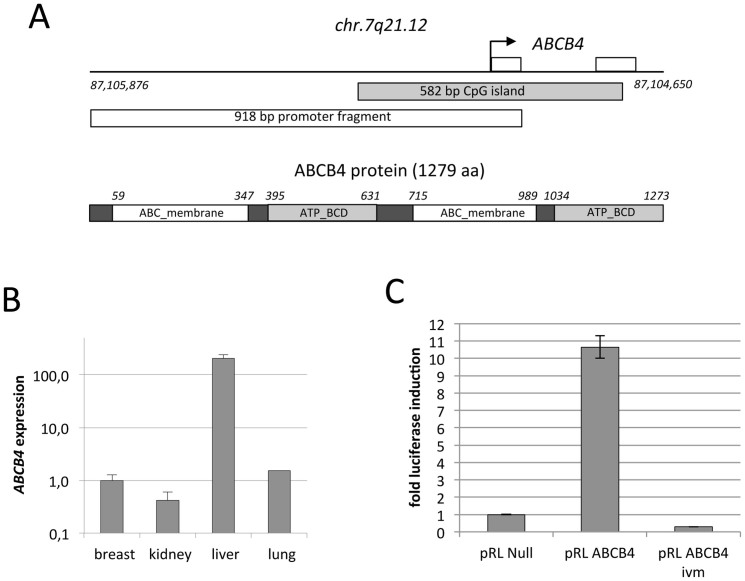
Figure 1. Epigenetic regulation of human ABCB4.
(A). Structure of ABCB4 CpG island promoter on chromosome 7q21.12 and the ACBC4 protein. Arrow marks transcriptional (+1) start site and boxes mark exons of ABCB4. The ABCB4 protein consists of two ABC transporter transmembrane regions (ABC_membrane) and two ATP-binding cassette domains (ATB_BCD). (B). ABCB4 levels in normal tissues. Expression of ABCB4 was analyzed in RNA isolated from normal lung, breast, liver and kidney samples by real-time RTPCR and normalized to GAPDH mRNA levels. (C). Promoter assay. HEK293 cells were transfected with 1 μg of pRL-null, pRL-ABCB4 and in vitro methylated (ivm) pRL-ABCB4 promoter construct and 0.35 μg of pGL3. Cells were isolated 24 h after transfection and studied using a dual-luciferase reporter assay.
Results
Methylation of ABCB4 occurred in distinct human cancer entities
We have performed a genome wide methylation screen (Infinium HumanMethylation450 BeadChip) in three lung cancer cell lines (A549, A427, H322) and normal human bronchial epithelial cells (NHBEC) and found a hypermethylation of ABCB4 at six CpG sites in its CpG island promoter in A549, A427 and H322 compared to NHBEC (21%, 48% and 88% compared to 7%, respectively). Subsequently, we have analyzed the RNA levels of ABCB4 in normal lung, breast, kidney and liver samples and detected expression of ABCB4 in all four tissues (Fig. 1B). Expression of ABCB4 in liver was found at much higher rate compared to the other three tissues. To analyze the impact of aberrant methylation at the ABCB4 promoter, we have cloned a 918 bp promoter fragment into a luciferase reporter system and transfected it in HeLa and HEK293 cells. In vitro methylation of ABCB4 drastically reduced the activity (36-fold reduction) of the promoter compared to the unmethylated promoter (Fig. 1C).
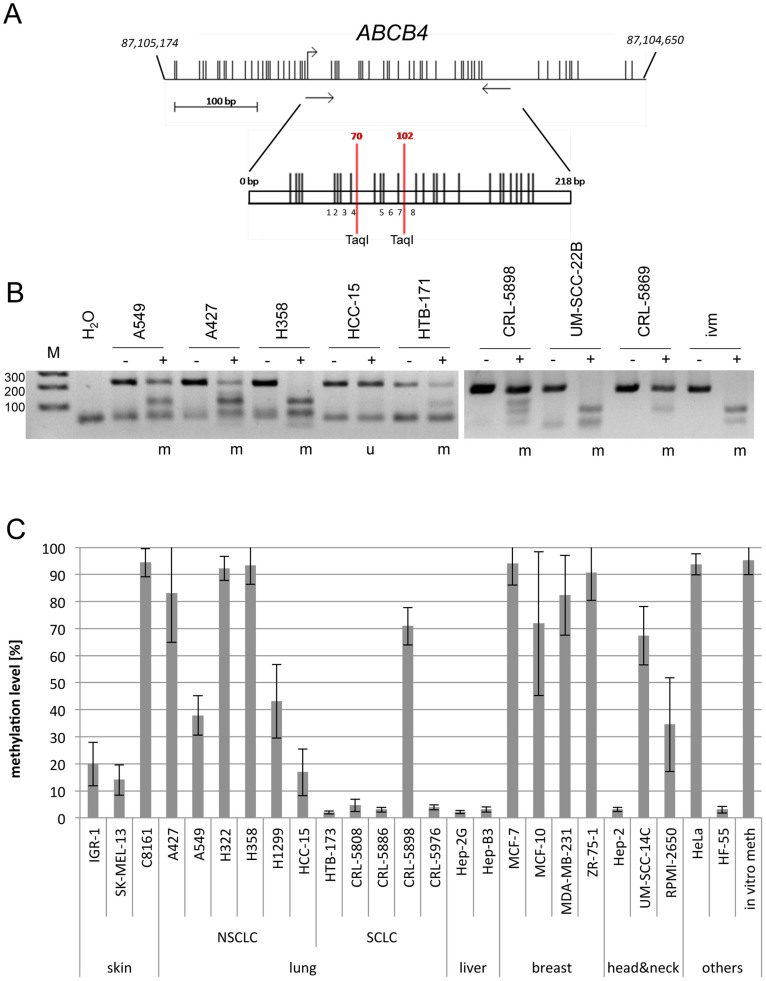
To investigate the epigenetic status of ABCB4 in human cancers in more details, we have analyzed its aberrant methylation in six non small cell lung cancers (A427, A549, H322, H358, HCC-15 and H1299), seven small cell lung cancers (HTB-173, CRL-5808, CRL-5976, CRL-5886, CRL-5898, CRL-5869, HTB-171), three breast cancers (MCF-7, ZR-75-1 and MDA-MB-231), skin cancer (IGR-1, SK-MEL-13, C8161), four head and neck (HN) cancers (Hep-2, UM-SCC-14C, UM-SCC-22B and RPMI-2650), two liver cancer (Hep-B3 and Hep-2G) cell lines, HeLa and human fibroblast (HF-55) by combined bisulfite restriction analysis (COBRA) and bisulfite pyrosequencing of eight CpG sites (Fig. 2). Fragmentation of the PCR product by TaqI indicates an underlying methylated ABCB4 (Fig. 2B). In vitro methylated genomic DNA (ivm) served as a methylated control for COBRA and pyrosequencing (Fig. 2B and C). Normal human fibroblast (HF-55) and liver cancer cells (Hep-2G and Hep-B3) were unmethylated. For lung cancer, five (A427, A549, H322, H358 and H1299) out of six NSCLC cell lines were hypermethylated (>20% methylation) and three (HTB-171, CRL-5898 and CRL-5896) out of seven SCLC cell lines were methylated (Fig. 2B and 2C). Also three breast cancer cell lines (MCF-7, ZR-75-1 and MDA-MB-231), three HN cancers (UM-SCC-14C, UM-SCC-22B and RPMI-2650), skin cancer C8161 and HeLa exhibited ABCB4 hypermethylation (>20%; Fig. 2B and C). Thus, 26 cancer cell lines were analyzed, of which 16 (62%) were methylated for ABCB4 and its hypermethylation was found in different human cancer entities including lung, breast, skin and HN cancers.
Figure 2. Methylation of ABCB4 in human cancer cell lines.
(A). CpG sites at the ABCB4 promoter regions are indicated as vertical lines. The PCR product (218 bp) with two TaqI sites (pos. 70 and 102) and eight CpG sites that were analyzed by pyrosequencing are depicted. (B). Combined bisulfite restriction analysis of ABCB4. Bisulfite-treated DNA from the indicated cancer cell lines and in vitro methylated DNA (ivm) were amplified, digested with TaqI (+) or mock digested (−) and resolved on 2% gels with a 100 bp marker (M); (m = methylated, u = unmethylated). (C). Bisulfite pyrosequencing of ABCB4. Methylation levels of eight CpGs of the bisulfite modified PCR products were analyzed by pyrosequencing.
Tumor specific ABCB4 hypermethylation in primary tumors and reduced ABCB4 expression in primary lung cancer
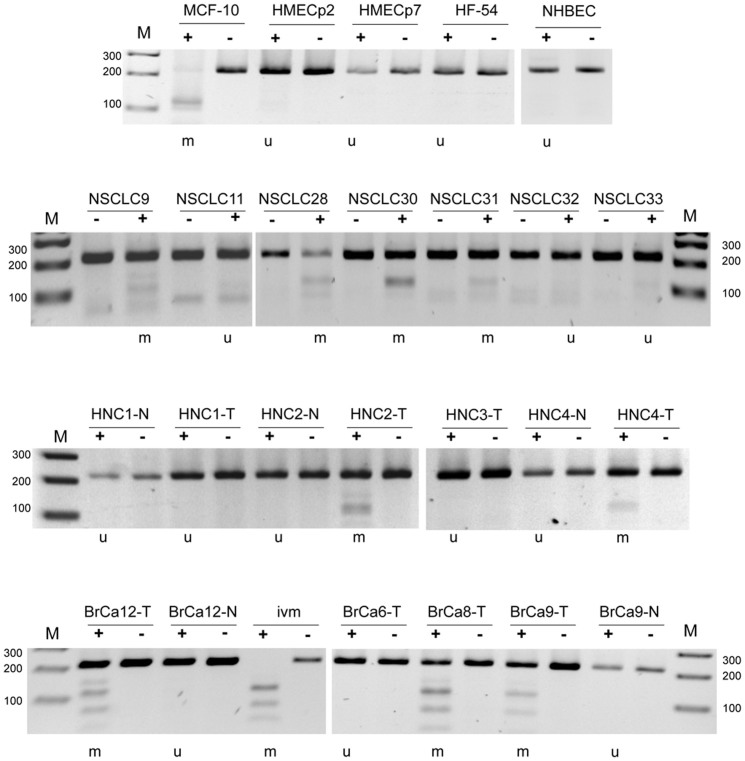
Next we have analyzed the methylation levels of ABCB4 in normal human bronchial epithelial cells (NHBEC) and human mammary epithelial cells (HMEC) by COBRA (Fig. 3). HMEC, NHBEC and normal human fibroblast (HF-54) show no promoter methylation of ABCB4 (Fig. 3). It is interesting to note that the MCF-10 cell line that served as a non-tumorigenic epithelial cell line also exhibited ABCB4 hypermethylation (Fig. 2 and 3). However, MCF10 proliferates indefinitely in cell culture, since it was immortalized spontaneously20. To analyze the impact of epigenetic silencing of ABCB4 in primary cancer tissues we investigated its hypermethylation in 46 primary NSCLC, 27 breast cancers and 10 HN cancers and several matching controls by COBRA. Representative results are shown in Figure 3. In HN cancer only two out of ten (20%) tumors (e.g. HNC2-T) exhibited ABCB4 hypermethylation (Fig. 3). 18 out of 46 (39%) NSCLC (e.g. NSCLC30) showed partial methylation of ABCB4 (Fig. 3). Moreover, 11 out of 27 (41%) breast cancer tissues (e.g. BrCa12-T) were methylated for ABCB4 (Fig. 3). All 20 analyzed matching breast control tissue were unmethylated (e.g. BrCa12-N) (Fig. 3). For breast cancer a significant tumor specific methylation of ABCB4 was found (p = 0.002, two tailed Fisher exact test).
Figure 3. Methylation of ABCB4 in primary tumors and normal tissues.
Representative COBRA analysis is shown for normal human bronchial epithelial cells (NHBEC), human mammary epithelial cells (HMEC passage 2 and 7) and normal human fibroblast (HF-54) and primary non small cell lung cancer (NSCLC), head and neck cancer (HNC) and breast cancer (BrCa). Mock digest (−) and TaqI digest (+) are shown. Products were resolved on 2% gel with 100 bp marker (M). (T = tumor, N = corresponding normal tissue, ivm = in vitro methylated control, m = methylated, u = unmethylated).
Expression of ABCB4 was also analyzed on a lung cancer tissue array (data not shown). 44 out of 70 (63%) of primary lung cancers showed staining of ABCB4 and its expression was absent in 37% of cases. The highest expression of ABCB4 was observed in SCLC, while no staining was observed in mesotheliomas. ABCB4 staining was slightly higher in squamous cell carcinoma (18/30 or 60%) than in adenocarcinoma (8/14 or 57%). ABCB4 staining was dominantly cytoplasmic, but about 24% of the cases (mainly squamous cell cancer and SCLC) also showed nuclear staining. Neither absence of nuclear staining nor the cytoplasmic staining correlated with the tumor grade or clinical stage. In concordance with the methylation frequency (39%) of ABCB4 in non small cell lung cancer, absence of its expression was observed in 41% (18/44) of primary NSCLC.
Hypermethylation of ABCB4 is associated with its downregulation in human cancer cell lines
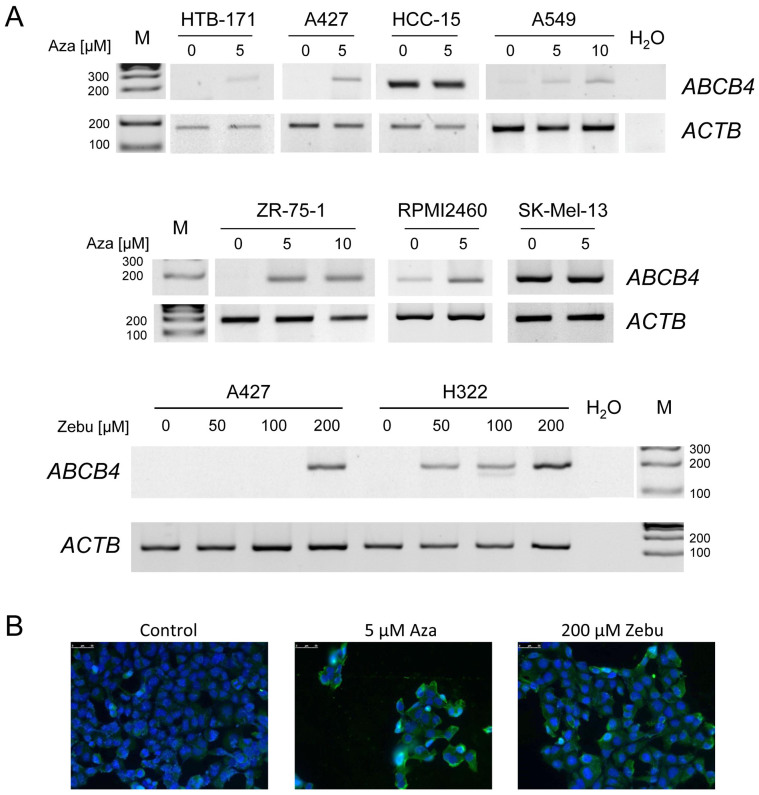
5-Aza-2'-deoxycytidine (Aza) and zebularine (Zebu) inhibit de novo methylation21,22 and are known to reverse hypermethylation of tumor suppressor genes, inducing their reexpression2. We therefore treated the lung cancer cell lines A427, A549, H322, HCC-15, HTB171 and H1299, the breast cancer cells ZR-75-1, the HN cancer cell RPMI-2650 and the melanoma cells SK-Mel-13 with Aza or Zebu and analyzed the ABCB4 expression (Fig. 4). A427, A549, H322, HTB-171, ZR-75-1 and H1299 harbor a hypermethylated ABCB4 promoter (>20% methylation Fig. 2) and show no endogenous ABCB4 expression (Fig. 4). Treatment with 5 μM Aza or 200 μM Zebu lead to ABCB4 reexpression on RNA level (Fig. 4A). Similar results were revealed for RPMI-2650, which are partially methylated (35% in Fig. 2) and ABCB4 expression is increased upon Aza treatment (Fig. 4A). HCC-15 and SK-Mel-13 are unmethylated for ABCB4 (<20% methylation; Fig. 2) and its expression is independent of the Aza treatment (Fig. 4A). Aza treatment of the lung cancer cells H1299 lead to ABCB4 reexpression on protein level (Figure 4B). In addition, reexpression of ABCB4 in lung tumor cell lines A549, A427 and H322 was accompanied by ABCB4 demethylation (data not shown). In cancer, we observed an epigenetic silencing of ABCB4, which was reversed by Aza or Zebu treatment.
Figure 4. ABCB4 expression in cancer cells, reexpression under DNMT inhibitor treatment.
(A). Expression analysis of ABCB4 is shown after 5-aza-2’-deoxycytidine (Aza) and zebularine (Zebu) treatments in lung cancer cell lines A427, A549, H322, HCC-15 and HTB-171, breast cancer ZR-75-1, head and neck cancer RPMI-2650 and melanoma SK-Mel-13. ABCB4 (185 bp) and ACTB (226 bp) expression was analyzed by RT-PCR after four days of Aza or Zebu treatment on 2% gel with 100 bp marker (M). (B). Lung cancer H1299 cells treated with either 5 μM Aza or 200 μM Zebu for 72 h. Cells were blocked with 5% BSA for 1 h, followed by incubation with anti-ABCB4 antibody overnight at 4°C. Finally, cells were incubated with Alexa Fluor 488-conjugated secondary antibody and nuclei were stained with DAPI. Representative images are shown.
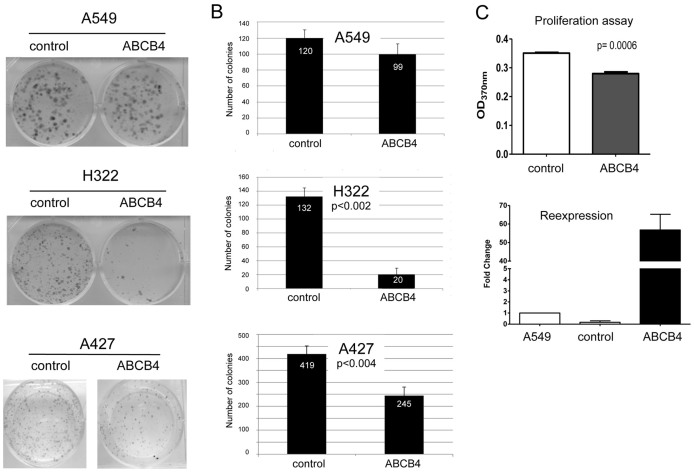
ABCB4 expression reduces colony formation and proliferation
To functionally test ABCB4 and its ability to suppress tumor formation, we performed colony formation and proliferation assays (Fig. 5). Therefore, we transfected A427, A549 and H322 with an ABCB4 expression- or empty control construct (pEGFP) and selected with G418 for three weeks. Colonies that formed were Giemsa stained and representative pictures are shown (Fig. 5A). In all three cancer cell lines ABCB4 is methylated (Fig. 2). Ectopic expression of ABCB4 in A549, H322 and A427 reduces the number of colonies (Fig. 5B). For H322 and A427 this growth suppressive effect was significant (p < 0.002 and p < 0.004, respectively; two tailed t-test). In addition, proliferation was also significantly reduced in A549 cells (p < 0.0006; Fig. 5C). These data suggest that ABCB4 exhibits a tumor suppressive function in human lung cancer cells.
Figure 5. Overexpression of ABCB4 reduces colony formation and proliferation in lung cancer cell lines.
(A). A549, A427 and H322 lung cancer cell lines were transfected with the ABCB4 expression construct or empty vector (pEGFP-C1), selected with G418 for 28 days and Giemsa stained. (B). Experiment was repeated three times and mean colony numbers with according SD are shown for A549, A427 and H322 cells. (C). Cell proliferation of A549 cells was evaluated by bromodeoxyuridine (BrdU) incorporation assay. BrdU incorporation was assessed by an ELISA reader. P values were calculated using two tailed paired t-test.
Epigenetic silencing of Abcb4 in mouse cancer
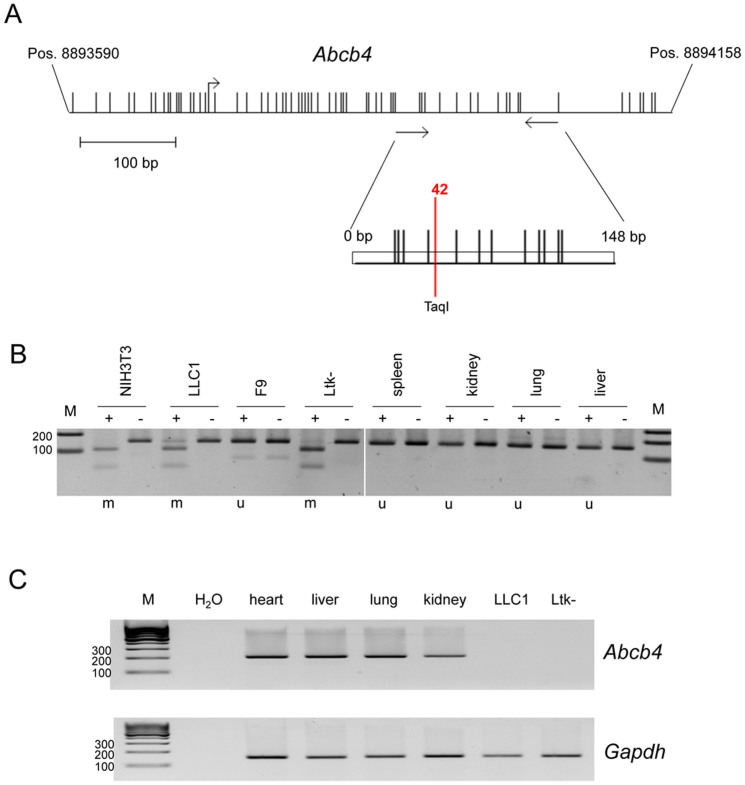
Subsequently, we have analyzed the methylation status and expression of the murine Abcb4 gene (Fig. 6). Abcb4 is localized on chromosome 5qA1 and is transcribed from a 402 bp long CpG island promoter at position 8893614 to 8894015 (Fig. 6A). We have analyzed the methylation in normal mouse tissues (spleen, kidney, lung and liver) and in the murine cell lines: Lewis lung cancer 1 (LLC1), NIH3T3, teratocarcinoma F9 and Ltk- (Fig. 6B). Interestingly, in normal tissues Abcb4 was unmethylated, but in three cell lines (LLC1, NIH3T3 and Ltk-) its hypermethylation was detected (Fig. 6B). NIH3T3 are primary mouse embryonic fibroblast cells, which were spontaneously immortalized and Ltk- was derived from the L929 murine fibrosarcoma cells. Moreover, we have analyzed the expression of Abcb4 by RT-PCR (Fig. 6C). In normal tissues Abcb4 expression was revealed, however in LLC1 and Ltk- expression of Abcb4 was silenced. Thus, our data suggest that epigenetic silencing of Abcb4 occurred also in pathogenesis of murine cancer.
Figure 6. Epigenetic regulation of murine Abcb4.
(A). Genomic organization of Abcb4 CpG island on chromosome 5qA1. Transcriptional start, CpG sites and COBRA primers are depicted. The PCR product (148 bp) with the TaqI site (pos. 42) is indicated. (B). Methylation analysis of Abcb4 by COBRA. Bisulfite converted DNA was amplified and PCR products were digested with TaqI (+) or mock-treated (−). Methylation status of Abcb4 is indicated (m = methylated, u = unmethylated). (C). Expression analysis of murine Abcb4 in heart, liver, lung and kidney and in murine cancer cell lines LLC1 and Ltk-. Expression of Abcb4 (200 bp) and Gapdh (156 bp) was analyzed by RT-PCR on a 2% gel.
Discussion
In our study, we have identified the ATP-binding cassette sub-family B member 4 (ABCB4) as novel epigenetically inactivated target gene in human cancer. Hypermethylation of ABCB4 was found in several epithelial cancer entities including lung, breast, head and neck (HN), skin and cervix cancer. We also observed that aberrant methylation of ABCB4 is also frequently found in primary NSCLC (39%), breast cancers (41%) and HN cancer (20%). In concordance with its methylation frequency absence of ABCB4 expression was observed in 41% primary NSCLC. To our knowledge hypermethylation of ABCB4 was not reported in cancer previously. Methylation of ABCB1 (MDR1) has been reported in breast cancer, HN cancer and other cancers23,24,25,26. Overexpression of several ABC transporter in cancer has been reported, however for ABCB4 and several other members (ABCA7, ABCA12, ABCB2, ABCB5 and ABCD1) downregulation in melanoma cell lines compared to normal melanocytes was revealed27. Thus, it will be interesting to analyze the methylation status of ABCB4 in primary melanomas.
We have analyzed the expression of ABCB4 in human tissues and observed the highest mRNA levels in liver. ABCB4 expression was also found in normal breast, lung and kidney. This is in accordance with the previous observation that ABCB4 is highly expressed in human liver, but ABCB4 expression was found in other normal samples (e.g. heart and muscle)13. In normal human bronchial epithelial cells, mammary epithelial cells and fibroblast ABCB4 was unmethylated, but in primary tumor tissues aberrant promoter methylation of ABCB4 was revealed (Fig. 3). In MCF10 cells that are utilized as a non-tumorigenic epithelial cell line, hypermethylation of ABCB4 was found. Since MCF10 cells immortalized spontaneously and therefore proliferate indefinitely in cell culture20, MCF10 cannot be considered as normal human mammary epithelial cells. Normal breast epithelial cells undergo cellular senescence after few passages20,28,29. In MCF10 cells promoter hypermethylation of several tumor repressive genes, including PTEN, p73, MGMT and cadherin (CDH1 and CDH13) has been reported30. Expression of mouse Abcb4 was also observed in all normal tissues including heart, liver, lung and kidney (Fig. 6). In these tissues the Abcb4 CpG island was unmethylated. We have also analyzed the methylation levels of ABCB4, in primary normal human liver tissues (data not shown). However, in these liver tissues and in the liver cancer cell lines Hep-B3 and Hep-2G no aberrant methylation of ABCB4 was found (Fig. 2 and data not shown). Considering the ABCB4 transporter function of phospholipids from liver hepatocytes into bile, a high tissue specific expression of ABCB4 in liver cells was expected and observed (Fig. 1). Mutations of ABCB4 are associated with progressive familial intrahepatic cholestasis type 3, low phospholipid associated cholelithiasis and found in women with intrahepatic cholestasis of pregnancy7,8,9,11,12. However, some patients with ABCB4 mutations also develop liver fibrosis, liver cirrhosis or cholangiocarcinoma10,31,32. Cholangiocarcinomas are malignant epithelial liver tumors with a poor prognosis and arise from the intra- and extra-hepatic bile ducts33. Since promoter hypermethylation of tumor associated genes has been reported in cholangiocarcinoma33,34, it could be important to investigate the methylation status of ABCB4 in this specific liver cancer entity. Abcb4 knock out mice develop chronic portal inflammation and bile duct proliferation with progression to liver fibrosis and hepatocellular carcinoma35. Other tumors in Abcb4 knock out mice have not been reported yet.
By using 5-aza-2'-deoxycytidine and zebularine treatments we were able to reexpress and demethylate ABCB4 (Fig. 4 and data not shown). Moreover, methylation of the ABCB4 promoter in vitro reduces its activity dramatically (Fig. 1). These results confirm that downregulation of ABCB4 in cancer cells is due to its promoter hypermethylation. Aza (Decitabine) and its derivatives (e.g. Vidaza) are used for therapies of blood cancers36.
To verify the ability of ABCB4 to suppress tumor growth like, we performed colony formation and proliferation assays in lung cancer cell lines (Fig. 5). Our results show that ABCB4 significantly suppresses colony growth in H322 and A427 cancer cells, in which ABCB4 is downregulated and inactivated by aberrant promoter methylation (Fig. 2 and Fig. 4). In A549 cells which show lower promoter hypermethylation levels compared to the other two cell lines growth suppression was less prominent, however proliferation was significantly reduced (Fig. 5C). Still, the exact function of ABCB4 in lung and breast epithelial cells has not been analyzed in detail. In ovarian cancer the resistance of ovarian carcinoma cells to cisplatin is not mediated by ABCB46. Thus it will be interesting to further dissect the role of ABCB4 in the pathogenesis of cancer.
In summary we demonstrate that the ATP-binding cassette sub-family B member 4 gene is frequently hypermethylated in human and murine cancers. Moreover aberrant methylation was frequently observed in primary lung and breast cancer samples. Demethylation of ABCB4 is accompanied by its reexpression in cancer cell lines. Furthermore, ectopic expression of ABCB4 suppresses colony formation in lung cancer cells. Future research will elucidate the exact function of ABCB4 during carcinogenesis. It will be interesting to evaluate if aberrant ABCB4 methylation may represent a novel biomarker for prognostic or diagnostic purposes in human cancer.
Methods
Tissue and cell lines
Primary cancer tissues and cancer cell lines were previously published15,16,17. All patients signed informed consent at initial clinical investigation. The study was approved by the local ethic committees (City of Hope Medical Center, Duarte, USA and Martin-Luther University, Halle, Germany). Mice tissues were obtained from C57/BL6 mouse strain. All cell lines were cultured in humidified atmosphere (37°C) with 5% CO2 and 1 × Penicillin/Streptomycin in according medium. Cells were transfected with 4 μg or 10 μg of constructs for 3.5 or 10 cm plates, respectively using Polyethylenimine (Sigma Aldrich) or Turbofect (Fermentas GmbH, St.Leon-Rot, Germany).
Cell Proliferation assay
Cell proliferation was evaluated by BrdU incorporation using Cell Proliferation ELISA colorimetric kit (Roche Diagnostics) according to the manufacturer’s protocol. A549 cells were transfected with ABCB4-overexpression vector were seeded at a density of 10 × 103/well in 96-well plates. After 24 h cells were starved in serum-free DMEM for further 24 h. BrdU was added to the cells and incubated for 4 h at 37°C. After removal of culture medium the cells were fixed and anti-BrdU antibody was added followed by the substrate. Finally BrdU incorporation was assessed by an ELISA reader.
Methylation analysis
DNA was isolated by phenol-chloroform extraction and then bisulfite treated prior to COBRA analysis and pyrosequencing18. 200 ng were subsequently used for PCR with primer ABCB4BSU1 (GAGTAAAGTTTAGGTTTTTTTGTTGTAG) and 5’-biotinylated primer ABCB4BSL1Bio (CCTCAAAACCAAATACACCCTCTCC). Products were digested with 0.5 μl TaqI (Fermentas GmbH, St.Leon-Rot, Germany) 1 h at 65°C and resolved on 2% TBE gel. Methylation status was quantified utilizing the primer ABCB4BSseq (TTTAGAGGTTTTGTTAGATA) and PyroMark Q24 (Qiagen, Hilden, Germany). Eight CpGs are included in the analyzed region of ABCB4 and mean methylation was calculated. For mouse Abcb4 the forward primer mABCB4BSU1 (GTGAAGTTTAGGTGAGGGAGGA) and the reverse primer mABCB4BSL1 (ACTACCTAAAAAAAAAAACCTCCAAA) were used and for in vitro methylation of genomic DNA M.SssI methylase was utilized (NEB, Frankfurt, Germany).
Expression analysis
RNA was isolated using the Isol-RNA lysis procedure (5 Prime, Hamburg, Germany). 25 μg of breast, kidney, liver and lung RNA of normal human samples were obtained from Agilent Technologies (Waldbronn, Germany). RNA was DNase (Fermentas GmbH, St.Leon-Rot, Germany) digested and then reversely transcribed19. RT-PCR was performed with primers: ABCB4RTF1: GCAGACGGTGGCCCTGGTTGG, ABCB4RTR1: TGGAAAACAGCACCGGCTCCTG,  ACTFW: CCTTCCTTCCTGGGCATGGAGTC and
ACTFW: CCTTCCTTCCTGGGCATGGAGTC and  ACTRV: CGGAGTACTTGCGCTCAGGAGGA. For mouse mABCB4RTF1: CCCTCCAGCCGGCTTTCTCCA, mABCB4RTR1: GGACCGGAGCCTTGTGGTGAGG, mGAPDHRTF1: GCCGCCTGGAGAAACCTGCC and mGAPDHRTF1: CCCCGGCATCGAAGGTGGAA were utilized. Quantitative PCR (qRT-PCR) was performed in triplicates with PerfeCTa SYBR® Green (Quanta BioSciences, Gaithersburg, USA) using Rotor-Gene 3000 (Corbett Research, Qiagen, Hilden, Germany).
ACTRV: CGGAGTACTTGCGCTCAGGAGGA. For mouse mABCB4RTF1: CCCTCCAGCCGGCTTTCTCCA, mABCB4RTR1: GGACCGGAGCCTTGTGGTGAGG, mGAPDHRTF1: GCCGCCTGGAGAAACCTGCC and mGAPDHRTF1: CCCCGGCATCGAAGGTGGAA were utilized. Quantitative PCR (qRT-PCR) was performed in triplicates with PerfeCTa SYBR® Green (Quanta BioSciences, Gaithersburg, USA) using Rotor-Gene 3000 (Corbett Research, Qiagen, Hilden, Germany).
Constructs
The cDNA of human ABCB4 was obtained as a full length cDNA vector IRATp970F09103D (Accession Number: BC042531; 4035 bp in pBluesriptR; imaGenes GmbH, Berlin, Germany) and the ORF of ABCB4 was cloned into the SacII and XmaI sites of pEGFP-C1. The ABCB4 promoter was amplified from genomic DNA and cloned into pRLnull (Promega, Mannheim, Germany). Therefore a 918 bp promoter fragment was obtained by PCR with primers ABCB4Bgl2U1: GAGTCTTTTGGGAAGAGTGTGGAGGAATTA and ABCB4EcoRIL1: GAATTCGCGCGTGTCTGGCAGG and cloned into the BglII and EcoRI sites of pRLnull. In vitro methylation of the promoter construct was done with M.SssI methylase (NEB, Frankfurt, Germany).
Promoter assay
HEK293 or HeLa cells were transfected with 1 μg of pRL-ABCB4 promoter construct and 0.35 μg of pGL3. Cells were isolated 24 h after transfection and studied using Dual-Luciferase Reporter Assay (Promega, Mannheim, Germany).
Immunocytochemistry
Cells were seeded on chamber slides and treated with either 5 μM azacytidine or 200 μM zebularine for 72 h. After washing with PBS, cells were fixed with acetone-methanol (1:1) for 20 min at −20°C. Then cells were blocked with 5% BSA for 1 h, followed by incubation with anti-ABCB4 antibody (LifeSpan LS-B5729, 1:200) at 4°C overnight. Finally, cells were incubated with Alexa Fluor 488-conjugated secondary antibody, nuclei were stained with DAPI and mounted with fluorescent mounting media (Dako Cytomation, Glostrup, Denmark).
Immunohistochemistry
Lung cancer tissue array were purchased from Pantomics, Inc. (Richmond, CA, USA). In brief, slides were treated with high pH antigen retrieval buffer. Rabbit anti-human ABCB4 (1:200; LS-B5729, lifespan biosciences, USA) and normal rabbit serum were used for stainings. Slides were incubated with ABCB4 antibody at 4°C overnight. After extensive washing, sections were incubate with secondary antibody, followed by staining with vector DAB (Vector Laboratories). The slides were washed and counterstained with hematoxylin for 5 min. All slides were analyzed under the Hamamatsu NDP slide scanner (Hamamatsu Nanozoomer 2.0HT) and its viewing platform (NDP. Viewer).
Author Contributions
R.H.D., W.S. and R.S. have created the study. S.K., R.S. and R.H.D. participated in the design of the study. S.K., S.H., L.F., N.E.N. and A.M.R. acquired data. S.K., S.H., A.M.R., N.E.N., R.S. and R.H.D. controlled analyzed and interpreted data. R.H.D., R.S. and S.K. prepared the manuscript. S.K., S.H., A.M.R., R.S., L.F., N.E.N., W.S. and R.H.D. read, corrected and approved the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
The work was supported by a LOEWE grant (UGMLC) from the Land Hessen. This organization had no involvement in the study design, acquisition, analysis, data interpretation, writing of the manuscript and in the decision to submit the manuscript for publication.
References
- Jones P. A. & Baylin S. B. The epigenomics of cancer. Cell 128, 683–692 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R. et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25, 315–319 (2000). [DOI] [PubMed] [Google Scholar]
- Richter A. M., Pfeifer G. P. & Dammann R. H. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta 1796, 114–128 (2009). [DOI] [PubMed] [Google Scholar]
- Fletcher J. I., Haber M., Henderson M. J. & Norris M. D. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer 10, 147–156, 10.1038/nrc2789 (2010). [DOI] [PubMed] [Google Scholar]
- Oude Elferink R. P. & Paulusma C. C. Function and pathophysiological importance of ABCB4 (MDR3 P-glycoprotein). Pflugers Arch 453, 601–610, 10.1007/s00424-006-0062-9 (2007). [DOI] [PubMed] [Google Scholar]
- Ren L., Xiao L. & Hu J. MDR1 and MDR3 genes and drug resistance to cisplatin of ovarian cancer cells. J Huazhong Univ Sci Technolog Med Sci 27, 721–724, 10.1007/s11596-007-0627-7 (2007). [DOI] [PubMed] [Google Scholar]
- Rosmorduc O. & Poupon R. Low phospholipid associated cholelithiasis: association with mutation in the MDR3/ABCB4 gene. Orphanet J Rare Dis 2, 29, 10.1186/1750-1172-2-29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floreani A. et al. Hepatobiliary phospholipid transporter ABCB4, MDR3 gene variants in a large cohort of Italian women with intrahepatic cholestasis of pregnancy. Dig Liver Dis 40, 366–370, 10.1016/j.dld.2007.10.016 (2008). [DOI] [PubMed] [Google Scholar]
- Denk G. U. et al. ABCB4 deficiency: A family saga of early onset cholelithiasis, sclerosing cholangitis and cirrhosis and a novel mutation in the ABCB4 gene. Hepatol Res 40, 937–941, 10.1111/j.1872-034X.2010.00698.x (2010). [DOI] [PubMed] [Google Scholar]
- Gotthardt D. et al. A mutation in the canalicular phospholipid transporter gene, ABCB4, is associated with cholestasis, ductopenia, and cirrhosis in adults. Hepatology 48, 1157–1166, 10.1002/hep.22485 (2008). [DOI] [PubMed] [Google Scholar]
- Mullenbach R. et al. ABCB4 gene sequence variation in women with intrahepatic cholestasis of pregnancy. J Med Genet 40, e70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vree J. M. et al. Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci U S A 95, 282–287 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J. J. et al. Characterization of the promoter region of the human MDR3 P-glycoprotein gene. Biochim Biophys Acta 1261, 44–56 (1995). [DOI] [PubMed] [Google Scholar]
- Lincke C. R., Smit J. J., van der Velde-Koerts T. & Borst P. Structure of the human MDR3 gene and physical mapping of the human MDR locus. J Biol Chem 266, 5303–5310 (1991). [PubMed] [Google Scholar]
- Schagdarsurengin U., Pfeifer G. P. & Dammann R. Frequent epigenetic inactivation of cystatin M in breast carcinoma. Oncogene 26, 3089–3094 (2007). [DOI] [PubMed] [Google Scholar]
- Dammann R. et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer 41, 1223–1236 (2005). [DOI] [PubMed] [Google Scholar]
- Steinmann K., Sandner A., Schagdarsurengin U. & Dammann R. H. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep 22, 1519–1526 (2009). [DOI] [PubMed] [Google Scholar]
- Dammann G. et al. Increased DNA methylation of neuropsychiatric genes occurs in borderline personality disorder. Epigenetics 6, 1454–1462, 10.4161/epi.6.12.18363 (2011). [DOI] [PubMed] [Google Scholar]
- Richter A. M., Walesch S. K., Wurl P., Taubert H. & Dammann R. H. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis 1, e18, 10.1038/oncsis.2012.18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule H. D. et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res 50, 6075–6086 (1990). [PubMed] [Google Scholar]
- Jones P. A. & Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol 321, 591–599 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebek G. et al. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum Mol Genet 21, 1557–1565, 10.1093/hmg/ddr593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D. & Vertino P. M. Epigenetic regulation of MDR1 gene in breast cancer: CpG methylation status dominates the stable maintenance of a silent gene. Cancer Biol Ther 3, 549–550 (2004). [DOI] [PubMed] [Google Scholar]
- Oberstadt M. C. et al. Epigenetic modulation of the drug resistance genes MGMT, ABCB1 and ABCG2 in glioblastoma multiforme. BMC Cancer 13, 617, 10.1186/1471-2407-13-617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggerud A. A. et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res 12, R3, 10.1186/bcr2466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimerl S., Bosserhoff A. K., Langmann T., Ecker J. & Schmitz G. Mapping ATP-binding cassette transporter gene expression profiles in melanocytes and melanoma cells. Melanoma Res 17, 265–273, 10.1097/CMR.0b013e3282a7e0b9 (2007). [DOI] [PubMed] [Google Scholar]
- Stampfer M. R. & Bartley J. C. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc Natl Acad Sci U S A 82, 2394–2398 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikova M. et al. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol 25, 3923–3933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro M. F., Saez-Ayala M., Pinero-Madrona A., Cabezas-Herrera J. & Rodriguez-Lopez J. N. Reactivation of the tumour suppressor RASSF1A in breast cancer by simultaneous targeting of DNA and E2F1 methylation. PLoS One 7, e52231, 10.1371/journal.pone.0052231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougeron D., Fotsing G., Barbu V. & Beauchant M. ABCB4/MDR3 gene mutations and cholangiocarcinomas. J Hepatol 57, 467–468, 10.1016/j.jhep.2012.01.025 (2012). [DOI] [PubMed] [Google Scholar]
- Wendum D. et al. Aspects of liver pathology in adult patients with MDR3/ABCB4 gene mutations. Virchows Arch 460, 291–298, 10.1007/s00428-012-1202-6 (2012). [DOI] [PubMed] [Google Scholar]
- Stutes M., Tran S. & DeMorrow S. Genetic and epigenetic changes associated with cholangiocarcinoma: from DNA methylation to microRNAs. World J Gastroenterol 13, 6465–6469 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim W. H., Jung H. Y., Yang M. H. & Kang G. H. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol 161, 1015–1022, 10.1016/S0002-9440(10)64262-9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauad T. H. et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol 145, 1237–1245 (1994). [PMC free article] [PubMed] [Google Scholar]
- Momparler R. L. A Perspective on the Comparative Antileukemic Activity of 5-Aza-2'-deoxycytidine (Decitabine) and 5-Azacytidine (Vidaza). Pharmaceuticals (Basel) 5, 875–881, 10.3390/ph5080875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information