Abstract
The National Database for Autism Research (NDAR) is a secure research data repository designed to promote scientific data sharing and collaboration among autism spectrum disorder investigators. The goal of the project is to accelerate scientific discovery through data sharing, data harmonization, and the reporting of research results. Data from over 25,000 research participants are available to qualified investigators through the NDAR portal. Summary information about the available data is available to everyone through that portal.
Keywords: Autism, database, common data elements, unique subject identifier, data federation, data repository
Introduction
Over the past decade, neuroscience research has increasingly relied on computation, informatics, and information technologies (Bjaalie, 2008; Huerta et al. 2006; Huerta and Koslow 1996; Van Horn and Ball, 2008). Concurrently, research addressing all aspects of autism spectrum disorders (ASD) has expanded significantly (Chen et al 2011; Kasari and Lawton 2010; McPheeters et al. 2011; Penagarikano et al. 2011; Scherer and Dawson 2011). The data collected in these ASD studies are highly heterogeneous, including those from clinical assessments, a wide variety of genetic and genomic measures, numerous different imaging modalities, and a range of quantitative behavioral assessments, such as eye movement tracking. Within each of these data types there also exists a large degree of diversity, as a given data type is collected from different laboratories using a plethora of protocols, a variety of instruments and differing experimental and operational conditions. Finally, ASD, itself, is characterized by a high degree of heterogeneity, with severity and symptom features varying widely across those affected.
Because of the multiple sources of heterogeneity, which compound and confound one another, no single research group has the resources, expertise, or funding to aggregate existing autism data together in a way that might allow a deeper understanding of the causes and treatment options for ASD. This heterogeneity also makes collaboration between members of the community difficult since groups would have to make a significant effort to standardize their lexicons and their data before collaboration could add value to such joint efforts.
A major objective of the National Database for Autism Research (http://ndar.nih.gov) is to explore ways to aggregate and curate existing autism data, measured in multiple laboratories, in order to speed research on the causes and treatment of ASD. When research using specific tools has sufficiently matured, this data aggregation should promote standard ways to collect and report in the autism research community. The validated, aggregated, and curated data allow researchers to explore hypotheses that would not be possible to formulate using data from a single laboratory. The database will also make it easier for researchers to reproduce and extend published results. If the database is truly successful, researchers who are not currently members of the autism research community will be able to explore what the community has deposited and will be able to suggest new ideas or approaches to gain a better understanding of autism.
As of May 2012, NDAR contains data from over 25,000 subjects. Forty of the clinical assessment or demographic data types defined in NDAR have more than 1,200 observations. Many of these are the behavioral, IQ, and cognitive data types that would be expected in an autism database. However, there are also significant data on a few environmental exposure types.
The genomics/sequencing data are just starting to arrive in the database. As of May 2012, there are microarray data available from more than 900 individuals and sequencing information is available from more than 400 individuals.
Much of the imaging data in NDAR comes from the NIH MRI Study of Normal Brain Development (Brain Development Cooperative Group, 2006), but there are images from 270 subjects with ASD, as well.
This article describes the basic structure of NDAR, the strategies that the database has used to try to overcome the issues of data heterogeneity, solutions to some of the problems in making data from human subjects widely available to the research community, and the results of these efforts so far. Readers are invited to apply for access to NDAR. Several ways to use NDAR are outlined in this paper.
NDAR Organization
The two key elements that form the basis of NDAR are a global unique identifier (GUID) for research subjects and researcher-defined data elements to describe experiments. Building on these two key components, it has been possible to develop a useful infrastructure for data sharing and to define phenotypes based on the defined data elements. Sharing data, associated tools, and methodologies, rather than just summaries or interpretations, can accelerate research progress by allowing re-analysis of data, as well as re-aggregation, integration, and rigorous comparison with other data, tools, and methods. However, this community-wide platform requires that common data definitions and standards, as well as comprehensive and coherent informatics approaches, be developed for - but more importantly, with - the active involvement of the research community (Belmonte et al. 2008).
Development of NDAR began in 2007 with support from five institutes and centers at the NIH (NIMH, NICHD, NINDS, NIEHS, and CIT). NDAR has the ability to store data itself, and many NIH awardees in the autism field are expected to submit their data to NDAR. When the project started in 2007 a great deal of ASD-related data had already been aggregated in privately funded data and bio-repositories such as the Autism Genetics Research Exchange (AGRE), the Autism Tissue Program (ATP), the Simons Foundation Autism Research Initiative (Fischbach and Lord 2010), and the Interactive Autism Network (IAN). As a result, NDAR chose to use a federation strategy linking with existing public (e.g. the NIMH Genetics Repository and dbGaP) and private repositories. The solution was much easier to implement than trying to persuade the established data repositories to donate their data to NDAR. One downside to a federation strategy is that researchers wanting to access data not held directly by NDAR need to be approved to access each data resource. While NDAR’s federation approach requires investigators to obtain permission for data access from each of its partners, NDAR allows users to browse the data available in each of the federated repositories to see if requesting data access is worthwhile.
Unique Subject Identifier – The NDAR GUID
A common subject identifier adopted by the community is essential to aggregate clinical research data involving human subjects when the personally identifiable information (PII) for each subject is not available to all researchers (Johnson et al. 2010). Research investigators who have collected a subject’s PII use NDAR-provided software to create a GUID for each subject in their study. The specific PII entered for each subject is drawn from the data available in the subject’s birth certificate. The birth certificate was chosen as the source for generating NDAR GUIDs because most subjects in ASD research are young and the information from the birth certificate is unchanging over time.
The NDAR GUID for a particular subject is the same regardless when or where it is generated. Thus, data obtained about a particular subject aggregates across laboratories, research projects, and data repositories, allowing for a consistent mechanism for the identification of a research subject across the entire research community over time. The software that creates the NDAR GUID is available only to those conducting research related to autism. This strategy offers protection against attempts to re-identify a research subject using the GUID. Using the entered information from the birth certificate, the software creates a series of one-way hash codes, encrypts them, and transmits them securely to NDAR (Johnson et al. 2010). No data other than these one-way hashes ever leaves the research site, so NDAR does not receive PII. If the same subject enrolls in research projects in different laboratories, or provides a biological specimen for a repository, the same information from his/her birth certificate is entered into the software by the second investigator and the same NDAR GUID will be generated. Since data submitted to NDAR must be associated with a GUID, these data aggregate for a given subject over space and time.
For subjects whose birth certificate data is not available, NDAR can generate a pseudo-GUID to accompany those research data. The pseudo-GUID is a random identifier and cannot be used to aggregate data between laboratories. However, the pseudo-GUID can be promoted to a standard GUID if the appropriate information is acquired. The pseudo-GUID does allow a specific laboratory to continue to deposit data about that subject at multiple time points.
The use of the NDAR GUID makes available data collected from a single participant across multiple studies by different investigators, saving effort and resources for both investigators and research subjects without redundant visits, blood draws, questionnaire administration, or imaging. Although no PII ever leaves the research site, data aggregation allows those who receive informed consent to register the same subject and see other data associated with that subject. For the majority of the community who do not have access to the NDAR GUID-generating software, there is no practical way to determine the identity of a subject. The utility of the NDAR GUID to researchers and the high level of privacy protection it offers have resulted in its wide acceptance in the autism research community. The NDAR GUID has become the standard as a patient identifier for autism research and serves as a model for similar standards in other research areas.
Data Definitions Overview
NDAR has established a data dictionary with over 250 clinical, imaging, and genomic research data definitions. These definitions were created in close collaboration with the autism research community who are using these data standards in their daily work. To contribute data to NDAR, a researcher is required to format their data in accord with an existing NDAR data definition or to define a new data definition which will be available for use by others. NDAR is written in Java, and Oracle is used for the database. In May 2012, NDAR contains over 35,000 discrete data elements and we have received over 200,000 records spread out across 250 separate data structures. NDAR hosts over 1 terabyte of imaging data and 14 terabytes of genomic data. Going forward, NDAR is planning to support roughly 100 terabytes of data. Once that point is reached, the usefulness of the deposited sequencing data will be evaluated.
The NDAR data validation tool, which is available as a community resource, enables a researcher to confirm that his or her data conforms to the existing definitions. The validation tool ensures that naming conventions are defined, NDAR GUIDs are properly registered, and the reported data are consistent with the value ranges defined in the dictionary and previously reported for that subject. The tool uses Java Web Start technology and runs on a local computer. Users submitting to NDAR must run the software prior to upload. The user specifies the data to be submitted, including any imaging or genomic files. The validation tool then downloads the NDAR data dictionary via a web service ensuring that every data cell conforms to the NDAR data standard. Once all data passes validation, only then can it be submitted to NDAR for inclusion into the database. Such a tool could easily be modified for use in other databases.
Furthermore NDAR has two additional tools that help a researcher translate their local data elements to those in the data dictionaries. There is an aliasing ability that allows the user to associate their local data element names with the NDAR data elements names. This allows the investigator to continue to use their own terms in their laboratory while at the same time allowing those variable names to be shared with the community in a consistent fashion. Second, a translate function allows the translation of values to the data standard, allowing NDAR to easily solve problems like the often described gender issue (e.g., where gender can be assigned in different laboratories as M or F, Male or Female, 0 or 1, and 1 or 2, etc.). These features, working on either the names of the data or on the data itself, permit laboratories to continue to use locally preferred terminology but allow simple translations into a single terminology for the autism community.
The data dictionary is maintained through a standard https web interface that can be opened to the community. After some trials with this open format, NDAR has found that it is better to manage changes to the data dictionary centrally. Specific instruction on the use of the data dictionary is available in the tutorials section of the NDAR website.
The NDAR approach to data definition and curation is pragmatic. All data contributed and shared with NDAR must pass validation before they are submitted. For new data structures or experiments, researchers can easily develop a new data definition, often using existing data structures as templates. Once provided, NDAR staff curate new data structures ensuring that new instruments, measures, and scientific experiments are properly defined and that common elements like gender, age, height and weight are reused. Still, the need to support heterogeneous studies, retrospective data, and other established repositories make it difficult to avoid duplication without either committing extensive resources to data migration or to require the community to adopt an established standard. Instead, NDAR’s approach is to negotiate simple accommodation with individual laboratories so most duplication is kept to a minimum.
NDAR has provided a resource to the autism research community to define, standardize and validate research data in a common way with minimal impact on the conduct of the research. We expect that this common data definition will prove to be of great value to the research community by easily promoting data sharing across labs and repositories.
This pragmatic approach does not permit the easy creation of an ontology, either between all of the data in the clinical assessments in NDAR or between the data in NDAR and other lexicons. We have chosen this strategy because of the rapid development of the ASD field. However, the ability to define phenotypes (see below) helps researchers search across data elements that are related to each other. When certain data collection instruments have become widely used in the field, we will explore the creation of a more formal lexicon.
Genomics Data Definition
NDAR’s first model of genomics data acquisition was based on Minimal Information About a Microarray Experiment (MIAME) format. While MIAME is an acceptable format for peer-to-peer data exchange, significant effort was needed by researchers to share their data with sufficient detail to ensure an experiment could be completely described.
To solve this problem, NDAR developed a genomics tool, allowing an investigator to easily provide the variables needed to define an experiment (e.g., molecule, technology, platform, extraction and processing kits). This approach provides a number of benefits. First, it allows for the easy definition of an experiment using relational database technology to ensure the reference data are defined. The tool captures the specific kits, technologies and protocols that are being used by the community. Once the experiment is defined, the laboratory can simply upload their samples and NDAR GUIDs into NDAR or an affiliated repository. Defining a genomics experiment, assuming the information is known by the researcher, takes less than 5 minutes using a one page https interface within NDAR. A tutorial explaining how to use this interface is available in the training section of the NDAR website.
While this tool makes it straightforward to upload experimental results, there are still significant bioinformatics challenges in interpreting this data (Fernald et al., 2011). NDAR is not the only data repository containing genomics and sequencing data, but it is one of the few that makes validated data available from multiple laboratories with a single data access request. As a result, NDAR should be useful in helping the community evaluate data processing pipelines.
Imaging Data Definition
A definition for genomics data has been successful largely because of the way that data are stored and made available by instrument manufacturers. Data from imaging experiments represent a much more difficult challenge (Belmonte et al. 2008). Proprietary data formats for equipment from different vendors is one problem (Bidgood et al. 1997), although standards like DICOM and NIFTI have resolved many of those issues (Cox et al. 2004, Law and Liu, 2009). In addition, correcting for variations between scanners also needs to be done to allow data sharing (Friedman et al. 2006; Mortamet et al. 2009). Furthermore, the software processing pipelines introduce significant additional complexity in trying to share imaging data (MacKenzie-Graham et al. 2008, Patel et al. 2010). For newer imaging modalities where there is less agreement in the community about how to collect or analyze data these concerns are further exacerbated.
Despite these problems, it has been possible to aggregate imaging data measured in different laboratories. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) defined a protocol for their imaging experiments and for quality control and have clearly demonstrated that it is possible to aggregate data from multiple laboratories (Weiner et al. 2010). The large number of papers that have resulted from the ADNI dataset has encouraged us to try to find ways to aggregate imaging data in NDAR even though specifying data collection protocols to achieve consistency is not yet appropriate for ASD.
Today, NDAR supports the receipt of unprocessed brain images in DICOM format and processed images in nearly all of the commonly used formats (e.g. DICOM, MINC 1.0 and 2.0, Analyze, NIfTI-1, AFNI and SPM). NDAR’s image submission tool is integrated with the NDAR data definition standard and is a freely available plug-in to the Medical Image Processing, Analysis, and Visualization application (McAuliffe et al. 2001). Using Java Web Start technologies, the image tool is downloaded locally to a user’s computer on virtually any platform. The imaging tool then consumes the NDAR image format and extracts out relevant information from the header and populates NDAR’s image data standard. For detailed information see the image tool section of the NDAR website and review the tutorial on this tool.
That tool extracts header data directly from the file headers of the many supported formats allowing embedded fields such as field strength, slice thickness, and scanner make and model to be automatically extracted and made available in the NDAR database for query access. Supporting documentation, expected with all structural and image data in NDAR, allows others to replicate results. For all images received, NDAR staff spot check the image files for PII before they are shared. Further checking of the header records for PII is not done, but the data submission agreement with users requires them to strip such information before sending the images to NDAR.
Data Federation
As discussed above, the autism research community has many important repositories of scientific data. Without the ability to search across all of these databases simultaneously, a large amount of information will be unavailable. For a disorder like ASD, missing blocks of information could easily result in an infrastructure that is not useful.
Each data repository is bound by existing policies, practices, ethical considerations, and legal obligations. While it is theoretically possible to make these practices consistent between a group of databases, that task is daunting. Similar issues would be involved in having a single database assume control of all of the data that currently exists in separate data infrastructures. Data federation overcomes these obstacles by allowing the data to remain in place, controlled independently by each organization, while at the same time providing significant scientific and operational conveniences for the user.
When connecting repositories, common data definitions and patient identifiers are prerequisites. Because of these requirements, federation inherently eases data aggregation over time. As a concrete example, all of the major autism data repositories are now using the NDAR GUID. Furthermore as investigators begin new studies, it should be easier for them to use the GUIDS and existing data standards. This will make data sharing much easier and will hopefully allow the field to find research areas that they would not have discovered otherwise.
The NDAR data federation only allows a one way transfer of information from the other data source. Currently, there is a point and click system that allows users to select the other databases that they want to include in their query. This is being replaced by a web service interface that enables system to system data retrieval and pipeline integration. When the web service interface has been implemented, users will be told how to apply for access to the data in other repositories that they do not already have approval to use.
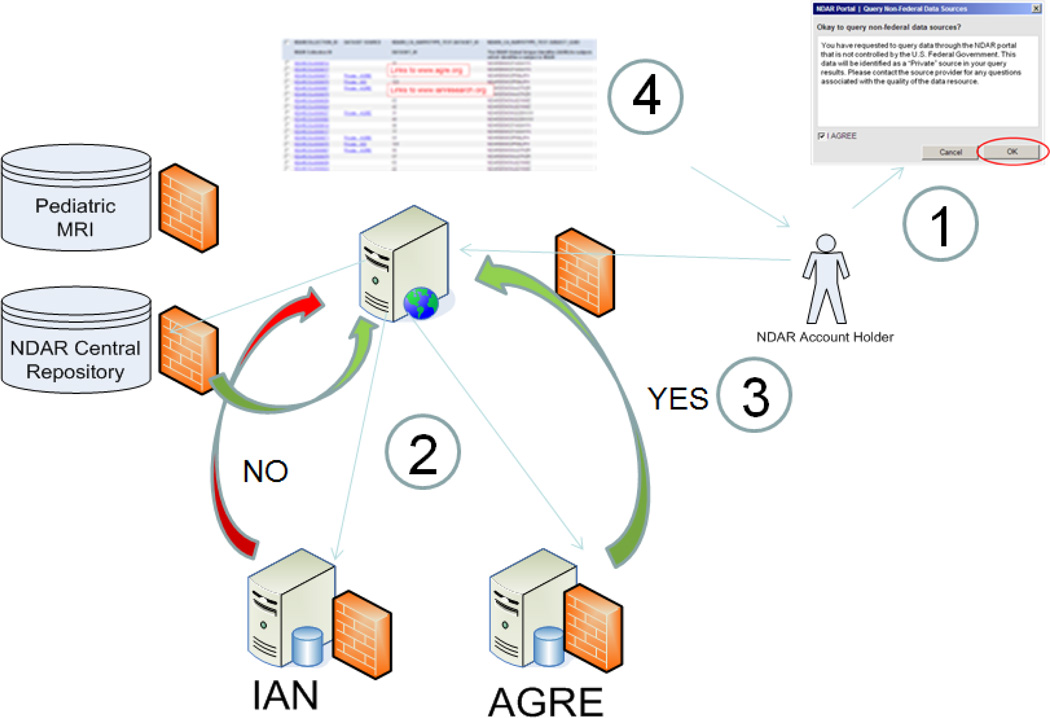
NDAR currently provides data from three private databases, the Autism Genetics Resource, the Autism Tissue Program, and the Interactive Autism Network. A schematic of how the data federation works is shown in Figure 1. NDAR is working to finalize federation agreements with other private databases. Any database with relevant information is encouraged to contact NDAR to begin the process of federating. Researchers with smaller amounts of data are encouraged to deposit that data directly into NDAR. NDAR is also a gateway to several federal databases.
Figure 1. Schematic of NDAR Data Federation.
This image shows the basic steps involved in delivering data to a user from all of the repositories federated with NDAR. In step 1, a user asks for data from multiple sites. This is currently requires the user to point and click, but this will be replaced by a web services interface shortly. In step 2, NDAR checks both the NDAR data repository as well as the data in the other repositories to see what is available. In step 3, the data available to that user is assembled. For this example, the user has been approved to receive data from AGRE but not from IAN. In step 4, the data is returned to the user.
The Autism Genetics Resource Exchange (AGRE)
The Autism Genetic Resource Exchange (http://agre.autismspeaks.org) is a collaborative gene bank for the study of autism spectrum disorders and is supported by Autism Speaks (Lajonchere et al. 2010). AGRE houses a collection of genomic and clinical data on over 1,300 well-characterized multiplex and simplex families made available to the greater scientific community.
Families with two or more members diagnosed with autism, pervasive developmental disorder not otherwise specified, or Asperger's syndrome may enroll in the AGRE program, provided they do not meet any of the exclusionary criteria. Families register for the program through the AGRE website or by completing a short registration form. AGRE recruitment staff members follow up with families to confirm eligibility and complete the intake process. Most of the data collection and blood draws take place in the family home in a standardized manner overseen by AGRE staff.
Biomaterial production and storage are located at the Rutgers University Cell and DNA Repository. A variety of phenotype data including clinical assessments such as the Autism Diagnostic Interview and the Autism Diagnostic Observation Schedule are available as well as medical histories. All of the data is available either directly on the AGRE web site or at the NDAR web site which has federated access to the AGRE database.
The Autism Tissue Program (ATP)
The Autism Tissue Program (http://www.autismtissueprogram.org) is a post-mortem brain donation program supported by Autism Speaks with the goal of accelerating the pace of neurobiological research in autism (Haroutunian and Pickett 2007). By making post-mortem brain tissue from individuals with ASD available to as many qualified scientists as possible, the ATP is able to facilitate a multidisciplinary understanding of the pathways and brain systems that are implicated in autism. It is the aim of this program to create the most biologically relevant tissue based resource for autism research by increasing the availability of brain tissue for the research community and providing scientists with the necessary resources to conduct their investigations.
As part of this effort, the ATP has obtained brain tissue from more than 170 individuals with autism, epilepsy, and from their family members and other normal controls across the lifespan (ages 4 - 64). Many of the brains in the collection have post mortem MRI images, and all have accompanying clinical phenotypic data obtained through chart review and family interviews. In addition, neuropathologic assessment, SNP analysis, microarray analysis, and epigenetic genetic data are available for many of the specimens in the collection.
The ATP Portal is a secure on-line informatics database that houses the ATP’s extensive collection of tissue information, clinical data and scientific information (i.e. genetic data) generated by researchers who accessed the tissue. The ATP Portal also functions as a web-based interface for researchers to check the inventory of available brain tissue, query the database, download clinical phenotype data and documents supportive of tissue based research, submit applications for tissue, track tissue distributions as well as curate the ATP’s dynamic tissue repository. The ATP has federated many of these data sources with NDAR, providing access to a variety of data sets including MRIs of the donor brain tissue.
The Interactive Autism Network (IAN)
The Interactive Autism Network (http://ianproject.org/), a project of the Kennedy Krieger Institute and funded by Autism Speaks, the Simons Foundation, and the National Institute of Mental Health, focuses on engaging the public in autism research and connecting families to the researcher community. IAN is made up of two interrelated components: IAN Community and IAN Research.
To engage the public in ASD research and facilitate research participation, retention, and literacy, IAN Community provides the public with family-friendly, evidence-based information about ASD and ASD research. During the past year, IAN Community received over 1.83 million visits, with an average of 1,248 visits per day.
IAN Research is a research database and a research registry. Since 2007, IAN has engaged over 38,000 participants in its innovative online research. These participants include more than 14,000 children with ASD; 800 adults with ASD; 9,000 siblings; and over 7,000 parents or legally authorized representatives. They have participated in a lengthy self-report protocol consisting of baseline questionnaires, standardized instruments, and one-time surveys.
The data that IAN has collected, along with the generous involvement of the IAN families, has enabled IAN to provide much-needed subject recruitment assistance to over 300 autism research projects that encompass the full range of ASD human subjects research in the United States. This means that many IAN Research participants have been recruited and have participated in non-IAN research studies throughout the United States.
Through NDAR, researchers have access to the rich phenotypic data provided by IAN and will have additional information about many of these IAN subjects that was gathered by other research projects. As of May, 2012, 8,500 IAN research subjects had consented to participate in NDAR, with more participants consenting daily.
Federal Databases
In addition to the private databases federated with NDAR, the system links research data with databases of the federal government that provide information about grant awards, about clinical trials, and about publications associated with the data. This information is being provided under the Data from Labs tab on the NDAR website as well as in the Autism Publications tab.
NDAR has become the data repository that houses the data measured in the NIH Study of Normal Brain Development (Evans et al. 2006). That study enrolled over 500 children and studied the development of those children using clinical/behavioral measures as well as structural MRI, diffusion tensor imaging, and MR spectroscopy. Researchers can apply to NDAR to access this data and they can use this data set for comparison with similar data from subjects with ASD.
NDAR is linking to dbGaP, dbVar, and the Sequence Read Archive, the federal data repositories containing genetic/genomic data. To ensure consistency in reporting the results from experiments performed in different laboratories, NDAR validates the data when it is deposited and holds the sequencing or microarray data. Entries are made in the other appropriate data base. Researchers interested in only this sort of data can apply to access the data from either NDAR or the other data repository. No matter where the approval came from, the data will be served from NDAR.
NDAR is in the process of linking to the data repository associated with the NIMH Human Genetics Initiative. When complete, NDAR users will be able to see when biosamples are available from the subjects that provided the clinical or genetic/genomic data in NDAR.
NDAR also provides information drawn from NLM’s PubMed for papers and the NIH Reporter for grant awards. The data available in NDAR associated with a paper listed in PubMed is made available using their LinkOut feature. This allows a reader to access the NDAR data with only a few key clicks.
Linking these federal databases with NDAR allows the research community to find a wealth of information about various aspects of autism in one location.
Using NDAR
NDAR currently offers a variety of different ways to search through the data base. The home page of the web site allows a user to enter a few variables (gender, age, phenotype, or verbal IQ) and see how many subjects are available in that cohort and what sort of data is available. The available federated repositories can be included or excluded from this search. The search feature allows a researcher who does not have access to NDAR (or to some of the federated repositories) to see if there is enough data to warrant applying for access. In the near future, NDAR will show the range of values in each data cell as well as the average value in the cell. This will allow users to find summary information very easily and will be a much simpler way to categorize and download data from NDAR and associated data repositories.
The number of phenotypes currently defined in NDAR is relatively small, but is growing with community input. The software logic involves defining a phenotype using an Oracle based rules engine. Through this rules engine it is possible to make new phenotypes based on clinical or demographic data (e.g. seizures, autism regression, gestational age) or on data collection parameters (e.g. image modality, RNA extraction kits) quickly, allowing the research community to find the data of interest to them. Turn-around time from request for a phenotype to implementation generally takes two business days.
Defining phenotypes both makes it easier to see what sort of data is available and helps establish quantitative standards in terms of the NDAR data dictionaries for subphenotypes like “mildly affected with autism-like developmental disorders”. With information from over 25,000 subjects in the database, it is possible to begin to empirically validate rules across many measures to define broad phenotypic categories. The exact data elements that define those sub-phenotypes are published on the NDAR web site. This approach allows researchers to easily search through the data and allows researchers to see what happens if specific parts of the sub-phenotype are altered. As more data become available and as the research community explores the data and contributes more granular data, the definitions of existing and yet-to-be-discovered subphenotypes should become more accurate.
As the ASD field develops, new phenotypes will be created. These phenotypes will be suggested by publications and by NDAR users asking for such search short cuts. We encourage the imaging community to help identify ways to define useful autism phenotypes that include imaging data. If such phenotypes require the use of specific data processing pipelines, NDAR will help implement those pipelines for community use.
A second way to search through the database is to find data associated with a particular laboratory. When data is received from a laboratory it is assembled in a collection listed in the Data from Labs section of the website. By clicking the button next to the collection, a user is prompted to login and can then quickly download the data. Before requesting access, a researcher can see the data dictionaries that were used to deposit the data, the number of observations, and the number of unique subjects. When the number of observations is different from the number of unique subjects, data were acquired on a subject more than one time. Most NIH awardees deposit their data twice a year.
A third way to search through the database is to use the Data from Papers tab. This tab allows users to see the exact data that are associated with a publication or any other study. Allowing easy download of the data associated with a publication facilitates the replication and extension of results. This feature need not be associated with a published paper. For example, users who wanted to calculate the volume of their favorite brain regions could do such calculations on all of the images in NDAR and upload that data in this section. With a relatively large number of images made available in one site, NDAR is a good source of raw data to compare results from different processing pipelines and to see which of those derived results correlate with data from the clinical and environmental assessments or with the demographic information about research subjects. Gorrindo et al. (2012) have used the Data by Papers feature in NDAR to associate the specific data that was used in the publication with the article. The data are available by searching in NDAR and by using the LinkOut feature in PubMed.
Although the amount of data in NDAR is just now reaching the point where it will be widely useful to the research community, the database has already been used to as a private collaboration space to allow a research team interested in Fragile X to assemble and analyze their data (Sansone, et al. 2011). After that paper was published, the data were shared with the entire community in NDAR.
Human Subjects
Sharing research data from human subjects is never simple. If such a database is supported by a private institution, the responsible Institutional Review Board often places significant limits on the way that data can be shared and even stronger limits on data re-distribution. As data re-analysis and the aggregation and analysis of data from multiple sites becomes more important, these limits on re-distributing data can place very serious constraints on the science that can be accomplished.
Databases operated by the federal government can be somewhat more flexible than those distributed by private institutions. The reason for this is that access to such a database can be tied to the Federal Wide Assurance agreement of the institution that employs the researcher (http://www.hhs.gov/ohrp/assurances/assurances/filasurt.html). Violation of the Federal Wide Assurance agreement could jeopardize the ability of the entire institution to conduct any research involving human subjects. That potential allows federal databases to be somewhat more open in sharing data with the research community.
Researchers who want access to NDAR must apply and have their institutional official co-sign the application. The request is evaluated by a data access committee composed of NIH program staff. If approved, access is granted to NDAR for one year. Investigators who want to access data in one of the federated databases need to complete an access agreement with that database. Once such an agreement is in place, the investigator can receive the information directly from NDAR as if the data were stored in a single location.
The data in NDAR are not shared as freely as the imaging data available in the 1000 Functional Connectomes Project (Milham, 2012). Those data have very little phenotypic information about the subject in contrast to the significant phenotypic data that can be associated with an image in NDAR. Because of the concerns about potential subject re-identification, NDAR follows the same guidelines as have been set forth for sharing GWAS data (http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html and http://gwas.nih.gov/).
Conclusions
As of May 2012, NDAR contained over 200,000 records from more than 25,000 research participants. Those records can be easily browsed by the public on the NDAR web site, and if a researcher finds that NDAR contains data that might be useful, the full data will be available after their request to access the data has been approved. The web site also provides easy data browsing by the investigator who submitted the data or by the paper that was published using the data. In addition, the NDAR data dictionaries describing the format for all of the submitted data can be browsed without requiring special access. The phenotype and subphenotype definitions used by NDAR can also be accessed via the web site.
The need for shared conventions and standards in the ASD research community has become increasingly urgent. NDAR’s ultimate goal is to enable the community to integrate and standardize data, analysis tools, and data collection methods to accelerate scientific discovery. Common standards such as the GUID and a variety of data definitions and data validation tools enable NDAR to organize heterogeneous research data across many laboratories and repositories. The current challenge for both the ASD community and the bioinformatics community is to see if the curated heterogeneous data that were not originally measured with the thought of deposition into a database will allow new hypotheses to be generated. An additional challenge for NDAR is to standardize data collection and description without stifling the innovation that is needed to understand the causes of and treatments for ASD.
Footnotes
Information Sharing Statement
The data in NDAR are available to researchers who are approved by the NDAR data access committee. Instructions for requesting access can be found at http://ndar.nih.gov. Many of the components of the NDAR software infrastructure are also freely available. Requests for software should be directed to ndarhelp@mail.nih.gov.
Contributor Information
Dan Hall, OMNITEC Solutions, Inc., 6001 Executive Boulevard, Suite 7161, Rockville, MD 20892-9640, 301-443-7156, halldan@mail.nih.gov.
Michael F. Huerta, Office of Health Information Programs Development, National Library of Medicine, 8600 Rockville Pike, Bethesda, MD 20894, 301-496-8834, mike.huerta@nih.gov
Matthew J. McAuliffe, Center for Information Technology, National Institute of Health, 12 South Drive, Room 2041, Bethesda, MD 20892, 301-594-2432, matthew.mcauliffe@nih.gov
Gregory K. Farber, Office of Technology Development and Coordination, National Institute of Mental Health, National Institutes of Health, 6001 Executive Boulevard, Suite 7162, Rockville, MD 20892-9640, 301-435-0778, farberg@mail.nih.gov
References
- Brain Development Cooperative Group. Evans AC. The NIH MRI Study of Normal Brain Development. NeuroImage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Mazziotta JC, et al. Offering to Share: How to Put Heads Together in Autism Neuroimaging. J. Autism and Dev. Disord. 2008;38(1):2–13. doi: 10.1007/s10803-006-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidgood WD, Horii SC, et al. Understanding and Using DICOM, the Data Interchange Standard for Biomedical Imaging. J. Am. Med. Inform. Assoc. 1997;4(3):199–212. doi: 10.1136/jamia.1997.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjaalie JG. Understanding the Brain Through Neuroinformatics. Front. Neurosci. 2008;2(1):19–21. doi: 10.3389/neuro.01.022.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jaio Y, Herskovits EH. Structural MRI in Autism Spectrum Disorder. Pediatr. Res. 2011;69(5 Pt 2):63R–68R. doi: 10.1203/PDR.0b013e318212c2b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, Ashburner J, et al. A (Sort of) New Image Data Format Standard: NifTI-1. Abstrace, 10th Annual Meeting of the Organization for Human Brain Mapping; June 13–17; Budapest, Hungary. 2004. [Google Scholar]
- Evans AE the Brain Development Cooperative Group. The NIH MRI Study of Normal Brain Development. NeuroImage. 2006;30(1):184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Fernald GH, Capriotti E, et al. Bioinformatics Challenges for Personalized Medicine. Bioinformatics. 2011;27(13):1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Friedman L, Glover GH, et al. Reducing Interscanner Variability of Activation in an Multicenter fMRI Study: Controlling for Signal-to-Fluctuation-Noise-Ratio (SFNR) Differences. Neuroimage. 2006;33(2):471–481. doi: 10.1016/j.neuroimage.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Gorrindo P, Williams KC, et al. Gastrointestinal Dysfunction in Autism: Parental Report, Clinical Evaluation, and Associated Factors. Autism Res. 2012;5(2):101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V, Pickett J. Autism Brain Tissue Banking. Brain Pathol. 2007;17(4):412–421. doi: 10.1111/j.1750-3639.2007.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Szelinger S, et al. Resolving Individuals Contributing Trace Amounts of DNA to Highly complex Mixtures Using High-Density SNP Genotyping Microarrays. PLoS Gene. 2008;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta MF, Koslow SH. Neuroinformatics: Opportunities across Disciplinary and National borders. Neuroimage. 1996;4(3):S4–S6. doi: 10.1006/nimg.1996.0040. [DOI] [PubMed] [Google Scholar]
- Huerta MF, Liu Y, et al. A view of the Digital Landscape for Neuroscience at NIH. Neuroinformatics. 2006;4(2):131–137. doi: 10.1385/NI:4:2:131. [DOI] [PubMed] [Google Scholar]
- Johnson SB, Whitney G, et al. Using Global Unique Identifiers to Link Autism Collections. J. Am. Med. Inform. Assoc. 2010;17(6):689–695. doi: 10.1136/jamia.2009.002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasari C, Lawton K. New Directions in Behavioral Treatment of Autism Spectrum Disorders. Curr. Opin. Neurol. 2010;23(2):137–143. doi: 10.1097/WCO.0b013e32833775cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MYY, Liu B. DICOM-RT and Its Utilization in Radiation Therapy. Radiographics. 2009;29(3):655–667. doi: 10.1148/rg.293075172. [DOI] [PubMed] [Google Scholar]
- Lajonchere CM the AGRE Consortium. Changing the Landscape of Autism Research: The Autism Genetic Resource Exchange. Neuron. 2010;68:187–191. doi: 10.1016/j.neuron.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Graham AJ, Van Horn JD, et al. Provenance in Neuroimaging. Neuroimage. 2008;42(1):178–195. doi: 10.1016/j.neuroimage.2008.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe MJ, Lalonde FM, et al. Medical Image Processing, Analysis & Visualization in Clinical Research. 14th IEEE Symposium on Computer-Based Medical Systems (CBMS); 2001. pp. 381–386. [Google Scholar]
- McPheeters ML, Warren Z, et al. A Systematic Review of Medical Treatments for Children with Autism Spectrum Disorders. Pediatrics. 2011;127(5):e1312–e1321. doi: 10.1542/peds.2011-0427. [DOI] [PubMed] [Google Scholar]
- Milham MP. Open Neuroscience Solutions for the Connectome-wide Association Era. Neuron. 2012;73(2):214–218. doi: 10.1016/j.neuron.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Mortamet B, Bernstein MA, et al. Automatic Quality Assessment in Structural Brain Magnetic Resonance Imaging. Magn. Reson. Med. 2009;62(2):365–372. doi: 10.1002/mrm.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Dinov ID, et al. LONI MiND: Metadata in NIfTI for DWI. Neuroimage. 2010;51(2):665–676. doi: 10.1016/j.neuroimage.2010.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, et al. Absence of CNTNAP2 Leads to Epilepsy, Neuronal Migration Abnormalities, and Core Autism-Related Deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone SM, Widaman KF, et al. Psychometric Study of the Aberrant Behavior Checklist in Fragile X Syndrome and Implications for Targeted Treatment. J. Autism. Dev. Disord. 2011 doi: 10.1007/s10803-011-1370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Dawson G. Risk Factors for Autism: Translating Genomic Discoveries into Diagnostics. Hum. Genet. 2011;130(1):123–148. doi: 10.1007/s00439-011-1037-2. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Ball CA. Domain-Specific Data Sharing in Neuroscience: What Do We Have to Learn from Each Other? Neuroinform. 2008;6(2):117–121. doi: 10.1007/s12021-008-9019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Aisne PS, et al. The Alzheimer’s Disease Neuroimaging Initiative: Progress Report and Future Plans. Alzheimers Dement. 2010;6(3):202–211. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]