Abstract
Significant advances in our understanding of continued androgen receptor (AR) signaling in castration-resistant prostate cancer have led to the development and FDA approval of two next-generation androgen-directed therapies, abiraterone and enzalutamide. These new therapies heralded a new era of prostate cancer therapy. However, disease progression during androgen-directed therapies remains the most critical challenge in the clinical management of prostate cancer. Accumulating evidence points to an important contribution of constitutively-active AR splice variants to AR-driven tumor progression during androgen-directed therapies. In this review, we will focus on the structure, activity, detection, clinical relevance, and mechanisms of production of AR splice variants.
Keywords: Androgen receptor, AR splice variants, castration resistance, prostate cancer
Introduction
Prostate cancer is the most common non-skin cancer and the second leading cause of cancer mortality in men in the United States. Androgen deprivation therapy, which disrupts androgen receptor (AR) signaling by reducing androgen levels through surgical or chemical castration, or by administration of anti-androgens that compete with androgens for binding to AR [2], is the first-line treatment for metastatic and locally advanced prostate cancer. While this regimen is effective initially, progression to the presently incurable and lethal stage, termed castration-resistant prostate cancer (CRPC), invariably occurs [1,2]. With a median survival of ~16-18 months [3], CRPC accounts for the majority of disease-related mortality. Mounting evidence suggests that resurgent AR drives therapeutic failure and castration-resistant progression [1,2]. A number of ligand-dependent and -independent mechanisms have been proposed to underlie AR reactivation during androgen deprivation therapy [1,2]. While these mechanisms are thoroughly reviewed by many, this review is focused on the discussion of the role of constitutively-active AR splice variants that lack the functional ligand-binding domain (LBD) in AR-signaling reactivation.
Structure and activity of the AR splice variants
The canonical AR protein is 919 amino acids long, encoded by 8 exons on the X-chromosome (Xq12) [4]. Structurally, the full-length AR (AR-FL) resembles other members of the steroid receptor family, consisting of 4 domains (Figure 1). The N-terminal domain (NTD) contains an activation function 1 (AF1) domain that functions as a ligand-independent transcriptional activation domain. Another important function of NTD is recruitment of coregulators. The DNA-binding domain (DBD) contains two zinc fingers that are involved in DNA recognition, dimerization, and stabilization. The hinge domain (H) provides flexibility to the protein and regulates the nuclear translocation of the receptor through a canonical nuclear localization signal. The C-terminal LBD contains a ligand-binding pocket that mediates high affinity ligand-binding. A second activation function domain (AF2) is located in the LBD and regulates transcriptional activation in a ligand-dependent manner. All hormonal therapies currently accepted in the clinic for treatment of prostate cancer, including the recent FDA-approved abiraterone [5] and enzalutamide [6], target the LBD de facto.
Figure 1.
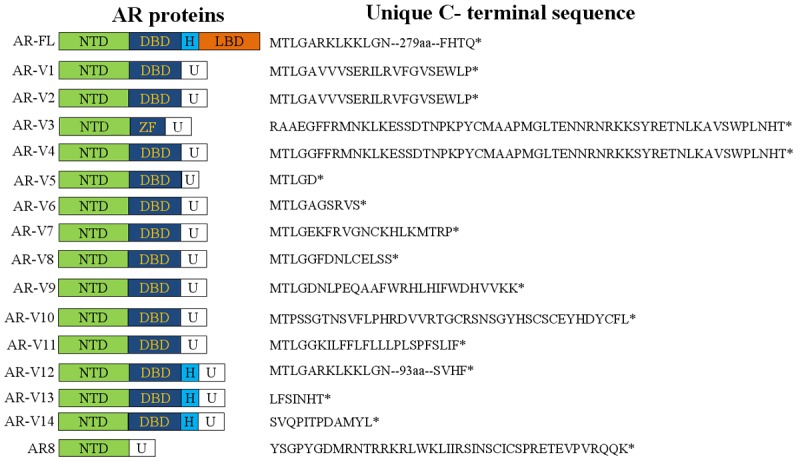
Schematic representation of the structure of AR-FL and AR-V proteins. U, unique sequence; ZF, zinc finger.
Recently, a cadre of AR splice variants (AR-Vs) that are devoid of a functional LBD have been identified (Figure 1). Structurally, these variants either have insertions of cryptic exons immediately downstream of the exons encoding the DBD or have deletions of the exons encoding the LBD, resulting in a disrupted AR open reading frame and the expression of truncated proteins [7-12]. Since the NTD and DBD remain intact in the majority of the AR-Vs identified to date, many variants display constitutive activity [7-12]. Others are considered conditionally active because these variants display ligand-independent activity only in certain cell models [12]. Two major AR-Vs, AR-V7 (also known as AR3) and ARv567es (a.k.a. AR-V12), have been shown to be capable of regulating target gene expression in the absence of the AR-FL signaling [8-10,13]. Gene expression profiling showed that AR-V7 and ARv567es regulate the expression of both canonical androgen-responsive genes and a distinct set of targets enriched for cell-cycle function [9,10,13]. However, there exists significant difference in the AR-V transcriptome identified by different studies [8-10,13], possibly due to the use of different model systems.
Notably, not all AR-Vs function as a transcription factor. For example, AR8 lacks a functional DBD, and has been shown to localize mainly to the plasma membrane [14]. AR8 may play a role in mediating Src kinase activation as well as AR-FL tyrosine phosphorylation and subsequent nuclear translocation in response to EGF treatment, possibly by forming a membrane-associated signaling complex that includes AR8, AR-FL, Src kinase, and EGFR [14]. Depletion of AR8 by RNA interference compromised EGF-induced Src activation and AR phosphorylation, as well as inhibited cell proliferation and induced cell death [14]. Thus, AR8 activates the AR signaling pathway and promotes cell survival via a nongenomic mechanism.
Potentiation of AR-FL activity is not limited to the transcriptionally inactive AR8. In cells co-expressing ARv567es and AR-FL, ARv567es could bind to AR-FL and facilitate the nuclear translocation of AR-FL in the absence of ligand [10]. Similarly, AR-V7 could also facilitate the nuclear translocation of AR-FL in an androgen depleted condition or in the presence of the potent AR antagonist enzalutamide (Cao et al., manuscript under review).
Detection of AR-V expression
The majority of AR-V transcripts can be detected by reverse transcription polymerase chain reaction (RT-PCR), taking advantage of their unique exon compositions and exon-exon junctions. A collation of published PCR primers for AR-Vs is presented in Table 1. Although RT-PCR provides a sensitive and specific assay for the mRNA, the results do not always correlate with protein expression. For example, the transcript, but not the protein product, of AR-V7 has been detected in the LNCaP cell line [15]. To date, isoform-specific antibodies have only been reported for AR-V7 [8,9] and AR8 [14], and the only commercially available antibody is for AR-V7 (Precision Antibodies, Columbia, MD). To overcome this limitation, Zhang et al. developed an immunohistochemical assay to detect the expression of AR-Vs by using two antibodies recognizing the N- and C-terminus of AR, respectively [16]. A decrease of AR nuclear staining by the C-terminal antibody when compared to that by the N-terminal antibody was used to indicate the presence of LBD-truncated, transcriptionally active AR-Vs. The authors reported a significant loss in AR C-terminal nuclear immunoreactivity in CRPC specimens, but not in tissues from primary cancers, suggesting the prevalence of AR-Vs in CRPC. The results were further corroborated by RT-PCR [16]. Despite the demonstrated success, this approach requires significant efforts in assay optimization and standardization. There is a great demand for the development of additional isoform-specific antibodies for AR-Vs. The unique C-terminal peptide sequences of AR-Vs are presented in Figure 1.
Table 1.
Isoform-specific PCR primers for detecting AR-Vs
| AR-Vs | Alias | PCR Primers | Primer locations | Product size (bp) | Ref. |
|---|---|---|---|---|---|
| AR-V1 | AR4 | F: 5’-CCATCTTGTCGTCTTCGGAAATGTTATGAAGC-3’ | Exon 3 | 149 | [8] |
| R: 5’-CTGTTGTGGATGAGCAGCTGAGAGTCT-3’ | CE1 | ||||
| F: 5’-CTACTCCGGACCTTACGGGGACATGCG-3’ | Exon 1 | 322 | [9] | ||
| R: 5’-GATTCTTTCAGAAACAACAACAGCTGCT-3’ | Exon 3/CE1 | ||||
| AR-V2 | N/A | [8] | |||
| AR-V3 | AR1/2/2b | N/A | [7,8] | ||
| AR-V4 | AR1/2/3/2b, AR5 | F: 5’-CTACTCCGGACCTTACGGGGACATGCG-3’ | Exon 1 | 323 | [9] |
| R: 5’-CTTTTAATTTGTTCATTCTGAAAAATCCTC-3’ | CE2 | ||||
| AR-V5 | N/A | [8] | |||
| AR-V6 | N/A | [8] | |||
| AR-V7 | AR3 | F: 5’-CCATCTTGTCGTCTTCGGAAATGTTATGAAGC-3’ | Exon 3 | 125 | [8] |
| F: 5’-TTTGAATGAGGCAAGTCAGCCTTTCT-3’ | CE3 | ||||
| F: 5’-CTACTCCGGACCTTACGGGGACATGCG-3’ | Exon 1 | 314 | [9] | ||
| R: 5’-TGCCAACCCGGAATTTTTCTCCC-3’ | Exon 3/CE3 | ||||
| AR-V8 | N/A | [11] | |||
| AR-V9 | F: 5’-CCATCTTGTCGTCTTCGGAAATGTTATGAAGC-3’ | Exon 3 | 128 | [12] | |
| R: 5’-TTAGTTCTACTTCTTAACAACGTGATCCCA-3’ | CE5 | ||||
| AR-V10 | N/A | [11] | |||
| AR-V11 | N/A | [11] | |||
| AR-V12 | ARv567es | F: 5-GCCTTGCCTGATTGCGAG | Exons 4/8 | 64 | [12] |
| R: 5’-CATGTGTGACTTGATTAGCAGGTCAAA | Exon 8 | ||||
| F: 5’-CCAAGGCCTTGCCTGATTGC-3’ | Exons 4/8 | 124 | [10] | ||
| R: 5’-TTGGGCACTTGCACAGAGAT-3’ | Exon 8 | ||||
| AR-V13 | N/A | [12] | |||
| AR-V14 | N/A | [12] | |||
| AR8 | F: 5’-CGACTTCACCGCACCTGATG-3’ | Exon 1 | 150 | [14] | |
| R: 5’-CTCTTTCTTCGGGTATTTCGCATG-3’ | Exons 1/3’ |
Clinical relevance of AR-Vs
AR-Vs are prevalently upregulated in CRPC compared to hormone-naïve cancers, and may emerge as an adaptive response to therapies targeting the androgen signaling axis [8-11,16,17]. It is important to recognize the existence of discrepancy between the relative abundance of AR-V mRNAs and that of AR-V proteins in clinical specimens. Although the levels of AR-V mRNAs in metastatic CRPCs have been shown to constitute at most 7% of the AR-FL mRNA level [11,17], Western analyses of 13 CRPC bone metastases demonstrated that the levels of AR-V proteins could constitute a median of 32% of the AR-FL protein level (ranging from 0 to 95%) [17]. In fact, in 38% of these CRPC bone metastases, the AR-V proteins are expressed at a level comparable to that of the AR-FL protein [17]. The relative high abundance of AR-V proteins is also supported by data from immunohistochemistry analysis. With the use of an antibody specific to AR-V7, several groups show that AR-V7 is readily detectable in prostate cancer specimens [9,13,18]. Using the two-antibodies approach described above, Zhang and colleagues analyzed 50 primary prostate cancer and 162 metastatic CRPC tissues and found that 24% of these CRPC tissues display a staining pattern similar to that of the LuCaP86.2 xenograft, which predominantly expresses AR-V [16].
It is also important to recognize that the absolute levels of AR-Vs may not be as important as that of AR-FL for their respective activity. This is because AR-FL is located in the cytoplasm in the absence of ligand and translocates to the nucleus and activates target-gene expression upon ligand binding, whereas constitutively-active AR-Vs localize to the nucleus and activate target-gene expression in the absence of ligand [8-12,19]. Strikingly, higher expression of AR-V7 in hormone-naïve prostate tumors predicts increased risk of biochemical recurrence following radical prostatectomy [8,9], and patients with high levels of expression of AR-V7 or detectable expression of ARv567es have a significantly shorter survival than other CRPC patients [17], indicating an association between AR-V expression and a more lethal form of prostate cancer. Collectively, the existing data support that AR-V proteins are expressed at a significant level in clinical specimens and should not be trivialized simply based on their relative low mRNA abundance.
Preclinical studies have pointed to an important role of AR-Vs in mediating castration resistance. Ectopic expression of AR-V7 or ARv567es confers castration-resistant growth of LNCaP xenograft tumors [9-11]. Conversely, knockdown of AR-V7 attenuates the growth of castration-resistant 22Rv1 xenograft tumors [9]. Targeted expression of ARv567es in prostate epithelium induces de novo prostate cancer development and promotes castration-resistant progression of the tumors in transgenic mice [20]. Although only prostatic intraepithelial neoplasia lesions are observed in AR-V7 transgenic mice, the majority of AR-V7-positive cells in castrated AR-V7 transgenic mice are ck5+/ck8+ intermediate cells, indicating a role of AR-V7 in maintaining or expanding prostate progenitor cell population during androgen deprivation [21].
AR-Vs have also been indicated to confer resistance to abiraterone and enzalutamide in preclinical studies. AR-Vs are increased in CRPC xenografts that recurred after abiraterone [22] or enzalutamide treatment (Cao et al., manuscript under review). Knockdown of AR-Vs sensitizes 22Rv1 cells and NFκB p52-transfected LNCaP cells to enzalutamide inhibition of growth [23,24]. Reducing AR-V levels with small-molecule drugs improves enzalutamide efficacy against the growth of 22Rv1 cells and xenografts [25]. Intriguingly, ectopic expression of AR-V7 in AR-FL-overexpressing LNCaP xenografts does not affect the growth inhibitory efficacy of enzalutamide [11]. A plausible explanation for the discrepancy is that, in the context of AR overexpression, the growth of LNCaP tumors may be driven mainly by the AR-FL signaling, making enzalutamide highly effective irrespective of AR-V expression. Nonetheless, our data showed that, when the ectopically-expressed AR-FL is lost in these xenografts, they can become resistant to enzalutamide, and the resistance is accompanied by increased expression of AR-Vs (Cao et al., manuscript under review). Thus, prostate cancers may evade all androgen-directed therapies through shifting towards AR-V-mediated signaling.
Mechanisms of AR-V production
Two mechanisms have been proposed for AR-V production, AR gene rearrangement [26,27] and increased pre-mRNA splicing [23]. Modeling gene rearrangement in prostate cancer cells showed expression of ARv567es without AR-FL in clonally selected cells [27]. While AR gene rearrangement could contribute to AR-V production in a subset of prostate cancers, AR-V production at the expense of AR-FL appears to be inconsistent with the tight correlation between AR-V and AR-FL mRNAs observed in individual clinical specimens and in xenograft [11,17] or co-expression of AR-FL and AR-V7 in CRPC specimens, as indicated by overlapping AR-FL and AR-V7 immunohistochemistry staining of adjacent tumor sections [13]. Moreover, while a clonal selection process is required for gene-rearrangement-mediated AR-V production to be manifested at the level of tumor tissues, change in AR-V levels in response to androgen deprivation was rather rapid in xenograft tumors [10,11]. Further, different AR-Vs can be expressed in the same tissues. Clonal expansion of cells with one type of gene arrangement could lead to expression of one specific AR-V but may not be able to account for the expression of different AR-Vs. Compared to gene arrangement, increased splicing appears to be more generalizable. RNA splicing is closely coupled with gene transcription [28]. Androgen deprivation was shown to enhance the rate of AR-gene transcription and thereby indirectly contribute to increased AR pre-mRNA splicing to produce both AR-FL and AR-V7 [23]. A comprehension of the mechanisms of AR-V production is paramount for developing effective means to suppress AR-V expression.
Conclusion
AR signaling is active in CRPC although the cancer is no longer responsive to androgen deprivation therapy. LBD-truncated AR splice variants not only may play a role in maintaining the canonical AR transcriptome in a genuine ligand-independent manner, but may also regulate a unique subset of target genes. Accumulating clinical and preclinical data suggest that AR-Vs are critically involved in the treatment failure of first- and second-line hormonal therapies. Therefore, targeting the AR-Vs appears to an important concept and a fruitful direction of therapeutic development. To this end, several natural or synthetic compounds have been shown pre-clinically to inhibit AR-V actions [18,29-34], and proof of efficacy in clinical trials is keenly awaited. Furthermore, the expression of constitutively active AR-Vs could serve as a prognostic and response biomarker to guide treatment decisions.
Acknowledgements
We are grateful to Dr. Stephen Plymate for in-depth discussions and communications on this subject. This work was supported by the following grants: DOD W81XWH-12-1-0112, National Natural Science Foundation of China 81272851 (Y.D.) W81XWH-12-1-0275, Louisiana Board of Regents Grant LEQSF (2012-15)-RD-A-25 (H.Z.), Louisiana Cancer Research Consortium Fund (Y.D. and H.Z.), and the Laborde endowed professorship (O.S.).
References
- 1.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: Adaptive responses in the androgen axis. Cancer Treat Rev. 2013 Sep 14; doi: 10.1016/j.ctrv.2013.09.011. [Epub ahead of print] . Available at: http://www.sciencedirect.com/science/article/pii/S0305737213002004. [DOI] [PubMed] [Google Scholar]
- 2.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–71. doi: 10.1158/1078-0432.CCR-06-0067. [DOI] [PubMed] [Google Scholar]
- 4.Bennett NC, Gardiner RA, Hooper JD, Johnson DW, Gobe GC. Molecular cell biology of androgen receptor signalling. Int J Biochem Cell Biol. 2010;42:813–27. doi: 10.1016/j.biocel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 5.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB Jr, Saad F, Staffurth JN, Mainwaring P, Harland S, Flaig TW, Hutson TE, Cheng T, Patterson H, Hainsworth JD, Ryan CJ, Sternberg CN, Ellard SL, Fléchon A, Saleh M, Scholz M, Efstathiou E, Zivi A, Bianchini D, Loriot Y, Chieffo N, Kheoh T, Haqq CM, Scher HI COU-AA-301 Investigators. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Fléchon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS AFFIRM Investigators. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, Bova GS, Luo J. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ, Brodie AM, Edwards J, Qiu Y. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, Nelson PS, Plymate SR. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–65. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–67. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, Plymate SR, Luo J. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H, Chen M, Brodie AM, Chen H, Xiao Z, Veenstra TD, Qiu Y. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem. 2011;286:36152–60. doi: 10.1074/jbc.M111.265124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haile S, Sadar MD. Androgen receptor and its splice variants in prostate cancer. Cell Mol Life Sci. 2011;68:3971–81. doi: 10.1007/s00018-011-0766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, Vakar-Lopez F, Vessella RL, Plymate SR. Androgen Receptor Variants Occur Frequently in Castration Resistant Prostate Cancer Metastases. PLoS One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hörnberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, Bergh A, Wikström P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita S, Lai KP, Chuang KL, Xu D, Miyamoto H, Tochigi T, Pang ST, Li L, Arai Y, Kung HJ, Yeh S, Chang C. ASC-J9 Suppresses Castration- Resistant Prostate Cancer Growth through Degradation of Full-length and Splice Variant Androgen Receptors. Neoplasia. 2012;14:74–83. doi: 10.1593/neo.111436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan SC, Li Y, Dehm SM. Androgen Receptor Splice Variants Activate Androgen Receptor Target Genes and Support Aberrant Prostate Cancer Cell Growth Independent of Canonical Androgen Receptor Nuclear Localization Signal. J Biol Chem. 2012;287:19736–49. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G, Sprenger C, Sun S, Epilepsia KS, Haugk K, Zhang X, Coleman I, Nelson PS, Plymate S. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia. 2013;15:1009–17. doi: 10.1593/neo.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun F, Chen HG, Li W, Yang X, Wang X, Jiang R, Guo Z, Chen H, Huang J, Borowsky AD, Qiu Y. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem. 2013 Dec 2; doi: 10.1074/jbc.M113.492140. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, Nelson PS, Montgomery RB. Resistance to CYP17A1 inhibition with abiraterone in castration resistant prostate cancer: Induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17:5913–25. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2013 doi: 10.1038/onc.2013.284. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP, Gao AC. NF-κB2/p52 Induces Resistance to Enzalutamide in Prostate Cancer: Role of Androgen Receptor and Its Variants. Mol Cancer Ther. 2013;12:1629–37. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan Y, Cao B, Qi Y, Liu S, Zhang Q, Zhou W, Xu D, Lu H, Sartor O, Kong W, Zhang H, Dong Y. Methylselenol prodrug enhances MDV3100 efficacy for treatment of castration-resistant prostate cancer. Int J Cancer. 2013;133:2225–33. doi: 10.1002/ijc.28202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, Hirsch B, Beckman KB, Silverstein KA, Dehm SM. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, Voytas DF, Dehm SM. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 29.Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, Watt K, Tam T, Yang YC, Bañuelos CA, Williams DE, McEwan IJ, Wang Y, Sadar MD. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–46. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Cao B, Liu X, Fu X, Xiong Z, Chen L, Sartor O, Dong Y, Zhang H. Berberine suppresses androgen receptor signaling in prostate cancer. Mol Cancer Ther. 2011;10:1346–56. doi: 10.1158/1535-7163.MCT-10-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao B, Liu X, Li J, Liu S, Qi Y, Xiong Z, Zhang A, Wiese T, Fu X, Gu J, Rennie PS, Sartor O, Lee BR, Ip C, Zhao L, Zhang H, Dong Y. 20(S)-protopanaxadiol-aglycone downregulation of the full-length and splice variants of androgen receptor. Int J Cancer. 2013;132:1277–87. doi: 10.1002/ijc.27754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Liu Z, Xu X, Blair CA, Sun Z, Xie J, Lilly MB, Zi X. Kava components down-regulate expression of AR and AR splice variants and reduce growth in patient-derived prostate cancer xenografts in mice. PLoS One. 2012;7:e31213. doi: 10.1371/journal.pone.0031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashima T, Okabe S, Seimiya H. Pharmacological targeting of constitutively active truncated androgen receptor by nigericin and suppression of hormone-refractory prostate cancer cell growth. Mol Pharmacol. 2010;78:46–54. doi: 10.1124/mol.110.064790. [DOI] [PubMed] [Google Scholar]
- 34.Narizhneva NV, Tararova ND, Ryabokon P, Shyshynova I, Prokvolit A, Komarov PG, Purmal AA, Gudkov AV, Gurova KV. Small molecule screening reveals a transcription-independent pro-survival function of androgen receptor in castration-resistant prostate cancer. Cell Cycle. 2009;8:4155–67. doi: 10.4161/cc.8.24.10316. [DOI] [PMC free article] [PubMed] [Google Scholar]


