Abstract
The great complexity of many human pathologies such as cancer, diabetes, and neurodegenerative diseases requires new tools for studies of biological processes on the whole organism level. The discovery of novel biocompatible reactions has tremendously advanced our understanding of basic biology, however, no efficient tools exist for real-time non-invasive imaging of many human proteases that play very important roles in multiple human disorders. We recently reported that “split luciferin” biocompatible reaction represents a valuable tool for evaluation of protease activity directly in living animals using bioluminescence imaging (BLI). Since BLI is the most sensitive in vivo imaging modality known to date, this method can be widely applied for the evaluation of multiple proteases activity as well as identification of their new peptide-specific substrates. In this protocol we describe several applications of this “split luciferin” reaction for quantification of protease activities in test tube assays and living animals.
Keywords: Protease activity, bioocompatible reaction, non-invasive, bioluminescent imaging, in vivo
INTRODUCTION
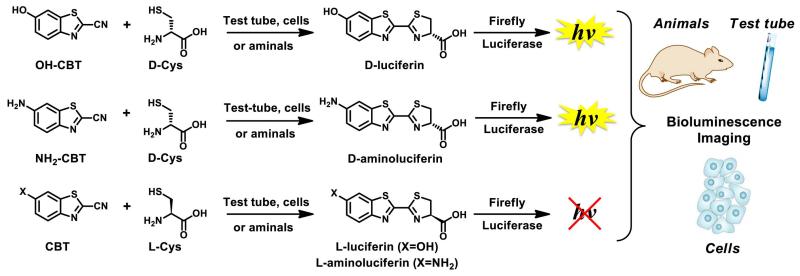
The discovery of biocompatible reactions had a significant impact on chemical biology and allowed studies of many biological processes directly in living systems. However, despite the fact that multiple biocompatible reactions have been developed in the past decade, very few work efficiently in complex living organisms. We previously reported that D-cysteine and 2-cyanobenzothiazole (CBT) can selectively react with each other in vivo to generate luciferin substrates for firefly luciferase (“split luciferin reaction”, Fig. 1) (Godinat et al., 2013).
Figure 1. “Split luciferin” reaction.
Overall schematic of the split luciferin ligation reaction between D- or L-cysteine and hydroxy- or amino-cyanobenzothiazole derivatives (OH-CBT and NH2-CBT) in various biological environments. Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
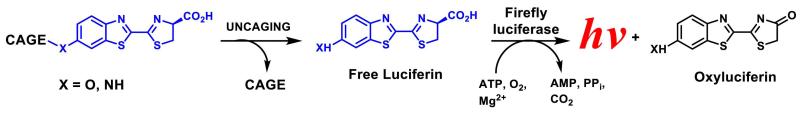
Since the production of luciferin substrates can be visualized and quantified directly in genetically modified living animals by bioluminescence imaging (BLI), it has immediate application for studies of many biological processes. For example, this reaction is well suited for interrogation of targeted tissues using a “caged” luciferin approach. This technique is based on the fact that luciferins caged on the phenolic oxygen or aryl nitrogen, do not lead to light production (Cohen et al., 2010; Henkin et al., 2012; van de Bittner et al., 2010; Wehrman et al., 2006) (Fig. 2). Despite the fact that caged luciferin compounds has been previously generated to quantify activities of several biological processes, (Cohen et al., 2010; Henkin et al., 2012; van de Bittner et al., 2010; Wehrman et al., 2006; Scabini et al., 2011; Hickson et al., 2010; Shah et al., 2005; Cosby et al., 2007; Biserni et al., 2010; Dragulescu-Andrasi et al., 2009; Yao et al., 2007; Goun et al., 2006) the synthesis of their scaffolds involves much more complex and low yielding synthetic procedures, representing major limitation of this technology (Cohen et al., 2010; Prescher et al., 2010; McCaffrey et al., 2003; Massoud and Gambhir, 2003; Henkin et al., 2012; van de Bittner et al., 2010; Wehrman et al., 2006; Shah et al., 2005; Dragulescu-Andrasi et al., 2009; Yao et al., 2007; Reddy et al., 2010; Harwood et al., 2011; Conley et al., 2012; McCutcheon et al., 2012; Liu et al., 2005; Liang et al., 2012).
Figure 2. Probing molecular signatures of target tissues through the use of caged luciferin substrates.
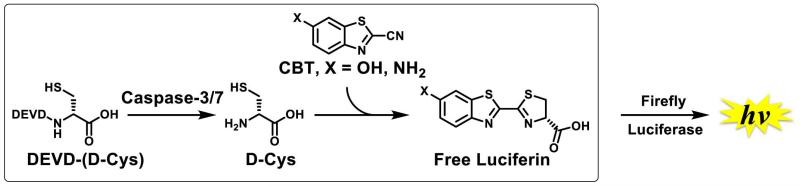
We previously reported application of this “split luciferin reaction” for the real-time and non-invasive imaging of apoptosis, associated with activation of caspase 3/7 (Godinat et al., 2013). Caspase-dependent release of free D-cysteine from the caspase 3/7-specific peptidic substrate Asp-Glu-Val-Asp-D-Cys (DEVD-(D-Cys)) allowed further reaction with 6-amino-2-cyanobenzothiazole (NH2-CBT) in vivo and subsequent formation of 6-amino-D-luciferin. Since this compound is known to be an excellent substrate for luciferase enzyme, its formation leads to the generation of light that is proportional to the activity of caspase 3/7. Importantly, this “split luciferin” strategy was found to be more sensitive when compared to the commercially available DEVD-aminoluciferin substrate (Godinat et al., 2013), in which the entire luciferin is caged with the same peptidic sequence. The underlining principle of the use of “split luciferin reaction” for proteases imaging is depicted in Fig. 3 using caspase 3/7 peptide-specific sequence (DEVD) as an example.
Figure 3. Imaging of Caspase 3/7 activity using “split luciferin” reaction.
Modification on the D-Cysteine moiety with DEVD sequence allows selective formation of D-luciferin upon Caspase-3/7 cleavage and subsequent production of light, proportional to activity of Caspase 3/7. Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
This result was particularly exciting because the “split luciferin” approach can now be applied for imaging and quantification of many other proteases that are known to cleave at the end of specific amino acid sequences (Yao et al., 2007; Zhou et al., 2006; Zhou and Valley et al., 2006; Cali et al., 2006; Wilson et al., 2002), as well as identification of new protease specific peptides associated with various human diseases. Immediate examples of mammalian proteases include dipeptidyl peptidase 4 (GP and VP), tryptase (PRNK), and various caspases such as caspase 2 (VDVAD), caspase 6 (VEID), caspase 8 (LETD), caspase 9 (LHTD), and caspase 12 (ATAD) (Ren et al., 2009; Geiger et al., 2006; O’Brian et al., 2005; Zhou et al., 2006; Zhou and Valley et al., 2006; Cali et al., 2006). In addition to mammalian proteases, this “split luciferin” methodology could also be used for studies of a wide variety of viral, parasite, and bacterial proteases that play very important role in the replication and the spread of infectious diseases (Wilson et al., 2002). They include SARS protease (TSAVLQ), caspase-like (nLPnLD), and trypsin-like (LRR) proteases (Geiger et al., 2006; Zhou et al., 2006; Zhou and Valley et al., 2006; Cali et al., 2006).
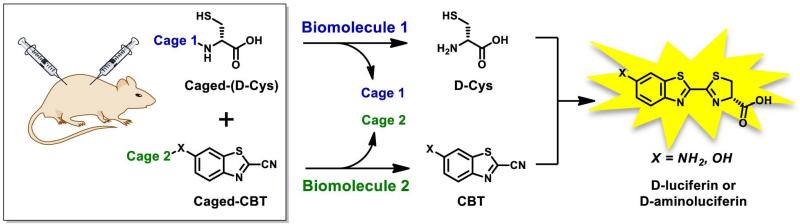
Importantly, the synthesis of short peptide sequences with C-terminal D-cysteine can be easily performed with the help of automated peptide synthesis, which is a widely available and versatile technique (Merrifield and Stewart, 1965; Merrifield, 1965). On the contrary note, the synthesis of caged luciferin substrates like DEVD-aminoluciferin represents a major synthetic challenge, that is reflected by a very high cost of commercially available probes. Moreover, the new approach suits itself perfectly for multiplex imaging based on the modular construction of bioluminogenic sensors, where either or both reaction partners (D-Cys and CBT) are caged to report on multiple biological events (Fig. 4).
Figure 4. Overall representation of the dual imaging concept for luciferin ligation.
Both luciferin ligation precursors could be caged as sensors for two different biomolecules. Only when both become uncaged, D-luciferin or D-aminoluciferin is formed as the result of split luciferin ligation reaction, allowing the production of light by luciferase enzyme. Reprinted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
Therefore, we believe that this protocol can be applied to evaluation of activity of multiple biologically important proteases that are known to cleave at the end of corresponding specific amino acid sequences and allow sensitive imaging and quantification of their activities directly in animal models of disease.
BASIC PROTOCOL 1 describes the methodology for imaging and quantifying protease activity in a cell-free in vitro experimental setup. The procedure is detailed for caspase-3 imaging. Thrombin protease activity imaging is also described in the ALTERNATE PROTOCOL. Finally, the BASIC PROTOCOL 2 describes in detail the use of “split luciferin” based bioluminescent probes for real time non-invasive caspase-3/7 imaging in luciferase expressing mice.
BASIC PROTOCOL 1
IN VITRO (CELL-FREE) BIOLUMINESCENCE IMAGING OF PROTEASES ACTIVITY USING “SPLIT LUCIFERIN” REACTION
Introductory paragraph
The following procedure describes in detail all the steps necessary for evaluation of activity of thrombin and caspase 3 proteases. However, the same approach can be adapted to studies of activity of other proteases that are known to cleave their peptide-specific sequences on the C-terminal of the cleavage site.
The development of novel probes for evaluation of protease activities should start from the selection of protease-specific amino-acid sequence that can be selectively recognized and cleaved by a protease of interest (POI). When such sequence is selected, a D-Cysteine (D-Cys) residue should be introduced at the C-terminal position using common and widely used automated or manual peptide synthesis techniques (Merrifield and Stewart, 1965; Merrifield, 1965). If evaluation of thrombin or caspase 3/7 proteases are intended, the exact protocol described below should be followed. To test the viability of this approach with the desired POI, the selected peptide containing the D-Cys on the C-term will be incubated with the purified POI. After addition of CBT and luciferase enzyme, light emission will be acquired.
Example with Caspase 3
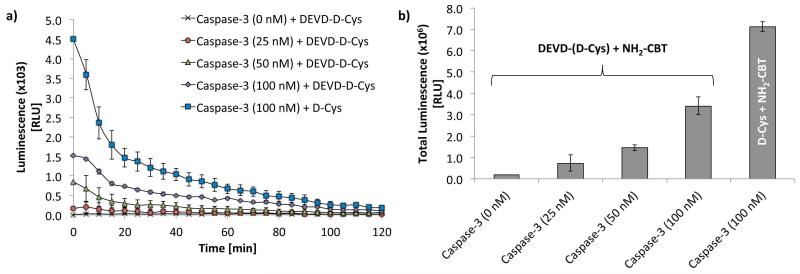
In the first step, activated caspase-3 enzyme is incubated with DEVD-(D-Cys) peptide. Once cleaved by the protease, the resulting D-Cys reacts with NH2-CBT leading to the formation of amino-D-luciferin and further produce light, proportional to the activity of caspase 3 (Fig. 3). An example of the data that was acquired for quantification of caspase-3 activity in vitro using split luciferin approach is depicted in Fig. 5 (Godinat et al., 2013). In this experiments, the DEVD-(D-Cys) probe was incubated with increasing concentration of caspase-3 followed by addition of NH2-CBT. The resulting light emission was acquired over 2 h and plotted as a function of time (Fig. 5 a). Quantification of the total light produced over this period of time was performed by integrating the area under the curves and plotted in the form of bar-graph (Fig. 5 b).
Figure 5. Quantification of caspase-3 activity in test-tube assay using “split luciferin approach”.
(a) Bioluminescent signal as a function of time from DEVD-(D-Cys) peptide (200 μM) or D-cysteine (200 μM) after incubation with different concentrations of Caspase-3 (25, 50 and 100nM) over 3 h at 37°C, followed by addition of NH2-CBT (400 μM in MeOH), and incubation for additional 1 h at 37°C. The signal acquisition was started immediately after addition of 5 μL of the pre-made luciferase buffer (see recipe) and acquired for the duration of 2 h using IVIS Spectrum (Perkin Elmer, USA). (Error bars are ± SD for three measurements). (b) Total light output collected over the period of 2 h (performed by integrating the area under the curves in panel A and plotted in the form of bar-graph. Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
Materials
200 nM of purified Caspase-3 solution (see recipe)
800 μM DEVD-(D-Cys) peptide solution (see recipe)
800 μM D-Cysteine solution (see recipe)
400 μM 6-Amino-2-cyanobenzothiazole (Intrace Medical®) solution (see recipe)
60 μg/mL Firefly luciferase enzyme solution (Sigma-Aldrich, St. Louis, MO) (see recipe)
Caspase buffer (see recipe)
Luciferase buffer (see recipe)
Black 96-well plate (BD Falcon, ref 353219)
V-shaped 96-well plate (Vitaris, 3897-COR)
Bioluminescence plate reader or imager (Spectramax M5, Molecular Devices, Sunnyvale, CA)
Protocol steps
Caspase-3 was purified and characterized following the reported procedure (Stennicke and Salvesen, 1999).
- Prepare different solutions of caspase-3 in caspase buffer by diluting 200 nM caspase-3 stock solution.
-
-0 nM: only caspase buffer
-
-50 nM: dilute 100 μL of 200 nM caspase-3 stock solution to 400 μL using caspase buffer
-
-100 nM: dilute 200 μL of 200 nM caspase-3 stock solution to 400 μL using caspase buffer
-
-
Aliquot caspase-3 solutions (0, 50, 100 nM) in triplicates in a v-shaped 96-well plate with 50 μL per well.
Aliquot 200 nM caspase-3 stock solution in six wells of the same plate with 50 μL per well.
Cover the plate with a lid to avoid evaporation and incubate at 37°C for 15 min in order to activate the caspase-3 enzyme.
Add 50 μL of 800 μM D-Cysteine solution in three of the wells containing the 200 nM caspase-3 solution.
Add 50 μL of 800 μM DEVD-(D-Cys) solution to all the other wells.
Cover the plate with a lid to avoid evaporation and incubate at 37°C for 3h.
Add 100 μL of the 400 μM NH2-CBT solution to each wells. Solution is mixed with a few up and down pipetting.
Protect the plate from light and incubated for 1 h at 37°C.
Prepare a second 96-well plate by adding 115 μL of the 60 μg/mL luciferase solution per well.
Step annotations
The type of 96-well plate that should be used here is dependent on the Bioluminescence imaging system used. If an imager such as IVIS Spectrum (PerkinElmer, Alameda, CA) is used, a Black 96-well plate is necessary. Please follow recommendations of plate reader/imager manufacturer.
-
11
Immediately before reading bioluminescence emission, quickly transfer 5 μL of the reaction solutions in the plate containing luciferase. Bioluminescence signal from the plate should be measured every 5 min for a duration of 2 h.
Step annotations
As soon as the incubated solutions are added to the luciferase buffer, light emission will be produced. As this process is very fast, it is important to add the luciferase buffer at the same time to all the wells and start data acquisition right after addition. Therefore, we suggest to use multichannel pipets.
If necessary, the delay between measurements can be shorten in order to have more data points.
If the plate reader is equipped with a liquid handling systems, a different procedure could be used. This would consist of adding 5 μL of the caspase-3 containing solutions to a 96-well plate and performing addition of 115 μL luciferase buffer using automated set up on the liquid handling system.
ALTERNATE PROTOCOL 1 (optional)
IN VITRO (CELL-FREE) IMAGING OF THROMBIN PROTEASE ACTIVITY
This alternate protocol focuses on the evaluation of thrombin protease activity using thrombin-specific Gly-Gly-Arg-(D-Cys) (GGR-(D-Cys)) peptide, that is designed to be cleaved by thrombin protease between Arg and D-Cys residues.
Thrombin enzyme was first incubated with GGR-(D-Cys) peptide. Upon enzymatic cleavage of peptidic sequence, D-Cys is liberated and react with NH2-CBT, resulting in the formation of D-aminoluciferin and further produce light that is proportional to the activity of thrombin enzyme.
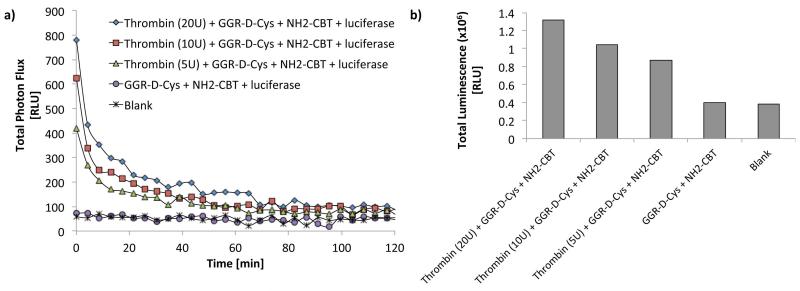
Figure 6 is an example of the data acquired for this experiment (Fig. 6) (Godinat et al., 2013). In this study, GGR-(D-Cys) peptide was first incubated with increasing concentration of thrombin enzyme, followed by addition of NH2-CBT. Subsequent light emission was acquired over 2 h (Fig. 6a). Integration of the area under each curve represent total photon flux produced over this time period and plotted in the form of bar-graphs in Fig. 6b.
Figure 6. Evaluation of thrombin activity using “split luciferin approach”.
(a) Bioluminescent signal in function of the time from GGR-(D-Cys) peptide incubated with thrombin protease at different concentrations (5, 10, and 20 U) over 3 h at 37°C before addition of NH2-CBT and additional incubation for another 1 h at rt. Signal acquisition was started right after addition of 5 μL of the pre-made luciferase buffer (see recipe) and continued for about 2 h using IVIS Spectrum (Perkin Elmer, USA). The blank represent the signal from luciferase buffer. (b) Total light output collected over the period of 2 h obtained by integrating the area under the curves in A and plotted in the form of bar-graph. Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
Materials
Thrombin enzyme (Sigma-Aldrich, St. Louis, MO, ref. T1063-1KU)
100 Units/mL Thrombin stock solutions (see recipe)
500 μM GGR-(D-Cys) peptide solutions (see recipe)
500 μM NH2-CBT (Intrace Medical®) solution (see recipe)
60 μg/mL Firefly luciferase enzyme (Sigma-Aldrich, St. Louis, MO) solution (see recipe)
Thrombin buffer (see recipe)
Luciferase buffer (see recipe)
MeOH
Black 96-well plate (BD Falcon, ref 353219)
V-shaped 96-well plate (Vitaris, 3897-COR)
Bioluminescence plate reader (Spectramax Gemini, Molecular Devices, Sunnyvale, CA) or camera like IVIS Spectrum (Perkin Elmer, USA).
Protocol steps
- Prepare the different solutions of thrombin enzyme in thrombin buffer by diluting the 1000 U/mL thrombin stock solution.
-
-0 U/mL: only thrombin buffer
-
-250 U/ml: dilute 20 μl of 1000 U/ml thrombin stock solution to 80 μl using thrombin buffer (1:4 dilution)
-
-500 U/ml: dilute 40 μl of 1000 U/ml thrombin stock solution to 80 μl using thrombin buffer (1:4 dilution)
-
-
Aliquot thrombin solutions (0, 250, 500 U/mL) in triplicates in a v-shaped 96-well plate with 20 μL per well.
Add 1000 U/ml thrombin stock solution into six wells of the same plate with 20 μl per well.
Add 80 μL of 500 μM D-Cysteine solution in three wells containing the 250 U/mL thrombin stock solution
Add 80 μL of 500 μM solutions of GGR-(D-Cys) peptide solution to all the other wells.
Cover the plate with a lid to avoid evaporation and incubate at 37°C for 3 h.
Add 100 μL of 400 μM NH2-CBT solution to all the wells. Solution is mixed by “up and down” pipetting.
Protect plates from light and incubate for 1 h at room temperature.
Prepare a second 96 well-plate by adding 115 μL of the 60 μg/mL luciferase solution per well.
Step annotations
The type of 96-well plate used for experiments depends on the Bioluminescence imaging system used. If a camera such as IVIS Spectrum (PerkinElmer, Alameda, CA) is used, a Black 96-well plate is recommended. Please follow recommendations of plate reader/imager manufacturer.
-
11
Immediately before reading bioluminescence light output, quickly transfer 5 μL of the reaction solutions in the plate containing luciferase. It is recommended to acquire bioluminescence signal every 5 min for 2 h.
Step annotations
As soon as samples solutions are added to the luciferase buffer, light emission will be produced. As this process is very fast, it is important to add the solution to all the wells at the same time and start imaging right after addition.
If necessary, the delay between measurements points can be shorten in order to have more data points.
If the plate reader is equipped with a liquid handling systems a different procedure could be used. This would consist on adding 5 μL of the caspase-3 containing solutions on a 96-well plate and doing the addition of the 115 μL luciferase solution using the liquid handling system.
BASIC PROTOCOL 2 (optional)
BASIC PROTOCOL TITLE
REAL TIME NON-INVASIVE IMAGING OF CASPASE-3/7 ACTIVITIES IN TRANSGENIC REPORTER MICE (FVB-Luc+)
Introductory paragraph
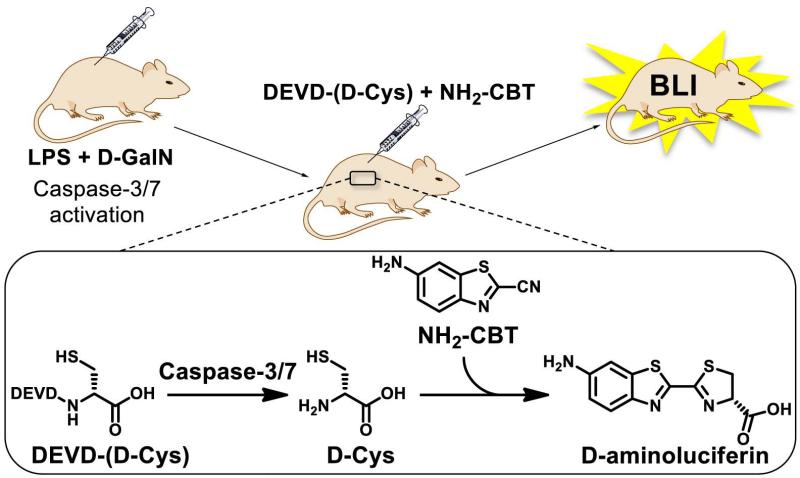
In this protocol, the procedure for evaluation of caspase-3/7 activity in living animals is described in details. It consists of two major parts: 1) Induction of caspase-3/7 in live animals; 2) Imaging steps using “split luciferin” approach (Fig. 7). A commercial kit is now available for the second step based on this technology (“Caspase 3 and Caspase 7 mouse kit (z-DEVD-D-Cys)”, Intrace Medical®)
Figure 7. Imaging of caspase-3 activity in live FVB+luc animals using “split luciferin” approach.
Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
In this study we used transgenic mice ubiquitously producing luciferase in every cell of their body under actin promoter (Cao et al., 2004). These and several other types of transgenic mice with luciferase expression in different organs are commercially available (from Taconic or The Jackson Laboratory).
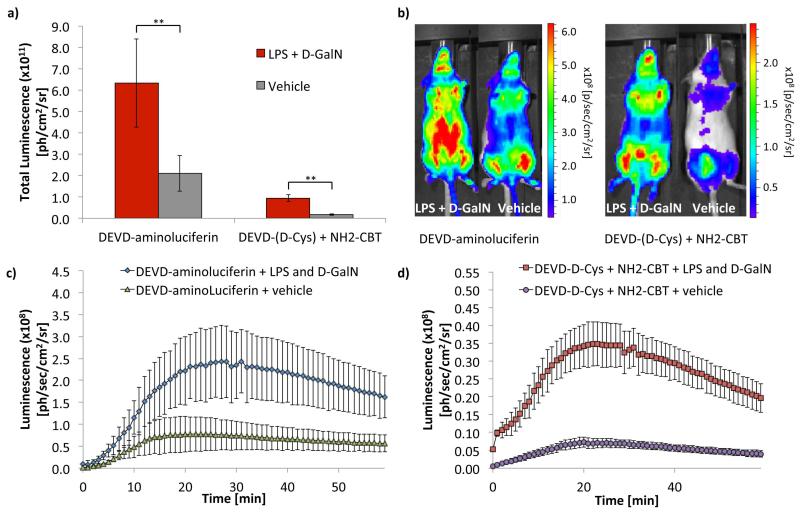
Caspase 3/7 was induced in the liver of FVB-luc+ mice by injecting the animals with lipopolysaccharide (LPS) and d-galactosamine (d-GalN), following a procedure that was previously established for bioluminescence imaging of caspase-3/7 (Biserni et al., 2010). Six hours post administration of LPS and d-GalN, animals were injected with “split luciferin” substrates (DEVD-(D-Cys) and NH2-CBT, Intrace Medical®) and the signal was acquired using IVIS Spectrum camera (Perkin Elmer California) (Godinat et al., 2013). Commercially available DEVD-aminoluciferin compound was used as a positive control and for comparison. The data resulting from this experiment are shown on Fig. 8.
Figure 8. Evaluation of Caspase-3/7 activity using “split luciferin” reaction in living transgenic reporter mice (FVB+luc).
(a) Total luminescence over 1 h from FVB+luc mice treated with either PBS (control group) or combination of LPS (100 μg/kg in 50 μL of PBS) and D-GalN (267 mg/kg in 50 μL of PBS). Six hours post-treatment, the animals received IP injections of either DEVD-aminoluciferin (34 mg/kg in 100 μL of PBS) or a combination of DEVD-(D-Cys) peptide (22.6 mg/kg in 100 μL of PBS) and NH2-CBT (6.8 mg/kg in 20 μL of DMSO). Statistical analyses were performed with a two-tailed Student’s t test. **P < 0.01 (n=8 for DEVD-aminoluciferin groups and n=4 for combination of DEVD-(D-Cys) and NH2-CBT reagents). Error bars are ± SD for eight and four measurements respectively. (b) Representative image of mice, 15 min post-injection of DEVD-aminoluciferin or a combination treatment with DEVD-(D-Cys) and NH2-CBT reagents. (c) Bioluminescent signal produced over 60 min following IP injection of DEVD-aminoluciferin in LPS/D-GalN (n=8) or vehicle (n=8) treated mice. (d) Luminescence emission over 60 min following IP injection of DEVD-(D-Cys) peptide and NH2-CBT in LPS/D-GalN (n=4) or vehicle (n=4) treated mice. Error bars are ± SD for eight or four measurements. Adapted with permission from Godinat, A., Park, H. M., Miller, S. C., Cheng, K., Hanahan, D., Sanman, L. E., Bogyo, M., Yu, A., Nikitin, G. N., Stahl, A., Dubikovskaya, E. A. 2013 A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice, ACS Chem. Biol. 8, 987–999. Copyright 2014 American Chemical Society.
Taking into account that the current protocol could be potentially used in different type of mice or animal diseases models, we thought to add a few important points. Expression of firefly luciferase is mandatory in order to obtain readout using this approach. The animals can express luciferase ubiquitously like it is the case in our study, or only in certain organs or tissues (like xenografts models or spontaneous cancer models where tumor cells express luciferase. It is also recommended that mice selected for the experiment should preferably be of approximately the same sex, age and weight.
The amount of light output depends on the amount/level of luciferase expression that becomes particularly important if tumor models are used. In order to account for the difference in levels of luciferase expression between each animal, the experiment should start with quantification of “basal” level of light resulting from administration of D-luciferin. This data set is important and can be later used for “calibration” of resulting signal from protease activation to the basal level of luciferase expression in each mouse.
Materials
The following procedure is described for an average 25 g FVB-luc+ mouse. If the mice used have different weight, it is advised to adjust the concentrations of solutions in order to keep the doses constant.
FVB-Luc+ mice (FVB-Tg(CAG-luc,-GFP)L2G85Chco/J, The Jackson Laboratory, Cao et al., 2004)
3.1 mg/mL D-luciferin potassium salt (Intrace Medical®) solution (see recipe)
50 ug/mL Lipopolysaccharide (LPS) (from Salmonella typhosa, Sigma-aldrich, ref. L7895) solution (see recipe)
133.5 mg/mL D-(+)-Galactosamine hydrochloride (D-GalN) (Applichem GmbH, Germany, ref. A6859) solution (see recipe)
5.65 mg/mL DEVD-(D-Cys) peptide solution (see recipe)
8.5 mg/mL 6-Amino-2-cyanobenzothiazole (NH2-CBT) (Intrace Medical®) solution (see recipe)
Phosphate-buffered saline (PBS; Life Technologies, cat. no. 20012-068), sterile
Dimethyl sulfoxide (DMSO), sterile
Bioluminescence imaging system for small laboratory animals (IVIS Spectrum, PerkinElmer, Alameda, CA) or similar device.
0.5-ml insulin syringes (U-100 insulin syringe, 28-G × ½-in. needle; BD, cat. no. 329465)
Isoflurane for anesthesia (Phoenix).
Protocol steps
-
1
Prepare D-luciferin solution in order to inject 12.4 mg/kg of mouse body weight in 100 μL of sterile PBS.
perform D-luciferin injections
For a typical mouse weighing 25 g, this represents 0.31 mg of D-luciferin potassium salt in 100 μL of sterile PBS.
-
2
Preload syringes with 100 μL of the Luciferin solution.
-
3
Place mice into a clear Plexiglas anesthesia box (3% isoflurane) that allows unimpeded visual monitoring of the animals.
Depending on the imaging system used as well as the legislation that apply in the country where experiments are performed, other anesthesia methods could be used
-
4
Once mice are fully anesthetized, inject mice intraperitoneally (IP) with 100 μL of the corresponding D-luciferin solutions.
-
5
Place the mice on the heating pad (37°C) in the imaging chamber and keep them under anesthesia during the entire imaging period (it is advised to decrease the overall level of anesthesia to equivalent of 1.5% isoflurane for imaging sessions over 1 h).
-
6
Acquire bioluminescent signal for about 1h, recording measurements every 1-5 minutes.
Step annotations
In order to minimize the time between D-luciferin injection and the beginning of data acquisition, it is advised to preload syringes with corresponding solutions prior to anesthesia step. Once all the mice are anesthetized, they should be injected sequentially and placed in the imaging chamber right after.
-
7
After imaging, turn off the isoflurane manifold to 0 and put the animals back to cages when awaken.
-
8
At least 48h of rest for the animals are respected before continuing this experiment.
Step annotations
The resting time between two imaging sessions might vary depending on the legislation that apply in the country where experiments are performed. It is also important to assure that no residual signal is left from previous imaging with luciferin before starting step 10. Therefore, it is advised to perform quick imaging of mice before proceeding with next imaging step. Prepare two experimental groups of animals defined as “treatment group” and “control group”. Label each group of mouse according to the legislation that applies in the country where experiment is performed (at least four mice per group is recommended).
Activate caspase-3
-
9
Prepare 300 μL of a sterile solution of lipopolysaccharides (LPS) in PBS at 50 μg/mL.
Step annotations
The total volume of the above LPS solution should be sufficient to treat all the mice of the “treatment group”. See step 15 for dosage.
-
10
Prepare 300 μL of a sterile solution of D-(+)-Galactosamine hydrochloride (D-GalN) in PBS at 133.5 mg/mL.
Step annotations
The total volume the above D-GalN solution should be sufficient to treat all the mice of the “treatment group”. See step 16 for dosage.
-
11
Prepare 500 μL of a sterile solution of PBS.
-
12
Preload syringes with 50 μL of sterile LPS solution (one for each animal in the “treatment group”).
-
13
Preload four syringes with 50 μL of sterile D-GalN solution (one for each animal in the “treatment group”).
-
14
Preload four syringes with 100 μL of sterile PBS (one for each animal in the “control group”).
-
15
Inject the four mice from “treatment group” intraperitoneally (IP) with 100 μg/kg of LPS in 50 μL (one for each animal in the “control group”).
Step annotations
An average 25 g mouse should receive a total amount of 2.5 μg LPS injected in 50 μL of sterile PBS solution (50 μg/mL).
-
16
Inject the same four mice from “treatment group” intraperitoneally (IP) with 267 mg/kg of D-GalN in 50 μL of sterile PBS.
Step annotations
A typical 25 g mouse should receive a total amount of 6.7 mg D-GalN in 50 μL of sterile PBS solution (133.5 mg/mL).
-
17
Inject the “control group” of mice intraperitoneally (IP) with 100 μL of sterile PBS.
-
18
All the mice are returned to their cages and a 6 h delay is respected before the next step.
Image caspase-3/7 activity
-
19
Prepare 500 μL of DEVD-(D-Cys) peptide solution in sterile PBS at 5.65 mg/mL.
-
20
Prepare 500 μL of NH2-CBT solution in sterile DMSO at 8.5 mg/mL.
-
21
Preload syringes with 100 μL of sterile DEVD-(D-Cys) solution (one for each mouse)
-
22
Inject the animals from “treatment group” intraperitoneally (IP) with 22.6 mg/kg dose of DEVD-(D-Cys) peptide solution in 100 μL of PBS.
Step annotations
An average 25 g mouse should receive a total amount of 0.565 mg of DEVD-(D-Cys) peptide in 100 μL of sterile PBS (5.65 mg/mL solution of the peptide in sterile PBS). A delay of 10 min is respected before the next step.
-
23
Place the animals from the “treatment group” into a clear Plexiglas anesthesia box (3% isoflurane) that allows unimpeded visual monitoring of the animals.
-
25
Preload syringes with 20 μL of sterile NH2-CBT (one for each mouse in the “treatment” group).
-
26
After the mice are fully anesthetized, inject the mice intraperitoneally (IP) with a 6.8 mg/kg dose of NH2-CBT solutions in 20 μL in sterile DMSO.
Step annotations
An average 25 g mouse should receive a total amount of 0.17 mg of NH2-CBT in 20 μL sterile DMSO (8.5 mg/mL solution)
-
27
Place the mice on the heating pad (37°C) in the imaging chamber and keep them under anesthesia during the entire imaging period (it is advised to decrease the overall level of anesthesia to equivalent of 1.5% isoflurane for imaging sessions over 1 h).
-
28
Acquire bioluminescent signal for about 1 h, recording measurements every 1-5 minutes.
-
29
After the end of data acquisition, the mice from “treatment group” have to be sacrificed according to the legislation that applies in the country where experiments are performed.
-
30
Repeat the experiment from entry 18 to 28 with the “control group” of animals
-
31
After the end of data acquisition, turn off the instruments return the mice from “control group” back to their cages (they normally do not have to be sacrificed due to non-invasive nature of control experiment).
-
32
Perform data analysis. Data analysis is performed using Living Image In Vivo Software (PerkinElmer). Regions of interest (ROI) are defined around each animal. Mice tails are not taken into account in the ROI. This is performed for every time point and quantified values are plotted in function of time (Fig. 8C,D). To calculate the overall light emission per mouse during the complete imaging time, areas under the curves are calculated and plotted in a bar graph (see Fig. 8A).
REAGENTS AND SOLUTIONS
Use Milli-Q purified water or equivalent in all recipes and protocol steps.
Caspase buffer
100 mM HEPES pH 7.4
0.1% CHAPS
1 mM EDTA
10 mM DTT
1% sucrose
Storage: Caspase buffer can be stored at 4 °C for 3 months.
200 nM solution of purifed Caspase-3
Caspase-3 can be purified and characterized following the reported procedure (Stennicke and Salvesen, 1999).
A caspase-3 solution is prepared freshly by dissolving 0.2 nmol of purified caspase-3 in 1 mL of caspase-3 buffer (see above for buffer formulation and preparation).
800 μM solution of DEVD-(D-Cys) peptide
800 μM DEVD-(D-Cys) solution is prepared by dissolving 0.5 mg of peptide in 1.08 mL of caspase buffer.
Fresh solution should be prepared before experiment.
800 μM solution of D-Cysteine
1 mg of D-Cysteine (Mw 121.15 g/mol) is dissolved in 10.318 mL of degased caspase buffer.
A fresh solution should be prepared before every experiment. If possible, before dissolving the D-Cysteine, the caspase buffer should be degased by bubbling inert gas (e. g. Nitrogen or Argon) through the solution for 30 min.
400 μM solution of 6-Amino-2-cyanobenzothiazole
1 mg of 6-Amino-2-cyanobenzothiazole (Mw 175.20 g/mol) is dissolved in 14.269 mL of pure methanol.
The solution should be protected from light and stored at −20 °C. A fresh solution should be prepared before every experiment.
Thrombin buffer
0.02 M Tris-HCl pH 8.4
0.15 M NaCl
2.5 mM CaCl2
Thrombin buffer can be stored at room temperature.
Thrombin stock solution
Thrombin enzyme is used without additional purification steps.
1000 U/mL Thrombin stock solution is prepared by diluting 1000 U of Thrombin enzyme in 1 mL in storage buffer (50 mM sodium citrate, pH 6.5, 200 mM NaCl, 0.1% PEG-8000, 50% glycerol).
Thrombin stock solution should be stored at −20 °C and multiple freeze/thaw cycles should be avoided.
500 μM solution of GGR-(D-Cys) peptide
500 μM Gly-Gly-Arg-(D-Cys) peptide solution is prepared by diluting 0.5 mg of peptide in 2.555 mL of Thrombin buffer.
500 μM solution of D-Cysteine
1 mg of D-Cysteine (Mw 121.15 g/mol) is dissolved in 16.508 mL of degased Thrombin buffer.
A fresh solution should be prepared before every experiment. If possible, before dissolving the D-Cysteine, the Thrombin buffer should be degased by bubbling inert gas (e. g. Nitrogen or Argon) in the solution for 30 min.
400 μM solution of 6-Amino-2-cyanobenzothiazole
1 mg of 6-Amino-2-cyanobenzothiazole (Mw 175.20 g/mol) is dissolved in 14.269 mL of pure methanol.
The solution should be protected from light and stored at −20° A fresh solution should be prepared before every experiment.
Luciferase buffer
0.1M Tris-HCl pH 8
2mM ATP
5 mM MgSO4
0.1M tris-HCl supplemented with 5 mM MgSO4 can be stored at 4 °C for several weeks but fresh ATP should be supplemented just before performing the experiments. It’s not recommended to store the luciferase buffer once ATP has been supplemented.
60 μg/mL solution of Firefly luciferase enzyme
1 mg of firefly luciferase is dissolved in 16.7 mL of luciferase buffer. The luciferase buffer (see above) should be freshly prepared.
Alternatively, a 2 mg/mL stock solution of Firefly luciferase can be prepared in PBS. 100 μL of the stock solution will then be diluted up to 3.333 mL with luciferase buffer in order to obtain the desired 60 μg/mL luciferase solution.
Freeze-thaw cycle should be avoided on the luciferase enzyme and if possible, used batch of Luciferase that have the same number of Freeze-thaw cycle.
Material for experiments in live mice
All the imaging reagents are also currently available in the form of a commercial kit “Caspase 3 and Caspase 7 mouse kit (z-DEVD-D-Cys)” from Intrace Medical®, Switzerland.
3.1 mg/mL solution of D-luciferin potassium salt
3.1 mg of D-luciferin potassium salt (Mw: 318.4 g/mol) is dissolved in 1 mL of sterile PBS.
The dosage of D-luciferin potassium salt is 12.4 mg/kg of mouse body weight and the resulting solution should be injected in approximately 100 μL. For a typical mouse weighing 25 g, prepare 3.1 mg/mL solution of D-luciferin in sterile PBS and inject 100 μL of the solution.
50 μg/mL Lipopolysaccharide (LPS) solution
50 μg of LPS is dissolved in 1 mL of sterile PBS.
The dosage of LPS is 100 μg/kg of mouse body weight and should be injected in approximately 50 μL volume. For a typical mouse weighing 25 g, prepare a 50 μg/mL solution of LPS in sterile PBS and inject 50 μL of the solution.
133.5 mg/mL solution of D-(+)-Galactosamine hydrochloride (D-GalN)
133.5 mg of D-GalN is dissolved in 1 mL of sterile PBS.
The dosage of D-GalN is 267 mg/kg of mouse body weight and should be injected in approximately 50 μL volume. For a typical mouse weighing 25g, prepare a 133.5 mg/mL solution of D-GalN in sterile PBS and inject 50 μL of the solution.
5.65 mg/mL solution of DEVD-(D-Cys) peptide
5.65 mg of DEVD-(D-Cys) peptide is dissolved in 1 mL of sterile PBS.
The dosage of DEVD-(D-Cys) peptide is 22.6 mg/kg of mouse body weight and should be injected in approximately 100 μL. For a typical mouse weighing 25 g, prepare a 5.65 mg/mL solution of DEVD-(D-Cys) peptide in sterile PBS and inject 100 μL of the solution.
8.5 mg/mL solution of 6-Amino-2-cyanobenzothiazole (NH2-CBT)
8.5 mg of NH2-CBT is dissolved in 1 mL of Sterile DMSO.
The dosage of NH2-CBT is 6.8 mg/kg of mouse body weight and should be injected in approximately 20 μL volume. For a typical mouse weighing 25 g, prepare a 8.5 mg/mL solution of NH2-CBT in sterile DMSO and inject 20 μL of the solution.
COMMENTARY
Background Information
In the past 10 years, several biocompatible “click” reactions have been developed and successfully applied for various study of biological processes (Sletten and Bertozzi, 2009). These reactions include copper-free cyclooctyne-type cycloaddition (Agard et al., 2006; Sletten et al., 2010; Jewett et al., 2010; Chang et al., 2010; Ning et al., 2008; Neves et al., 2011), Staudinger ligation (Lin et al., 2005; Saxon and Bertozzi, 2000; Saxon et al., 2000; Presher et al., 2004; Dube et al., 2006; Nilson et al., 2000; Chang et al., 2007; Hangauer and Bertozzi, 2008), and alkene-tetrazine reactions (Devaraj et al., 2008; Blackman et al., 2008; Budin et al. 2011; Yang et al., 2012; Lang et al., 2012; Liang et al., 2012; Devaraj et al., 2012). However, most of these reactions only work in cell lysates or live cells, and the great complexity of human pathologies requires tools that allow studies of biochemical transformations on the level of the whole organism.
Since multiple animal models of various human pathologies have been successfully established, development of new biocompatible reactions applicable for studies of biological processes on the level of a live animal plays a crucial role in biology and medical research (Rogers, 2012; Laferla and Green, 2012; Peters et al., 2012; Langdon, 2012; Fedele et al., 2012; Li et al., 2012).
Only few biocompatible reactions have been shown to work efficiently in living animals and one of them is known as Staudinger Ligation (Saxon and Bertozzi, 2000; Prescher et al., 2004; Neves et al., 2011; van Berkel et al., 2011). Recently, [4 + 2] tetrazine/trans-cyclooctene cycloaddition has also been used in living mice, despite the fact that the tetrazine reagent was bound to a polymeric support to improve its pharmacokinetic properties of the reagent in vivo settings (Devaraj et al., 2012).
It has been previously reported that D-cysteine amino acid efficiently reacts with 6-hydroxy-2-cyanobenzothiazole (OH-CBT) in physiological solutions (Ren et al., 2009; Ye et al., 2011; Liang et al., 2010) and was first reported by White and co-workers in 1963 as final step in the synthesis of D-luciferin (Fig. 1) (White et al., 1963). Recently, others have reported novel applications of this reaction for the selective labeling of proteins on N-terminal cysteines (Nguyen et al., 2011; Ren et al., 2009), as well as the controlled assembly of polymers in physiological solutions and living cells (Ye et al., 2011; Liang et al., 2010). Remarkably, the rate of this reaction was found to be three orders of magnitude faster than Staudinger ligation (Ren et al., 2009; Yuana and Liang, 2014).
We have recently reported that this reaction can also occur directly in living animals and efficiently used in combination with bioluminescence imaging (Godinat et al., 2013) that is the most sensitive in vivo imaging technique currently available in living animals up to date (Prescher and Contag, 2010; McCaffrey et al., 2003; Massoud and Gambhir, 2003). Indeed, in the past BLI has been extensively used for tracking luciferase-expressing cells in living animals and visualization of transcriptional activation (Geiger et al., 2006). More recent applications of BLI include probing of molecular signatures of target tissues through the use of “caged” luciferin probes that are only uncaged by the activity of specific biological molecules. The underlying principle in the design of all these caged luciferin substrates is based on the fact that luciferins substituted on the phenolic oxygen or aryl nitrogen are not capable of light emission (Cohen et al., 2010; Henkin et al., 2012; van de Bittner et al., 2010; Wehrman et al., 2006). This approach was reported for the design of probes to sense enzymatic activities directly in vivo such as those of beta-galactosidase (Wehrman et al., 2006) and caspases (Scabini et al., 2011; Hickson et al., 2010; Shah et al., 2005; Cosby et al., 2007; Biserni et al., 2010). Furin (Dragulescu-Andrasi et al., 2009) as well as beta-lactamases (Yao et al., 2007) were also sensed using a similar methodology. Previously, we successfully reported the use of this concept for real-time imaging and quantification of fatty acids uptake (Henkin et al., 2012), cell surface glycosylation (Cohen et al., 2010), hydrogen peroxide fluxes (van de Bittner et al., 2010), as well as studies of efficiency of delivery, linker release, and biodistribution of cell-penetrating peptide conjugates (Goun et al., 2006). Recently, the use of new red-shifted luciferin derivatives and their corresponding luciferase enzymes for multi-color application were reported (Reddy et al., 2010; Harwood et al., 2011; Conley et al., 2012; McCutcheon et al., 2012; Woodroofe et al., 2008).
In addition, to prove that “split luciferin” reaction can occur directly in living mice to form luciferin, we used this new methodology to image caspase-3/7 activation directly in living mice by “caging” the D-Cys component with the sequence of amino acids that can be specifically recognized by caspase 3/7 protease (Godinat et al., 2013). This novel technology opens up new ways for imaging many biological processes and can even be used for dual analyte detection, as reported recently for simultaneous imaging of caspase-8 and hydrogen peroxide (van de Bittner et al., 2013). Moreover, this novel technology could be applied for studies of many other proteases that are known to cleave at the end of peptidic sequences using sensitive bioluminescent imaging.
Critical Parameters
Before starting the experiments described in this protocol, careful consideration must be taken.
Multi-well plates
Even though several types of 96-well or similar plates could be used in this experiment, it is essential that the chosen plates are compatible with bioluminescent imaging and have been validated with the equipment available. For example, we propose to first measure the background signal from the plate with PBS solution before starting the real measurements. Careful attention must be paid to those parameters in order to maximize assay sensitivity.
Choice of animal model
Many animal models can be used for this study. However, in order to obtain light emission, the animals should express luciferase either ubiquitously or in certain organs, or tissues. For example, animals ubiquitously expressing luciferase under control of beta-actin promoter as well as mice expressing luciferase in certain organs are now commercially available (The Jackson Laboratory). Alternatively, animals can be injected with luciferase expressing cells subcutaneously, intraperitoneally, intravenously, by an intracardiac route or orthotopically.
In order to quantify the signal from protease activation, it is important to determine basic level of expression of luciferase enzyme, as it will greatly influence the amount of light produced. It is particularly important when this experiment is performed in tumor models where the tumor size differs significantly from one animal to another. The basic level of luciferase expression should be measured before activation of protease step and these data can be used for calibration to the level of protease expression. For generation of these data please follow “Basic protocol” number 2 (Steps 1-8).
In addition, several controls are essential for distinguishing background from protease-specific signal, such as negative control without treatment (“Basic protocol” number 2, Steps 17-28 and 31).
If the animal model used or the protease of interest are different from the ones described in the protocol, we recommend conducting an initial pilot trial on a smaller group of animals. This should help to reduce number of animals and help to optimize the imaging conditions.
Reagent purity and storage
Probes used in assays have to be of the highest purity available to minimize presence of contaminating side products. Purification of cysteine-labeled peptide is generally performed by HPLC. Luciferin and CBTs derivatives should be kept protected from direct light during experiment and storage. In order to maximize possible signal production, buffers have to be degased when possible to avoid oxidation of cysteine and its derivatives. All the solutions that are injected in animals have to be filtered through sterile filter before using in animals or sterile buffers has to be used for preparation of solutions.
Acquisition of bioluminescence signal
For all the experiments (cell-free and in vivo), it is recommended to acquire sequence of data over time for at least one hour by taking measurements every 1-10 min. One acquisition per minute is usually a good starting point. It is also important to do the same for measuring the signal from luciferin alone as this signal changes several fold over time and therefore one-point measurement will certainly lead to significant mistake in calculations.
Caspase activation
Activation of caspase 3/7 in mice using LPS/D-GalN may result in pain and distress in animals. Therefore, this experiment should be performed in agreement with the legislation that applies in the country where experiments are realized.
Troubleshooting
A troubleshooting guide for “cell-free” and “live mice” protease activity imaging experiments are presented in Table 1 and Table 2 respectively.
Table 1. Troubleshooting table for cell-free experiments.
| Problem | Cause | Solution |
|---|---|---|
| Very low or non-existent signal | Inactive Luciferase enzyme | Run a control experiment by incubating D-luciferin with luciferase enzyme. If no light emission is observed, luciferase might be degraded. Use another batch of luciferase enzyme |
| Compromised luciferase buffer | Prepare a fresh luciferase buffer (with freshly added ATP) and control activity by incubating the luciferase enzyme with D-luciferin |
|
| Exposure (integration) time is too short |
Increase the exposure time on the bioluminescence reader/imager and repeat experiment again |
|
| Too low concentration of probes | Increase probes concentrations | |
| Delay between luciferase addition and the beginning of acquisition is too long |
Decrease the delay between luciferase addition and acquisition |
|
| Low signal to background ratio (no increase in light emission when the probe is incubated with protease enzyme). |
Inactive protease enzyme. | Verify enzymatic activity of the protease using commercial colorimetric or fluorescent kit (e.g. for caspase-3 : Sigma-Aldrich : NAc-Asp-Glu-Val-Asp-pNA, ref A2559) |
| Quality of protease-specific peptide is compromised |
Verify that the peptide probe is pure and not oxidized or degraded. HPLC/MS can be used for analysis |
|
| Low purity of peptide | Very high degree of purity is necessary since traces of Free D- Cysteine can result in unspecific formation of D-luciferin and significantly increase background signal. Verify peptide purity using HPLC and MS. If stored solution was used, prepare a fresh one. |
Table 2. Troubleshooting for in vivo experiments.
| Very low or non-existent signal | Low luciferase expression in the selected animal model |
Overall, light emission from protease-specific bioluminescent probe is expected to have a lower intensity that light resulted from D- luciferin injection. If light emission generated by D-Luciferin is already low, increase dosage of protease- specific peptide and CBT |
| Quality of reagent issue | Verify that the peptide probe is not oxidized or degraded using HPLC/MS analysis |
|
| Low signal to background ratio (no difference in light emission between treatment and control groups). |
Low level of protease activation | Increase drug/effector used to activate (or decrease) protease activity |
| Verify protease activation using commercial probe (if available) or use classical ex vivo methods | ||
| Quality of protease-specific peptide is compromised |
Verify that the peptide probe is pure and not oxidized or degraded. HPLC/MS can be used for analysis |
|
| Low purity of peptide | Very high degree of purity is necessary as traces of Free D- Cysteine can result in unspecific formation of D-luciferin that significant contribute to background. Verify peptide purity using HPLC and MS analysis. If stored solution was used, prepare a fresh one |
Anticipated Results
Detection of apoptosis has become an important factor in understanding tumor pathology and finding new antitumor treatment. This assay can be used to investigate the apoptotic profile of diseases and also to evaluate cytotoxicity of drugs in vitro and in vivo by measuring accurately cell death parameters.
Figure 5 and 6 demonstrate the examples of data expected from cell-free assays. Figure 8 represents an example of results that can be obtained from in vivo experiments.
The signal to background ratio obtained is the key parameter in these experiments. A significant difference in bioluminescent light emission between treated and untreated animals is expected for both types of assays.
Time Considerations
Once all the necessary reagents are obtained, the described procedure for cell-free test tube assays can be performed within a day. In practice, a few more days should be considered if optimization process is necessary.
Experiments in live animals described in the paper typically take three days. Although, if taking into account the initial pilot trial in small group of animals in order to optimize conditions, a week or two could be required.
ACKNOWLEDGEMENT
This work was supported by the grants from Neva Foundation and industrial grant from Intrace Medical S.A., Switzerland to E.A.D. National Institutes of Health grants R01DK089202-01A1 and R01DK066336-08 to A.S. and R01EB05011 to M.B.
LITERATURE CITED
- Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- Biserni A, Martorana F, Roncoroni C, Klaubert D, Maggi A, Ciana P. [Accessed November 2012];Identification of Apoptotic Cells in Reporter Mice Using Modified Luciferin, Promega Corporation Web site. 2010 http://www.promega.com/resources/articles/pubhub/identification-of-apoptotic-cells-in-reporter-mice-using-modified-luciferin.
- Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budin G, Yang KS, Reiner T, Weissleder R. Bioorthogonal Probes for Polo-Like Kinase 1 Imaging and Quantification. Angew. Chem. Int. Ed. 2011;50:9378–9381. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. Luminogenic cytochrome P450 assays. Expert Opin. Drug Metab. Toxicol. 2006;2:629–645. doi: 10.1517/17425255.2.4.629. [DOI] [PubMed] [Google Scholar]
- Cao Y, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Prescher JA, Hangauer MJ, Bertozzi CR. Imaging cell surface glycans with bioorthogonal chemical reporters. J. Am. Chem. Soc. 2007;129:8400–8401. doi: 10.1021/ja070238o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AS, Dubikovskaya EA, Rush JS, Bertozzi CR. Real-time bioluminescence imaging of glycans on live cells. J. Am. Chem. Soc. 2010;132:8563–8565. doi: 10.1021/ja101766r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley NR, Dragulescu-Andrasi A, Rao J, Moerner WE. A selenium analogue of firefly d-luciferin with red-shifted bioluminescence emission. Angew. Chem., Int. Ed. 2012;51:3350–3353. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby N, Scurria M, Daily W, Ugo T, Promega Corp., and Promega Biosciences INC. Custom enzyme substrates for luciferase-based assays. Cell Notes. 2007;18:9–11. [Google Scholar]
- Devaraj NK, Thurber GM, Keliher EJ, Marinelli B, Weissleder R. Reactive polymer enables efficient in vivo bioorthogonal chemistry. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4762–4767. doi: 10.1073/pnas.1113466109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjugate chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu-Andrasi A, Liang G, Rao J. In vivo bioluminescence imaging of furin activity in breast cancer cells using bioluminogenic substrates. Bioconjugate Chem. 2009;20:1660–1666. doi: 10.1021/bc9002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Prescher JA, Quang CN, Bertozzi CR. Probing mucin-type O-linked glycosylation in living animals. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4819–4824. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele M, Gualillo O, Vecchione A. Animal models of human pathology 2012. J. Biomed. Biotechnol. 2012:404130. doi: 10.1155/2012/404130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger GA, Parker SE, Beothy AP, Tucker JA, Mullins MC, Kao GD. Zebrafish as a “biosensor”? Effects of ionizing radiation and amifostine on embryonic viability and development. Cancer Res. 2006;66:8172–8181. doi: 10.1158/0008-5472.CAN-06-0466. [DOI] [PubMed] [Google Scholar]
- Godinat A, Park HM, Miller SC, Cheng K, Hanahan D, Sanman LE, Bogyo M, Yu A, Nikitin GN, Stahl A, Dubikovskaya EA. A Biocompatible In Vivo Ligation Reaction and its Application for Non-Invasive Bioluminescent Imaging of Protease Activity in Living Mice. ACS Chem. Biol. 2013;8:987–999. doi: 10.1021/cb3007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goun EA, Pillow TH, Jones LR, Rothbard JB, Wender PA. Molecular transporters: synthesis of oligoguanidinium transporters and their application to drug delivery and real-time imaging. Chembiochem. 2006;7:1497–1515. doi: 10.1002/cbic.200600171. [DOI] [PubMed] [Google Scholar]
- Hangauer MJ, Bertozzi CR. A FRET-based fluorogenic phosphine for live-cell imaging with the Staudinger ligation. Angew. Chem., Int. Ed. Engl. 2008;47:2394–2397. doi: 10.1002/anie.200704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood KR, Mofford DM, Reddy GR, Miller SC. Identification of mutant firefly luciferases that efficiently utilize aminoluciferins. Chem. Biol. 2011;18:1649–1657. doi: 10.1016/j.chembiol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin AH, Cohen AS, Dubikovskaya EA, Park HM, Nikitin GF, Auzias MG, Kazantzis M, Bertozzi CR, Stahl A. Real-Time Noninvasive Imaging of Fatty Acid Uptake in Vivo. ACS chem. Biol. 2012;7:1884–1891. doi: 10.1021/cb300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson J, Ackler S, Klaubert D, Bouska J, Ellis P, Foster K, Oleksijew A, Rodriguez L, Schlessinger S, Wang B, Frost D. Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin. Cell death differ. 2010;17:1003–1010. doi: 10.1038/cdd.2009.205. [DOI] [PubMed] [Google Scholar]
- Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferla FM, Green KN. Animal Models of Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012;2:a006320. doi: 10.1101/cshperspect.a006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J. Am. Chem. Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon SP. Molecular Pathology in Cancer Therapeutics: where are we now and where are we going? Animal modeling of cancer pathology and studying tumor response to therapy. Curr. Drug Targets. 2012;13:1535–1547. doi: 10.2174/138945012803530152. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao G, Qian S, Yang Z, Chen X, Chen J, Cai C, Liang X, Guo J. Cerebrovascular protection of β-asarone in Alzheimer’s disease rats: A behavioral, cerebral blood flow, biochemical and genic study. J. ethnopharmacology. 2012;144:305–312. doi: 10.1016/j.jep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Liang G, Ren H, Rao J. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. J. Nat. chem. 2010;2:54–60. doi: 10.1038/nchem.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Mackey JL, Lopez SA, Liu F, Houk KN. Control and Design of Mutual Orthogonality in Bioorthogonal Cycloadditions. J. Am. Chem. Soc. 2012;134:17904–17907. doi: 10.1021/ja309241e. 2012. [DOI] [PubMed] [Google Scholar]
- Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J. Biomed. Opt. 2012;17:016004. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Hoyt HM, van Halbeek H, Bergman RG, Bertozzi CR. Mechanistic investigation of the staudinger ligation. J. Am. Chem. Soc. 2005;127:2686–2695. doi: 10.1021/ja044461m. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Wang W, Dicker DT, El-Deiry WS. Bioluminescent imaging of TRAIL-induced apoptosis through detection of caspase activation following cleavage of DEVD-aminoluciferin. Cancer Biol. Ther. 2005;4:885–892. doi: 10.4161/cbt.4.8.2133. [DOI] [PubMed] [Google Scholar]
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- McCaffrey A, Kay MA, Contag CH. Advancing molecular therapies through in vivo bioluminescent imaging. Mol. Imaging. 2003;2:75–86. doi: 10.1162/15353500200303124. [DOI] [PubMed] [Google Scholar]
- McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA. Expedient synthesis of electronically modified luciferins for bioluminescence imaging. J. Am. Chem. Soc. 2012;134:7604–7607. doi: 10.1021/ja301493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield RB. Automated synthesis of peptides. Science. 1965;150:178–185. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- Merrifield RB, Stewart JM. Automated peptide synthesis. Nature. 1965;207:522–523. doi: 10.1038/207522a0. [DOI] [PubMed] [Google Scholar]
- Neves AA, Stöckmann H, Harmston RR, Pryor HJ, Alam IS, Ireland-Zecchini H, Lewis DY, Lyons SK, Leeper FJ, Brindle KM. Imaging sialylated tumor cell glycans in vivo. FASEB J. 2011;25:2528–2537. doi: 10.1096/fj.10-178590. [DOI] [PubMed] [Google Scholar]
- Neves AA, Stöckmann H, Stairs S, Ireland-Zecchini H, Brindle KM, Leeper FJ. Development and evaluation of new cyclooctynes for cell surface glycan imaging in cancer cells. Chem. Sci. 2011;2:932–936. doi: 10.1039/C0SC00631A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. Genetically encoded 1,2-aminothiols facilitate rapid and site-specific protein labeling via a bio-orthogonal cyanobenzothiazole condensation. J. Am. Chem. Soc. 2011;133:11418–11421. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- Nilsson BL, Kiessling LL, Raines RT. Staudinger ligation: a peptide from a thioester and azide. Org. Lett. 2000;2:1939–1941. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]
- Ning X, Guo J, Wolfert MA, Boons G. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem., Int. Ed. Engl. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV. Homogeneous, bioluminescent protease assays: caspase-3 as a model. J. Biomol. Screening. 2005;10:137–148. doi: 10.1177/1087057104271865. [DOI] [PubMed] [Google Scholar]
- Peters M, Trembovler V, Alexandrovich A, Parnas M, Birnbaumer L, Minke B, Shohami E. Carvacrol Together With TRPC1 Elimination Improve Functional Recovery After Traumatic Brain Injury in Mice. J. Neurotrauma. 2012;29:2831–2834. doi: 10.1089/neu.2012.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol. 2010;14:80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Prescher JA, Dube DH, Bertozzi CR. Chemical remodeling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- Reddy GR, Thompson WC, Miller SC. Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J. Am. Chem. Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Xiao F, Zhan K, Kim Y, Xie H, Xia Z, Rao J. A biocompatible condensation reaction for the labeling of terminal cysteine residues on proteins. Angew. Chem., Int. Ed. 2009;48:9658–9662. doi: 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AB. Gastric Helicobacter spp. in Animal Models: Pathogenesis and Modulation by Extragastric Coinfections. Methods Mol. Biol. 2012;921:175–188. doi: 10.1007/978-1-62703-005-2_21. 2012. [DOI] [PubMed] [Google Scholar]
- Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- Saxon E, Armstrong JI, Bertozzi CR. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org. Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- Scabini M, Stellari F, Cappella P, Rizzitano S, Texido G, Pesenti E. In vivo imaging of early stage apoptosis by measuring real-time caspase-3/7 activation. Apoptosis. 2011;16:198–207. doi: 10.1007/s10495-010-0553-1. [DOI] [PubMed] [Google Scholar]
- Shah K, Tung C, Breakefield XO, Weissleder R. In vivo imaging of S-TRAIL-mediated tumor regression and apoptosis. Mol. Ther. 2005;11:926–931. doi: 10.1016/j.ymthe.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem., Int. Ed. Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten EM, Nakamura H, Jewett JC, Bertozzi CR. Difluorobenzocyclooctyne: synthesis, reactivity, and stabilization by beta-cyclodextrin. J. Am. Chem. Soc. 2010;132:11799–11805. doi: 10.1021/ja105005t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- Van Berkel SS, van Eldijk MB, van Hest JC. Staudinger ligation as a method for bioconjugation. Angew. Chem., Int. Ed. 2011;50:8806–8827. doi: 10.1002/anie.201008102. [DOI] [PubMed] [Google Scholar]
- Van de Bittner GC, Bertozzi CR, Chang CJ. Strategy for Dual-Analyte Luciferin Imaging: In Vivo Bioluminescence Detection of Hydrogen Peroxide and Caspase Activity in a Murine Model of Acute Inflammation. J. Am. Chem. Soc. 2013;135:1783–1795. doi: 10.1021/ja309078t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21316–21321. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrman TS, Degenfeld, von G, Krutzik PO, Nolan GP, Blau HM. Luminescent imaging of beta-galactosidase activity in living subjects using sequential reporter-enzyme luminescence. Nat. Methods. 2006;3:295–301. doi: 10.1038/nmeth868. [DOI] [PubMed] [Google Scholar]
- White EH, McCapra F, Field GF. The Structure and Synthesis of Firefly Luciferin. J. Am. Chem. Soc. 1963;85:337–343. [Google Scholar]
- Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 2002;78:216–224. doi: 10.1136/pmj.78.918.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. N-Alkylated 6′-aminoluciferins are bioluminescent substrates for Ultra-Glo and QuantiLum luciferase: new potential scaffolds for bioluminescent assays. Biochemistry. 2008;47:10383–10393. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]
- Yang J, Šečkutė J, Cole CM, Devaraj NK. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem., Int. Ed. Engl. 2012;51:7476–7479. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, So M, Rao J. A bioluminogenic substrate for in vivo imaging of beta-lactamase activity. Angew. Chem., Int. Ed. Engl. 2007;46:7031–7034. doi: 10.1002/anie.200701931. [DOI] [PubMed] [Google Scholar]
- Ye D, Liang G, Ma ML, Rao J. Controlling intracellular macrocyclization for the imaging of protease activity. Angew. Chem., Int. Ed. 2011;50:2275–2279. doi: 10.1002/anie.201006140. [DOI] [PubMed] [Google Scholar]
- Yuana Y, Liang G. A biocompatible, highly efficient click reaction and its applications. Org. Biomol. Chem. 2014;12:865–871. doi: 10.1039/c3ob41241e. [DOI] [PubMed] [Google Scholar]
- Zhou W, Shultz JW, Murphy N, Hawkins EM, Bernad L, Good T, Moothart L, Frackman S, Klaubert DH, Bulleit RF, Wood KV. Electrophilic aromatic substituted luciferins as bioluminescent probes for glutathione S-transferase assays. Chem. Commun. 2006:4620–4622. doi: 10.1039/b610682j. [DOI] [PubMed] [Google Scholar]
- Zhou W, Valley MP, Shultz J, Hawkins EM, Bernad L, Good T, Good D, Riss TL, Klaubert DH, Wood KV. New bioluminogenic substrates for monoamine oxidase assays. J. Am. Chem. Soc. 2006;128:3122–3123. doi: 10.1021/ja058519o. [DOI] [PubMed] [Google Scholar]