Abstract
Objectives
To determine the current state of pathology resident training in genomic and molecular pathology.
Methods
The Training Residents in Genomics (TRIG) Working Group developed survey and knowledge questions for the 2013 Pathology Resident In-Service Examination (RISE). Sixteen demographic questions related to amount of training, current and predicted future use, and perceived ability in molecular pathology vs. genomic medicine were included along with five genomic pathology and 19 molecular pathology knowledge questions.
Results
A total of 2,506 pathology residents took the 2013 RISE with approximately 600 individuals per post-graduate year (PGY). For genomic medicine, 42% of PGY-4 respondents stated they had no training compared to 7% for molecular pathology (p<0.001). PGY-4 resident perceived ability in genomic medicine, comfort in discussing results, and predicted future use as a practicing pathologist were less than reported for molecular pathology (p<0.001). There was a greater increase by PGY in knowledge question scores for molecular than for genomic pathology.
Conclusions
The RISE is a powerful tool in assessing the state of resident training in genomic pathology and current results suggest a significant deficit. The results also provide a baseline to assess future initiatives to improve genomics education for pathology residents such as those developed by the TRIG Working Group.
Keywords: Genomics, next-generation sequencing, medical education, pathology, residency training
Genomic medicine is revolutionizing patient care. Next generation sequencing methods are already being applied to prenatal diagnosis and oncology and, with decreasing costs, such testing will only increase.1-3 To best translate this new technology to the clinic, there is a clear need for health care professionals to obtain formal education in genomic medicine.4-8 There are, however, only a handful of examples of published genomic medicine curricula.9-14
Pathologists, as experts in clinical diagnostics, must not only take a leading role in the development of genomic methods; they must also act as consultants to other medical colleagues for test selection and interpretation of data.15,16 This testing includes whole exome, transcriptome, and genome sequencing that goes far beyond the single gene analysis that comprises much of existing resident molecular pathology curricula.17,18 In a survey of 42 pathology residency programs conducted in 2010, only 31% had any genomics training.19
The Training Residents in Genomics (TRIG) Working Group was established to address this educational gap. Under the aegis of the Pathology Residency Program Directors Section (PRODS) of the Association of Pathology Chairs (APC), the TRIG Working Group has taken a leading role in promoting and facilitating pathology resident genomic training. Members include experts in medical education, molecular pathology, genetic counseling and medical genetics. In 2012, the chair of the TRIG Working Group was awarded a $1.3 million grant over five years from the National Institutes of Health to further develop and assess genomic medicine curricula, with educational design support from the American Society for Clinical Pathology (ASCP). These efforts have resulted in the development of lectures, as well as resident training workshops, at major pathology meetings in 2013-14.20
One key feature of the TRIG Working Group's approach is utilization of the ASCP's pathology Resident In-Service Examination (RISE) to determine the current status of genomics training among United States residents and to track the effects of educational interventions.21 Survey and knowledge questions created by the working group are being used to determine current attitudes towards, as well as perceived and actual abilities, related to genomic pathology. Since the RISE is taken by every pathology resident in the United States, this assessment of resident experience on such a large scale is unique in medical education.
In this manuscript, we report resident responses on the 2013 RISE to both the survey and knowledge questions created by the TRIG Working Group. The results are compared to the established molecular pathology section of the exam offering a comprehensive picture of genomics training in the United States and providing a baseline to assess future initiatives to improve genomics education for pathology residents.
Materials and Methods
The Resident In-Service Examination (RISE)
The ASCP RISE has been offered annually to pathology residency programs since 1983. The RISE is administered during a two-week period in the spring with all United States pathology residency programs participating, as well as a number of international training programs. The RISE has been used by program directors as an assessment tool for medical knowledge gained during pathology residency training. RISE reports have consistently demonstrated the overall progression of attainment of medical knowledge during pathology residency training as well as the ability to predict board exam performance.21
The RISE consists of an initial untimed demographic survey section followed by a timed six-hour exam with more than 350 multiple-choice questions in a “one-best-answer” format, and each year's RISE is unique. The RISE is divided into three sections, Anatomic Pathology (AP), Clinical Pathology (CP) and Special Topics (ST) common to AP and CP. In 2013, the ST section contained nineteen molecular pathology questions (5%).
Scaled scores (i.e. linear transformation of the raw measures that are comparable across years and examinees) are calculated with 999 being the highest and 100 the lowest reportable scores. Program Directors and residents receive scores (and percentiles) in comparison with their peer- training group (postgraduate year 1, 2, 3, or 4) for each of the 10 subsections and for the total examination.
Question Design Process
The TRIG working group, utilizing e-mails and monthly conference calls, developed both survey and knowledge genomic medicine questions for the RISE. Survey questions were finalized by the chair after vetting by other members and input from ASCP experts in survey design. For genomics knowledge questions, each TRIG Working Group member wrote approximately two questions using the standard RISE question submission form. To contrast with single gene “molecular pathology” questions, the goal was to develop questions related to testing of large portions of the genome. The submitted questions were then ranked by all members for inclusion on the exam. The most highly ranked were added to the RISE as ungraded questions.
For the 2013 RISE, there were sixteen questions added to the demographic section to assess attitudes and perceived ability in genomic medicine. The specific text of each question appears in the figure legend or table footnote for the figure or table describing the response to that question. The five top-ranked questions testing knowledge in genomic pathology were included un-scored with the 19 scored molecular pathology questions in that exam section. Examinees were not aware of the un-scored nature of the genomic pathology questions. The genomic pathology knowledge questions were related to genetic association studies, terminology, test interpretation and ethical issues.
Rasch and statistical analysis
Rasch modeling is a measurement methodology by which a survey can be assessed for accuracy by comparing expected with actual responses .22 This method can be used to understand the “fit” of survey responses by comparing the performance of individual respondents on survey construct(s). For example, if experts in genomic pathology provide similar answers to a series of questions regarding their perceived ability, the survey has accurately assessed that construct. The measure of this consistency for each question is the “fit score,” based on Chi-square analysis, with an optimal score of 1.0 indicating that individuals gave the responses predicted by the Rasch model. Reliability (precision) can also be calculated giving a value that may be interpreted in a similar manner as Cronbach's alpha. Rasch analysis can also be used to assess the quality of knowledge questions; however, with such a small number of genomics questions, the results are difficult to interpret.23
Two-tailed t-tests were used to compare average Likert scores for molecular pathology vs. genomic medicine survey questions related to training time and current/future use. A one-way ANOVA was used to compare responses to the survey questions related to perceived ability in six specific topic areas. For the knowledge questions, scaled scores, instead of percentage of correct responses, were used as this analysis results in scores that are comparable from year to year. A two-tailed t-test was used to compare scaled genomic pathology question scores of residents who, on the survey, indicated they had training in genomic medicine vs. those indicating they had no training. A one-way ANOVA was used to assess any difference in molecular and genomic pathology scores by PGY. A p value of less than 0.05 was considered statistically significant.
As the RISE Exam is administered toward the end of the academic year, unless otherwise indicated, the reported data will be limited to PGY4 AP/CP residents in order to gather information for the entirety of residency training. In addition, for all ability related questions, PGY-4 residents rated their ability higher than those at other years.
Results
Demographic and Rasch analysis of the 2013 TRIG survey
There were 2,506 AP/CP residents who took the pathology RISE in 2013 with approximately 600 at each of the four PGY levels. They represented 143 programs in the US and 22 in other countries. Rasch analysis demonstrated that the survey was comprised of three constructs: training time, perceived ability, and current/future use of genomic and molecular pathology. Upon removal of the training questions and the anticipated use questions, the application questions demonstrated excellent fit statistics at 1.01 for all PGYs and 0.99 for PGY4s (optimum = 1.00). Reliability was high at 1.00 for all PGYs and 0.99 for PGY4s. There were too few questions to analyze the fit of the other constructs.
Genomic Pathology: Training and Application
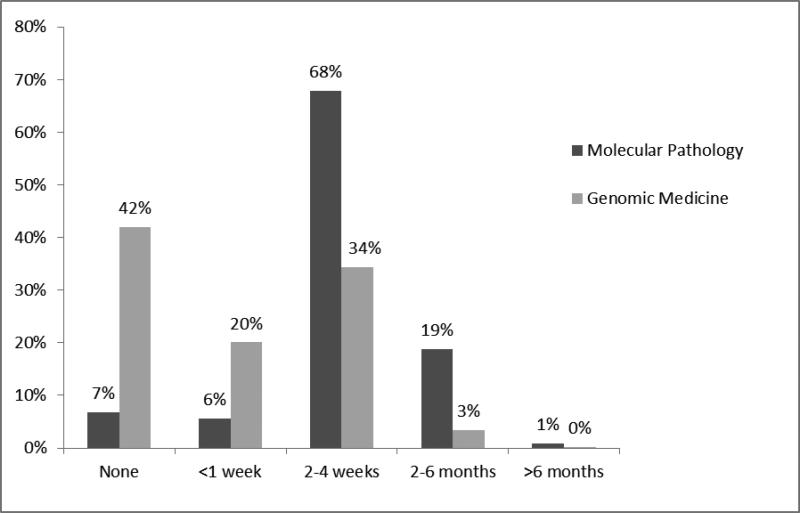
RISE Examinees were asked about the amount of training they received in molecular pathology vs. genomic medicine. There was a gradual increase by PGY in both subject areas but, by the fourth year of residency, over 90% had received some training in molecular pathology compared to only 58% for genomic medicine.(figure 1, p<0.001). When training did occur, PGY4s reported that it was typically for 2-4 weeks for both topic areas.
Figure 1.
PGY-4 resident reported training in molecular pathology and genomic medicine. Examinees were asked “please indicate how much training you have completed during your residency in (molecular pathology/genomic medicine).” Mean(molecular)= 2-4 weeks, Mean(genomics) =< 1 week; p <0.001, independent samples t-test.
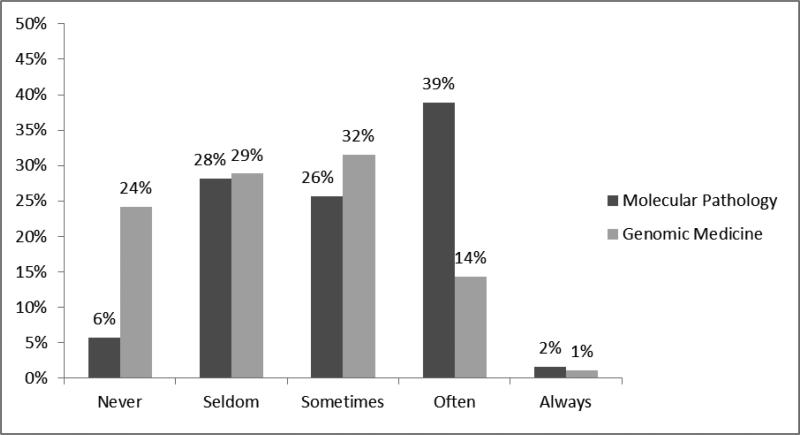
With regard to applications of training, 24% of PGY-4 residents responded that they “never use” genomic medicine as a resident, compared to only 6% for molecular pathology. (figure 2, p <0.001) In a similar vein, 39% responded they “often use” molecular pathology, compared to only 14% for genomic medicine. A similar result was noted when residents were asked their expected use of molecular pathology or genomic medicine as a practicing pathologist (figure 3, p <0.001). While there was less of a difference in those stating that they will never use genomic medicine vs. molecular pathology, (14% vs. 9%), almost 50% responded “often” for molecular pathology, as compared to only 28% for genomic medicine.
Figure 2.
PGY-4 resident reported use of molecular pathology vs. genomic medicine during their training. Examinees were asked “I am using (molecular pathology/genomic medicine) as a pathology resident.” Likert score mean(molecular)= 3.03, mean(genomics) =2.39; p<0.001, independent samples t-test
Figure 3.
PGY-4 resident predicted use of molecular pathology vs. genomic medicine as a practicing pathologist. Examinees were asked “I will use (molecular pathology/genomic medicine) as a practicing pathologist.” Likert score mean(molecular)= 3.23, mean(genomics) =2.82; p<0.001, independent samples t-test
Perceived Ability
Residents were asked to describe their ability, on a scale of 1 (poor) to 5 (excellent), to perform interpretative and consultative tasks related to molecular or genomic pathology. For PGY-4 residents, topics associated with genomic pathology such as interpreting next generation sequencing (NGS) data or genome wide association studies (GWAS), were rated lower than those associated with molecular pathology, such as interpreting sequencing or polymerase chain reaction (PCR) data. Topics with greater methodological overlap between molecular and genomic pathology, such as interpreting microarray data and the clinical significance of a single nucleotide polymorphism (SNP), had ratings intermediate to those more purely associated with the individual topic areas. There was a significant difference in response by topic area. (table 1, p<0.001)
Table 1.
PGY-4 resident ability ratings by topic area
| Subject Area* | Specific Topic | Percentage Responding (Excellent/Very Good) | Mean**+ |
|---|---|---|---|
| Molecular pathology | Polymerase Chain Reaction (PCR) | 25% | 2.85 |
| DNA Sequencing | 18% | 2.60 | |
| Combined topics | Single nucleotide polymorphism (SNP) | 11% | 2.25 |
| Microarray data | 11% | 2.24 | |
| Genomic medicine | Next- generation sequencing (NGS) | 9% | 2.08 |
| Genome- wide association studies (GWAS) | 6% | 1.94 |
Specific topics grouped into three subject areas by authors
Examinees were asked to rate their “ability to determine the clinical significance” of data related to each specific topic. Response options: (1) Poor, (2)Fair, (3)Good, (4)Very Good, (5)Excellent
P <0.001 (one-way ANOVA comparing topic areas)
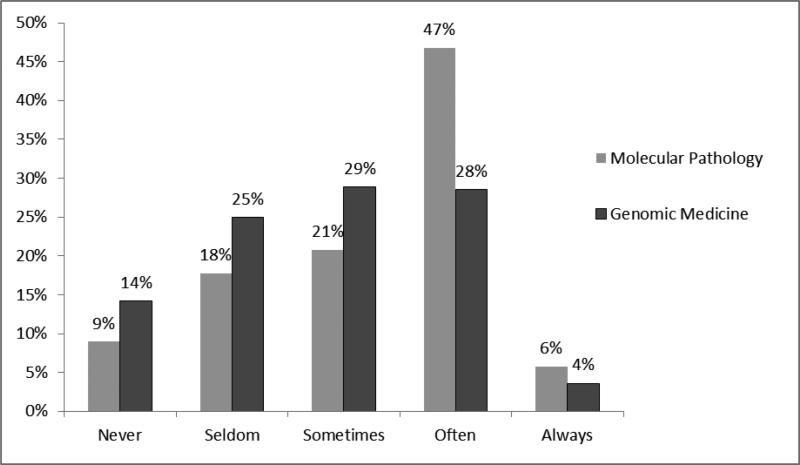
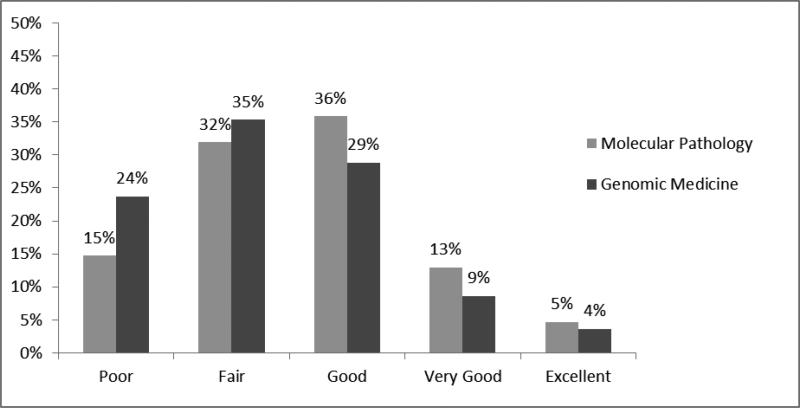
In regard to the ability to discuss results with other providers, 24% of PGY-4 residents selected a “poor” rating for genomic medicine compared to 15% for molecular pathology. (figure 4, p<0.001). While the ability rating was significantly lower for genomic medicine, almost 50% still rated their consultative ability in molecular pathology as only “poor” or “fair.” Similar results were obtained regarding resident perceived ability to discuss results with a patient (data not shown). Of note, for the 42% residents who reported no genomic medicine training, 25% still reported “good” to “excellent” ability to discuss results with a clinician. For the 58% with some genomic medicine training, this value was 53%.”
Figure 4.
PGY-4 residents’ reported ability to discuss molecular pathology vs. genomic medicine test results with a provider. Examinees were asked to rate their ability to “knowledgably discuss the results of (molecular pathology/genomic medicine) testing with a clinician.” Likert score mean(molecular)=2.61, mean(genomics) =2.33; p<0.001, t- test for independent samples.
Knowledge questions
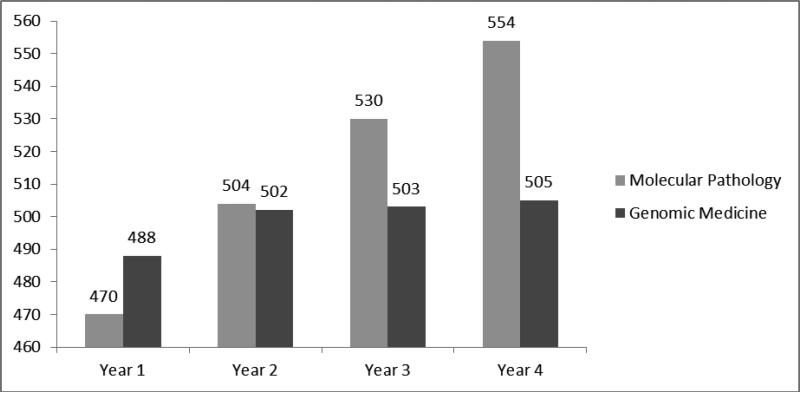
While there was a statistically significant difference in comparing scores by PGY for both molecular pathology (p<.001) and genomic pathology (p=0.03), molecular scores steadily increased from 470 in PGY-1 to 554 in PGY-4. In contrast, genomic pathology scores increased from 488 to 502 between PGY-1 and PGY-2 and then leveled off through the remainder of training with a PGY-4 average of 505.(figure 5) Individuals who, on the survey, indicated some training in genomic medicine had higher genomic pathology scores than those who indicated they had no training. (517 vs. 493; p<.001)
Figure 5.
2013 RISE molecular pathology vs. genomic medicine knowledge question score comparison by PGY. The scaled results for the 19 molecular pathology questions and 5 genomic medicine questions are presented by PGY.
Discussion
What do the 2013 RISE results tell us?
The 2013 RISE demonstrates that training in genomic pathology lags behind that of molecular pathology. Approximately 40% of PGY-4 residents have not received any instruction in genomic medicine as compared to 7% for molecular pathology. This lack of training translated to lower perceived ability with regard to genomic pathology related topics such as interpreting genome-wide association studies (GWAS) and next-generation sequencing data (NGS) as well as a greater increase by PGY in knowledge question scores for molecular than for genomic pathology.
Given that genomic pathology is less established as a clinical discipline than molecular pathology, the above results are not surprising nor is the finding that residents are more comfortable discussing test results related to molecular pathology than genomic medicine with other clinicians and patients. One unexpected result is that about a quarter of residents with no reported genomic training still felt comfortable (i.e., a rating of “good” or better) discussing genomic test results with a clinician. It is possible that residents may believe that they are able to apply some concepts from molecular pathology towards ability to discuss genomic medicine with providers. In addition, some residents may have had experience in genomics prior to residency or picked up information through their own reading or other venues outside of residency training. Regardless, the majority of residents have only a self-reported “poor” or “fair” ability explaining genomic testing to providers. Another unexpected result of our survey is that residents anticipated using genomic medicine less than molecular pathology in their future as practicing pathologists. These data suggest that the message regarding the future importance of genomic pathology in clinical practice has not yet reached many residents.
The TRIG Working Group and Improving Resident Genomic Pathology Training
The TRIG Working Group is taking action to improve training in genomic pathology. A daylong resident workshop was held at the 2013 ASCP annual meeting. The seminar used a “flipped classroom” approach; brief lectures with the majority of time spent on group-oriented application exercises.24,25 Working in teams, residents used online genomics tools to answer clinical questions about a theoretical breast cancer patient.
Similar workshops are planned for the 2014 United States and Canadian Academy of Pathology (USCAP) and Academy of Clinical Laboratory Physicians and Scientists (ACLPS) Annual Meetings. A three-hour course was also presented at the 2013 College of American Pathologists (CAP) Annual Meeting which converted the interactive nature of the workshop into a lecture based format. Session participants were encouraged to follow along using their tablets and laptops as speakers demonstrated online genomic tools.
A rigorous post-workshop survey analysis is being performed to hone the workshops with the goal of creating a “train-the-trainer” guide. The aim of this guide is that faculty at residency sites with expertise, but insufficient time to create a curriculum, can utilize this guide as a framework to teach their residents and then customize as needed for their program. For sites lacking faculty expertise, the TRIG Group, using NIH grant funding, plans to transform the workshop into online modules and to subsequently test the efficacy of these modules, using a validated genomic pathology exam, at four pathology residency sites.
Using the RISE: Limitations and Future Directions
The RISE is a powerful tool for assessing the current state of genomic pathology training. There are, of course, inherent limitations. Survey questions can be subjective such that interpretation will vary from resident to resident. In particular, there are many different types of genomic assays and the survey cannot pinpoint how each resident interpreted “genomic medicine training.” In this initial iteration of the survey, the goal was for the resident to use their own interpretation to focus specifically on perceptions as opposed to specific topic areas. In the future, survey questions can be added to determine actual thematic content and structure of resident training.
In addition, suggesting consistent interpretation, across all types of questions (i.e., training time, perceived ability, and comfort discussing results) PGY-4 resident responses for molecular pathology vs. genomic medicine demonstrated similar differences. In addition, Rasch analysis demonstrated good survey question “fit” and reliability. Interestingly, questions regarding the amount of training fell into a separate construct from the perceived ability questions. This suggests that that simply having training is not enough to translate to comfort with genomic pathology-related tasks; such training must be of high quality. Still, residents who reported some prior genomic medicine training did achieve significantly higher scores on the genomic pathology portion of the exam than those who reported no training. To get a better sense of the basis for perceived vs. actual ability, we plan on adding additional survey questions related to quality of training to future exams.
While a statistically significant difference by PGY was seen for both the molecular and genomic pathology average knowledge question scores, the magnitude of the difference for PGY-1 vs. PGY-4 residents was greater for molecular pathology. This analysis is clearly limited by the small number of genomics questions, and furthermore, these questions were created without a direct and structured relationship to TRIG Working Group curriculum objectives. As such, there was a limited foundation to develop genomic questions of greatest importance to assessing pathology resident knowledge. In future iterations of the RISE, we will employ a larger bank of questions derived from the TRIG curriculum, to better (and more relevantly) assess resident genomic pathology knowledge.
Despite the limitations mentioned, the RISE can provide objective educational data on a scale rarely seen in medical education. With these current results, we now have a baseline to build upon and truly assess if initiatives of the TRIG Working Group (and others) are improving genomic pathology education. Given the already existing expertise in molecular diagnostic pathology, we must train pathology residents to be leaders in genomics-related testing in order to provide the best patient care. The current data suggests we have considerable work to do and that the RISE will allow us to track progress in a comprehensive manner.
Acknowledgments
This work was supported by the National Institutes of Health (1R25CA168544-01).
References
- 1.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morain SM, Greene MF, Mello MM. A new era in noninvasive prenatal testing. New Engl J Med. 2013;369:499–501. doi: 10.1056/NEJMp1304843. [DOI] [PubMed] [Google Scholar]
- 4.Guttmacher AE, Porteous ME, McInerney JD. Educating health-care professionals about genetics and genomics. Nat Rev Genet. 2007;8:151–7. doi: 10.1038/nrg2007. [DOI] [PubMed] [Google Scholar]
- 5.Salari K. The dawning era of personalized medicine exposes a gap in medical education. PLoS Med. 2009;6:e1000138. doi: 10.1371/journal.pmed.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson EA, McGuire AL. The need for medical education reform: genomics and the changing nature of health information. Genome Medicine. 2010;2:18. doi: 10.1186/gm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feero WG, Green ED. Genomics education for health care professionals in the 21st century. JAMA. 2011;306:989–90. doi: 10.1001/jama.2011.1245. [DOI] [PubMed] [Google Scholar]
- 8.Haspel RL, Arnaout R, Briere L, et al. A call to action: training pathology residents in genomics and personalized medicine. Am J Clin Pathol. 2010;133:832–4. doi: 10.1309/AJCPN6Q1QKCLYKXM. [DOI] [PubMed] [Google Scholar]
- 9.Salari K, Pizzo PA, Prober CG. Commentary: to genotype or not to genotype? Addressing the debate through the development of a genomics and personalized medicine curriculum. Acad Med. 2011;86:925–7. doi: 10.1097/ACM.0b013e3182223acf. [DOI] [PubMed] [Google Scholar]
- 10.Dhar SU, Alford RL, Nelson EA, et al. Enhancing exposure to genetics and genomics through an innovative medical school curriculum. Genet Med. 2012;14:163–7. doi: 10.1038/gim.0b013e31822dd7d4. [DOI] [PubMed] [Google Scholar]
- 11.Walt DR, Kuhlik A, Epstein SK, et al. Lessons learned from the introduction of personalized genotyping into a medical school curriculum. Genet Med. 2011;13:63–6. doi: 10.1097/GIM.0b013e3181f872ac. [DOI] [PubMed] [Google Scholar]
- 12.Wiener CM, Thomas PA, Goodspeed E, et al. “Genes to society”--the logic and process of the new curriculum for the Johns Hopkins University School of Medicine. Acad Med. 2010;85:498–506. doi: 10.1097/ACM.0b013e3181ccbebf. [DOI] [PubMed] [Google Scholar]
- 13.Haspel RL, Arnaout R, Briere L, et al. A curriculum in genomics and personalized medicine for pathology residents. [1/28/14];Am J Clin Pathol. 2010 133(6) doi: 10.1309/AJCPN6Q1QKCLYKXM. online supplement. http://ajcp.ascpjournals.org/site/misc/pdf/Haspel_online.pdf. [DOI] [PubMed] [Google Scholar]
- 14.Schrijver I, Natkunam Y, Galli S, et al. Integration of genomic medicine into pathology residency training: the Stanford Open Curriculum. J Mol Diagn. 2013;15:141–8. doi: 10.1016/j.jmoldx.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Tonellato PJ, Crawford JM, Boguski MS, et al. A national agenda for the future of pathology in personalized medicine: report of the proceedings of a meeting at the Banbury Conference Center on genome-era pathology, precision diagnostics, and preemptive care: a stakeholder summit. Am J Clin Pathol. 2011;135:668–72. doi: 10.1309/AJCP9GDNLWB4GACI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross JS. Next-generation pathology. Am J Clin Pathol. 2011;135:663–5. doi: 10.1309/AJCPBMXETHAPAV1E. [DOI] [PubMed] [Google Scholar]
- 17.J Mol Diagn. 1999;Goals and objectives for molecular pathology education in residency programs. The Association for Molecular Pathology Training and Education Committee.1:5–15. doi: 10.1016/s1525-1578(10)60603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith BR, Wells A, Alexander CB, et al. Curriculum content and evaluation of resident competency in clinical pathology (laboratory medicine): a proposal. Hum Pathol. 2006;37:934–68. doi: 10.1016/j.humpath.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Haspel RL, Atkinson JB, Barr FG, et al. TRIG on track: educating pathology residents in genomic medicine. Personalized Medicine. 2012;9:287–293. doi: 10.2217/pme.12.6. [DOI] [PubMed] [Google Scholar]
- 20.TRIG Working Group Website [1/28/14];2013 Available via ascp.org/TRIG. [Google Scholar]
- 21.Rinder HM, Grimes MM, Wagner J, et al. Senior pathology resident in-service examination (RISE) scores correlate with outcomes of the American Board of Pathology (ABP) certifying examinations. Am J Clin Pathol. 2011;136:499–506. doi: 10.1309/AJCPA7O4BBUGLSWW. [DOI] [PubMed] [Google Scholar]
- 22.Wright Benjamin, Stone Mark. Measurement Essentials. Wide Range Press; Wilmington, DE: 1999. [Google Scholar]
- 23.De Champlain AF. A primer on classical test theory and item response theory for assessments in medical education. Medical Education. 2010;44:109–117. doi: 10.1111/j.1365-2923.2009.03425.x. [DOI] [PubMed] [Google Scholar]
- 24.Michaelsen LK, Sweet M. The essential elements of team based learning. In: Michaelsen LK, Sweet M, Parmalee DX, editors. Team-Based Learning: Small Group Learning's Next Big Step: New Directions for Teaching and Learning, Number 116. Wiley; Hoboken: 2008. [Google Scholar]
- 25.McLaughlin JE, Roth MT, Glatt DM, et al. The Flipped Classroom: A Course Redesign to Foster Learning and Engagement in a Health Professions School. Acad Med. 2013 Nov 21; doi: 10.1097/ACM.0000000000000086. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]