Abstract
One of the ultimate goals in plant systems biology is to elucidate the genotype-phenotype relationship in plant cellular systems. Integrated network analysis that combines omics data with mathematical models has received particular attention. Here we focus on the latest cutting-edge computational advances that facilitate their combination. We highlight (1) network visualization tools, (2) pathway analyses, (3) genome-scale metabolic reconstruction, and (4) the integration of high-throughput experimental data and mathematical models. Multi-omics data that contain the genome, transcriptome, proteome, and metabolome and mathematical models are expected to integrate and expand our knowledge of complex plant metabolisms.
Keywords: network visualization, pathway analysis, flux balance analysis, genome-scale metabolic reconstruction, plant metabolism
Introduction
Plants are a paramount source of food, energy, and valuable compounds. The developing field of plant systems biology has provided outstanding insights into how these products are synthesized; its ultimate goal is an understanding of the genotype-phenotype relationship in cellular systems (Kell, 2002; Benfey and Mitchell-Olds, 2008; Weckwerth, 2011). Recent technical advances in high-throughput sequencing and various analytical instruments have made it possible to comprehensively measure and analyze genes, transcripts, proteins, and metabolites (Fukushima et al., 2009; Lei et al., 2011; Lucas et al., 2011; Stitt, 2013). These omics technologies are not only platforms that monitor the cellular inventory, but they also provide the opportunity to evaluate cellular behaviors from a multi-level perspective and enhance our understanding of plant systems (Krouk et al., 2010; Saito and Matsuda, 2010; Dhondt et al., 2013).
Major effective and efficient approaches to analyze omics data are network- and pathway analysis (for example, see, Ramanan et al., 2012; Carter et al., 2013). The former is based on the network concept derived from mathematical graph theory and typically represents a biological component (e.g., a gene) as a node and physical-, genetic-, and/or functional interactions as a link in the network to visualize and interpret the omics data (“data-driven approach”). On the other hand, pathway analysis is a knowledge-based approach that involves the associated biochemical pathway. Enrichment analysis approaches can be combined with pathway analysis to evaluate whether a particular molecular group is significantly over-represented. Examples are gene set enrichment analysis (Hung et al., 2012), Metabolite Set Enrichment Analysis (MSEA) (Xia and Wishart, 2010), and other functional enrichment analyses using gene ontology (GO) and biochemical pathways (for comprehensive reviews see Chagoyen and Pazos, 2013 or Khatri et al., 2012).
For a holistic view of plant metabolisms, measuring the metabolic flux by experimental flux analysis, e.g., metabolic flux analysis (MFA) (Libourel and Shachar-Hill, 2008; Sweetlove et al., 2014) or in silico flux modeling, e.g., flux balance analysis (FBA) (Kruger and Ratcliffe, 2012; Junker, 2014) is also important. FBA is a constraint-based approach for predicting flux through reactions in a quantitative manner (Orth et al., 2010; Sweetlove and Ratcliffe, 2011); it complements experimental flux analysis. It does not use knowledge of kinetic parameters from metabolic reactions but relies solely on the stoichiometric balance assuming steady-state conditions. These models can be extended to a level that almost fully includes the metabolism. Indeed, the past few years have seen an increase in the use of genome-scale metabolic models in plants (Collakova et al., 2012; Seaver et al., 2012; De Oliveira Dal'molin and Nielsen, 2013). Integrated network analysis by combining omics data with mathematical models has become popular. In this review we focus on the latest cutting-edge computational advances for analyzing omics networks and performing pathway analysis. We highlight (1) network visualization tools, (2) pathway analyses, (3) genome-scale metabolic reconstruction, and (4) the integration of high-throughput experimental data with mathematical modeling. These topics correspond to interaction-based and constraint-based approaches to the mathematical modeling of cellular networks as classified by Stelling (2004), Lewis et al. (2012).
Network visualization and pathway analysis tools for interaction-based approaches
The relationship between the biological components of a biological network includes four types of interactions: physical interactions (e.g., drug targets Yildirim et al., 2007 and protein-protein interactions Brandao et al., 2009), genetic interactions (Costanzo et al., 2010), and functional interactions (e.g., biochemical/signaling pathways Caspi et al., 2012; Kanehisa et al., 2014). Interaction-based approaches such as topological analysis (e.g., shortest path search Yu et al., 2014, centrality analysis Carrera et al., 2009, and network module detection Altaf-Ul-Amin et al., 2006), correlation network analysis (Provart, 2012), or enrichment analysis (Hung et al., 2012) have been used to construct and analyze biological networks from omics data. For example, GeneMANIA (Montojo et al., 2010; Zuberi et al., 2013) is a web-based interaction network for the visualization of physical, genetic, and functional interactions. Network visualization tools (e.g., igraph, http://igraph.org/) can not only describe a biological network, but also calculate and perform computational analysis (for a comprehensive review see Gehlenborg et al., 2010). Furthermore, network visualization tools assist the database client and facilitate data integration (Table 1).
Table 1.
List of software discussed in this review: Network tools for metabolic system biology analysis and related data formats.
| Software name | Description | Supported data formats (built-in annotation data) | Interface/ Language | Reference/Author name | URL |
|---|---|---|---|---|---|
| NETWORK AND PATHWAY ANALYSIS TOOL | |||||
| igraph | Library for the analysis of networks. It supports topological- and centrality analysis. | (in most cases) adjacency list, edge list, GraphML | R, Python, C/C++ | http://igraph.org/index.html | |
| Networkx | A Python language software library for the analysis of networks. This package supports topological- and centrality analysis. | (in most cases) adjacency list, edge list, GraphML, json | Python | http://networkx.github.io/ | |
| CentiScaPe | Centralities analysis plug-in for Cytoscape. | Java | Scardoni et al. (2014) | http://apps.cytoscape.org/apps/centiscape | |
| CentiLib | Centralities analysis plug-in for VANTED. | Java | Gräßler et al. (2012) | http://centilib.ipk-gatersleben.de/ | |
| BiNGO | Gene ontology enrichment analysis plug-in for Cytoscape. | Java | Maere et al. (2005) | http://apps.cytoscape.org/apps/bingo | |
| FluxMap | FBA plug-in for VANTED. | Java | Rohn et al. (2012a) | http://vanted.ipk-gatersleben.de/addons/fluxmap/ | |
| Enrichment Map | A Cytoscape app/plug-in to perform and visualize pathway/gene set enrichment analysis. | Java | Merico et al. (2010) | http://apps.cytoscape.org/apps/enrichmentmap | |
| Network visualization | |||||
| Cytoscape | A widely used biological network analysis platform. This software supports many biological network data formats and its visual appearance is fully customizable. The 'NetworkAnalyzer' default plug-in computes basic properties of the whole network. A huge number of plug-ins is available from the Cytoscape App Store. In contrast to VANTED, this software does not support KGML by default. | SBML, BioPAX, edge list, PSI-MI | Java | Smoot et al. (2011) | http://www.cytoscape.org/ |
| KEGGscape | A Cytoscape plug-in for KGML import. | KGML | Java | Nishida et al. (2014) | http://apps.cytoscape.org/apps/keggscape |
| VANTED | Along with Cytoscape, a popular network analysis software that also supports many biological network data formats. Its visual appearance is fully customizable. | SBML, BioPAX, edge list, KGML, DOT | Java | Rohn et al. (2012b) | http://VANTED.ipk-gatersleben.de/ |
| Kappa-View | A web-based correlation network viewer on metabolic pathway maps. | User's own omics data (AraCyc version8) | Java | Sakurai et al. (2011) | http://kpv.kazusa.or.jp/ |
| MapMan | The view is not a network graph, but it is useful for displaying omics data onto diagrams of metabolic pathways or other processes. | User's own omics data (MapMan ontology) | Java | Usadel et al. (2005) | http://mapman.gabipd.org/ |
| Pathview | An R/Bioconductor package for pathway-based data integration and visualization. | Bioconductor compliant data | R | Luo and Brouwer (2013) | http://www.bioconductor.org/packages/release/bioc/html/pathview.html |
| GeneMANIA | A web-based interaction network (including physical and genetic interactions, and co-expression and prediction networks) viewer. Also accessible via a Cytoscape plug-in. | Gene list | Java | Montojo et al. (2014) | http://genemania.org/ |
| CellDesigner | A software for browsing and modifying existing SBML models. | SBML | Java | Funahashi et al. (2008) | http://www.celldesigner.org/ |
| PathVisio | Pathway drawing and analysis tool for WikiPathways (http://www.wikipathways.org). | GPML | Java | Van Iersel et al. (2008) | http://www.pathvisio.org/ |
| Data format | |||||
| KGML | XML for the KEGG PATHWAY database (available without the KEGG FTP academic subscription). Commonly used to reconstruct a KEGG pathway network layout. It can be used for the visualization of pathway maps of a multitude of different organisms. | Kanehisa et al. (2014) | http://www.kegg.jp/kegg/xml/ | ||
| BioPAX | General format for pathway data. It is defined in OWL and represented in the RDF/XML format. Its main focus is data exchange and integration. | Demir et al. (2010) | http://www.biopax.org/ | ||
| SBML | Tuned and commonly used format for mathematical models of biological networks/pathways. Almost all metabolic reconstructions are written in this format. | Hucka et al. (2003) | http://sbml.org/ | ||
| PSI-MI | Suitable format for representing details about molecular-, especially protein-protein interaction data. Arabidopsis Interactome Mapping Consortium data are distributed in this format. | Van Roey et al. (2013) | https://code.google.com/p/psimi/ | ||
Network visualization and pathway analysis tools
Data analysis of biological networks by graph representations includes topological analysis (for an example see, Toubiana et al., 2013). For functional networks, correlation and enrichment analyses can be used. Correlation analysis is based on associations between biological components (e.g., genes and metabolites). The Pearson correlation coefficient is a special case of association that evaluates linear relationships among molecular abundances (Kusano and Fukushima, 2013). Enrichment analysis uses a given molecular group such as gene ontology and biochemical pathways. Some network visualization tools implement these approaches while others involve independent, plug-in software modules (e.g., Cytoscape Smoot et al., 2011 and VANTED Rohn et al., 2012b). Cytoscape apps/plug-ins include BiNGO (Maere et al., 2005) for GO enrichment analysis and FluxMap for FBA (Rohn et al., 2012a). Network analysis platforms such as Enrichment map (Merico et al., 2010) feature system flexibility and expandability for omics data. Most network visualization tools manage and visualize network data that correspond to the type of interaction. For example, when performing a quality check of protein-protein interaction data generated from a high-throughput yeast two-hybrid screening system, these tools can visualize a giant network component from a large number of interactions (for example see Arabidopsis Interactome Mapping Consortium, 2011). For functional interactions, when mapping transcriptomics profiles onto metabolic pathways, a pathway-level representation of the gene expressions involved can be assessed (Usadel et al., 2005; Sakurai et al., 2011). The network visualization tool requires a sophisticated function, structured and controlled functional categories, and vocabularies, to inspect the profile data on a pathway. Two typical functional categories are the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2014) and GO, which can be used to evaluate perturbed pathways in the omics data. For physical interaction networks, concerted efforts are being made to share data formats that visualize biological networks such as the PSI-MI format (Van Roey et al., 2013) in IntAct (Kerrien et al., 2012).
Network/pathway data formats
There are many data formats for functional interaction networks, especially biochemical pathway databases. As KEGG XML (KGML) (Kanehisa et al., 2014), BioPAX (Demir et al., 2010), and SBML (Hucka et al., 2003) are available for pathway data exchanges, a network visualization tool that implements and supports these data formats as import and export functions is desirable. For example, AraCyc (Zhang et al., 2005) and Arabidopsis Reactome (Tsesmetzis et al., 2008) are also represented in the BioPAX format (Table 1). BioPAX is defined in Web Ontology Language (OWL); it contains the most comprehensive ontology for representing pathway knowledge. It can also serve as a Resource Description Framework (RDF) for describing information on the world-wide-web (Jupp et al., 2014) and it is expected to utilize semantic data integration. According to Strömbäck and Lambrix (2005), SBML (Hucka et al., 2003) is the most widely used and finely tuned format for mathematical models (e.g., the FBA model). KEGG pathways are manually drawn and the layout is created by domain experts. Because KGML includes all KEGG pathway layout information, it uses another SBML-based software/database to reconstruct a pathway map. Although it is a de facto standard network visualization tool and supports most data formats, Cytoscape (Smoot et al., 2011) cannot seamlessly integrate all data resources irrespective of the data schema and controlled ontology. There are considerable community-wide efforts in the sustainable development and integration of various database resources with RDF (for example see BioHackathon Katayama et al., 2014, the Rhea database Alcantara et al., 2012, and Path2Models Buchel et al., 2013). The number of Wiki-based databases (Arita, 2009) is also increasing; this community curation process includes WikiPathways (Hanumappa et al., 2013) with PathVisio (Van Iersel et al., 2008) and LipidBank (http://jcbl.jp/wiki/Category:LB). Currently, several pathway resources are often combined with SBML to use FBA (see Section Genome-scale metabolic reconstruction in plants and constraint-based approaches).
Visualization of omics data for exploring biological networks
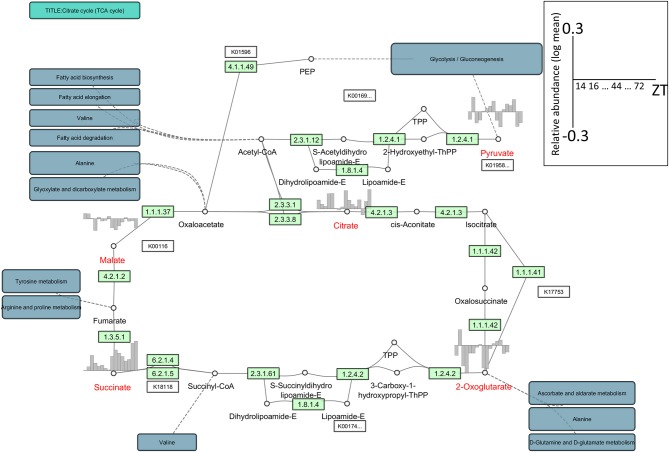
Optimal network visualization tools must allow the seamless integration of multiple data resources and their comparison, irrespective of differences in the data formats generated by primary data providers. However, with currently available network visualization tools, the integration of different data resources remains difficult although the visualization of omics data has been partially achieved with tools such as VANTED and Pathview (Luo and Brouwer, 2013). As an example, to visualize metabolomic data we used Cytoscape with its app/plug-in KEGGscape (http://apps.cytoscape.org/apps/keggscape) (Nishida et al., 2014) and VANTED. KEGGscape supports KEGG pathway files in KGML format and reproduces KEGG pathway diagrams as a standard network object in Cytoscape. Users can easily integrate their own datasets with biologist-friendly KEGG pathway diagrams. Figure 1 is a network representation of the time-series metabolome (Espinoza et al., 2010) in Arabidopsis thaliana using KEGGscape. We integrated the metabolite profiles and the tricarboxylic acid (TCA) cycle with the KEGG compound IDs as the keys. Although Cytoscape and VANTED are different in design, both tools can visualize the same figure (Supplemental Figure S1) and we posit that they will be widely used to visualize omics-profiles on pathway maps. Such network visualization allows users to consider pathway-related profile variations that cannot be inferred immediately from the profile.
Figure 1.
An example of the network representation of time-series metabolome data in Arabidopsis using KEGGscape (http://apps.cytoscape.org/apps/keggscape). The datasets were sampled with 4-h resolution under a 16 h/8 h light/dark cycle at 20°C (Espinoza et al., 2010). We used the KEGG pathway map (ath00020), the tricarboxylic acid (TCA) cycle, or the citrate cycle. We queried MetMask (Redestig et al., 2010) for a list of KEGG compound IDs associated with a list of predefined metabolite names and picked up the most pathway-mapped KEGG compound ID for each metabolite. Metabolite names shown in red represent detected compounds in the dataset. The diurnal changes were visualized on bar charts ranging from −0.3 to 0.3 in log-mean values. ZT, Zeitgeber time.
Genome-scale metabolic reconstruction in plants and constraint-based approaches
Both MFA and FBA use stoichiometric simulation to estimate and predict cellular metabolic flux. Although MFA with 13C labeling is the most promising approach to characterize metabolic phenotypes in a cell, technological issues prevent its application to complete metabolisms. In this section we focus on genome-scale metabolic reconstruction and FBA.
A genome-scale metabolic reconstruction
Due to the already extremely large and growing amount of genomic sequences yielded by high-throughput techniques, metabolism reconstruction from an organism's genome sequence has become possible (Thiele and Palsson, 2010). Although the metabolism has been reconstructed for only a few of the sequenced plant genomes, it has been modeled in some plants and crops (Seaver et al., 2012; De Oliveira Dal'molin and Nielsen, 2013). The first step in metabolic reconstruction from genome sequences involves the collection and integration of compounds, enzymes, genes, and curated published pathway databases. Subsequently, gene-protein-reaction (GPR) relationships in an organism (Fell et al., 2010) are identified and a stoichiometric matrix consisting of substances and reactions is generated. This draft metabolism requires further curation including metabolic gap filling and FBA. To explore flux states computationally, FBA uses the optimization of an objective function and predicts the growth rate of an organism or the production rate of industrially and medicinally important metabolites (Feist and Palsson, 2010). The collection of information on the biomass including proteins, amino acids, and lipid(s) as an objective function is required. Because these steps tend to be time-consuming, rapid algorithms for reconstructing genome-scale metabolisms have been developed (Chen et al., 2012; Kim et al., 2012). On SEED (Henry et al., 2010) and PlantSEED (Seaver et al., 2014) servers a significant number of genome-scale metabolisms in different organisms has already been reconstructed. In this review we do not present a comprehensive review of software tools/algorithms involved in reconstructing a genome-scale model and FBA.
Reconstructed plant metabolisms
The first genome-scale models in plants were designed and published for barley seeds and heterotrophic Arabidopsis cells. Grafahrend-Belau et al. (2009) constructed a compartmentalized barley seed metabolism model and performed mathematical simulations to investigate storage patterns that included responses to environmental and genetic perturbations (Grafahrend-Belau et al., 2009). Their comparison of published data for grain yields and growth rates with in silico data showed good reproducibility, indicating the usefulness of their model for predicting the seed storage metabolism. Poolman et al. (2009) generated an Arabidopsis genome-scale metabolic model using the AraCyc database (Poolman et al., 2009). They demonstrated that only 15% of the reactions in the reconstructed network (“minimal network”) were required to produce amino acids, nucleotides, and other biomass components. For the Arabidopsis metabolism, two other models are available, i.e., AraGEM (De Oliveira Dal'molin et al., 2010) and the model of Radrich (Radrich et al., 2010). In addition, 7 tissue-specific models for Arabidopsis have been presented (Mintz-Oron et al., 2012). The model of Poolman et al. (2009) was extended and updated to include more information on the subcellular localization of enzymes and transport reactions (Cheung et al., 2013) and to model the leaf metabolism over a day-night diel cycle (Cheung et al., 2014). The approach with MFA demonstrated a marked improvement in the quantitative match between predicted- and experimentally-estimated fluxes. To assess the central carbon partitioning and enzyme costs precisely, Arnold and Nikoloski (2014) newly reconstructed the Arabidopsis metabolism based on genomic and bibliomic data that included biochemical, genomic, and genetic information on compartmentalization and transport processes. Their model produced all amino acids and was able to estimate various cell performances (Arnold and Nikoloski, 2014). De Oliveira Dal'molin et al. (2010) constructed a genome-scale metabolic model for C4 plants (C4GEM) (Dal'molin et al., 2010), Saha et al. (2011) modeled the maize metabolism that contains maize-specific GPR (Saha et al., 2011), and Grafahrend-Belau et al. (2013) developed multi-scale metabolic modeling (MMM) for predicting the plant metabolism at the whole plant level; their barley model has provided significant insights into the metabolic capacity for yield stability and crop improvement (Grafahrend-Belau et al., 2013).
Integration of high-throughput experimental data with mathematical modeling
The integration of omics data and mathematical models is a promising approach to gain a better understanding of plant metabolisms (Bordbar et al., 2014; Saha et al., 2014). Integrated concepts involving FBA make it possible to predict genotype-phenotype relationships and to gain important insights into the metabolic network capacity of an organism (Blazier and Papin, 2012). For example, an integrated model in which gene expression was combined with a metabolic network (ME model) in Escherichia coli increased the accuracy for predicting feasible and computable phenotypes that respond to optimal growth conditions (Lerman et al., 2012). Karr et al. (2012) showed that a whole-cell model in Mycoplasma genitalium was useful for describing protein-DNA binding and correlations between DNA replication and its initiation. Their findings indicate that the integrated approach makes it possible to study previously unknown biological processes in a cell. These earlier studies demonstrated that high-throughput omics data are available as a constraint parameter for generating high-quality metabolic models. The model-building algorithm (MBA) developed by Jerby et al. (2010) is used to construct tissue-specific metabolisms from generic models and omics data (Jerby et al., 2010). Gene Inactivity Moderated by Metabolism and Expression (GIMME) (Becker and Palsson, 2008) is based on the premise that gene expression data correlate with metabolic fluxes and the user's pre-defined threshold of expression levels; GIMME removes reactions with expression levels lower than the threshold from the model and evaluates metabolic capacities. iMAT (Folger et al., 2011) is similar to GIMME; it is based on the discretization of input expression data and returns predictive optimal flux with confidence values over all network reactions. Metabolic Adjustment by Differential Expression (MADE) (Jensen and Papin, 2011) uses significant changes in transcript levels between two or more conditions classified into so-called “switch” approaches. This is then used to identify on/off reaction fluxes based on threshold expression levels in the constraint-based models (Hyduke et al., 2013; Saha et al., 2014).
Two other approaches exist, they are known as “valve” approaches and they allow the use of gene expression data to limit the maximum activity of an enzyme. The first, E-FLUX (Colijn et al., 2009), uses maximum flux constraints as a function of measured transcript levels without binalization of the expression data. The other approach is GIM3E, it does not apply arbitrary cutoffs for expression levels (Schmidt et al., 2013). Protein data can also be included. PROM (Chandrasekaran and Price, 2010) invokes a threshold to determine whether an enzyme is in its active or inactive state and uses information about regulatory interactions including transcription factor-target gene interactions. Integrative Omics-Metabolic Analysis (IOMA) (Yizhak et al., 2010) integrates proteomic and metabolomic data into a genome-scale metabolic model by evaluating kinetic rate equations subject to quantitative omics measurements. Machado and Herrgard (2014) who systematically evaluated different methods for the integration of transcriptome data into constraint-based models reported that no robust approaches worked well under all examinations (Machado and Herrgard, 2014). In plant science, Topfer et al. (2013) performed E-FLUX on the Arabidopsis genome-scale models created by Mintz-Oron et al. (2012) and used high-resolution time-series transcriptome data (Caldana et al., 2011) to investigate metabolic capacities in response to different environmental changes (Topfer et al., 2013). Their optimization-based approach was able to characterize many aspects of the metabolic behaviors and functions in response to a changing environment. In an attempt to integrate metabolome data with constraint-based mathematical models, Nagele and Weckwerth (2013) developed a complementary approach to obtain a comprehensive view of metabolic capacities in Arabidopsis leaves (Nagele and Weckwerth, 2013). Using experimentally accessible metabolites and the Mintz-Oron model (Mintz-Oron et al., 2012) they derived a metabolic model that yielded an overview of metabolic phenotypes perturbed by genetic and environmental differences.
Future perspectives
Metabolic network models have contributed to the study of metabolic capacity in response to environmental and genetic perturbations and to the identification of feasible metabolic networks in an organism. They provided important clues about genotype-phenotype relationships. Reconstruction of the metabolism from genome sequences is a non-trivial task that requires not only effective computational tools but also integrated knowledge-based systems. For comprehensive reconstructions, improved technologies, including more sophisticated algorithms and tools, better software frameworks for multiple omics data analyses, improved visualization of biological networks, and more effective integration of data with mathematical models are needed. Multi-omics data that include the genome, transcriptome, proteome, and metabolome plus mathematical modeling can be expected to deepen our knowledge of complex plant metabolisms and to illuminate unexplored biological processes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to authors whose work is not cited due to space limitations. The research activity of Atsushi Fukushima is partly supported by a Grant-in-Aid for Young Scientists (B; grant no. 26850024 to Atsushi Fukushima) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We thank Dr. Kansuporn Sriyudthsak for a critical reading of the manuscript and Ms. Ursula Petralia for editorial assistance.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00598/abstract
References
- Alcantara R., Axelsen K. B., Morgat A., Belda E., Coudert E., Bridge A., et al. (2012). Rhea–a manually curated resource of biochemical reactions. Nucleic Acids Res. 40, D754–D760. 10.1093/nar/gkr1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf-Ul-Amin M., Shinbo Y., Mihara K., Kurokawa K., Kanaya S. (2006). Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinformatics 7:207. 10.1186/1471-2105-7-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. 10.1126/science.1203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M. (2009). A pitfall of wiki solution for biological databases. Brief. Bioinform. 10, 295–296. 10.1093/bib/bbn053 [DOI] [PubMed] [Google Scholar]
- Arnold A., Nikoloski Z. (2014). Bottom-up metabolic reconstruction of Arabidopsis and its application to determining the metabolic costs of enzyme production. Plant Physiol. 165, 1380–1391. 10.1104/pp.114.235358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S. A., Palsson B. O. (2008). Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4:e1000082. 10.1371/journal.pcbi.1000082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P. N., Mitchell-Olds T. (2008). From genotype to phenotype: systems biology meets natural variation. Science 320, 495–497. 10.1126/science.1153716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier A. S., Papin J. A. (2012). Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 3:299. 10.3389/fphys.2012.00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A., Monk J. M., King Z. A., Palsson B. O. (2014). Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120. 10.1038/nrg3643 [DOI] [PubMed] [Google Scholar]
- Brandao M. M., Dantas L. L., Silva-Filho M. C. (2009). AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics 10:454. 10.1186/1471-2105-10-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel F., Rodriguez N., Swainston N., Wrzodek C., Czauderna T., Keller R., et al. (2013). Path2Models: large-scale generation of computational models from biochemical pathway maps. BMC Syst. Biol. 7:116. 10.1186/1752-0509-7-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., et al. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67, 869–884. 10.1111/j.1365-313X.2011.04640.x [DOI] [PubMed] [Google Scholar]
- Carrera J., Rodrigo G., Jaramillo A., Elena S. F. (2009). Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 10:R96. 10.1186/gb-2009-10-9-r96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H., Hofree M., Ideker T. (2013). Genotype to phenotype via network analysis. Curr. Opin. Genet. Dev. 23, 611–621. 10.1016/j.gde.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R., Altman T., Dreher K., Fulcher C. A., Subhraveti P., Keseler I. M., et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40, D742–D753. 10.1093/nar/gkr1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagoyen M., Pazos F. (2013). Tools for the functional interpretation of metabolomic experiments. Brief. Bioinform. 14, 737–744. 10.1093/bib/bbs055 [DOI] [PubMed] [Google Scholar]
- Chandrasekaran S., Price N. D. (2010). Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 107, 17845–17850. 10.1073/pnas.1005139107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Del Val I. J., Kyriakopoulos S., Polizzi K. M., Kontoravdi C. (2012). Metabolic network reconstruction: advances in in silico interpretation of analytical information. Curr. Opin. Biotechnol. 23, 77–82. 10.1016/j.copbio.2011.10.015 [DOI] [PubMed] [Google Scholar]
- Cheung C. Y., Poolman M. G., Fell D. A., Ratcliffe R. G., Sweetlove L. J. (2014). A diel flux balance model captures interactions between light and dark metabolism during Day-Night cycles in C3 and crassulacean acid metabolism leaves. Plant Physiol. 165, 917–929. 10.1104/pp.113.234468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C. Y., Williams T. C., Poolman M. G., Fell D. A., Ratcliffe R. G., Sweetlove L. J. (2013). A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J. 75, 1050–1061. 10.1111/tpj.12252 [DOI] [PubMed] [Google Scholar]
- Colijn C., Brandes A., Zucker J., Lun D. S., Weiner B., Farhat M. R., et al. (2009). Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput. Biol. 5:e1000489. 10.1371/journal.pcbi.1000489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E., Yen J. Y., Senger R. S. (2012). Are we ready for genome-scale modeling in plants? Plant Sci. 191–192, 53–70. 10.1016/j.plantsci.2012.04.010 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., et al. (2010). The genetic landscape of a cell. Science 327, 425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal'molin C. G., Quek L. E., Palfreyman R. W., Brumbley S. M., Nielsen L. K. (2010). C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 154, 1871–1885. 10.1104/pp.110.166488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E., Cary M. P., Paley S., Fukuda K., Lemer C., Vastrik I., et al. (2010). The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 28, 935–942. 10.1038/nbt.1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira Dal'molin C. G., Nielsen L. K. (2013). Plant genome-scale metabolic reconstruction and modelling. Curr. Opin. Biotechnol. 24, 271–277. 10.1016/j.copbio.2012.08.007 [DOI] [PubMed] [Google Scholar]
- De Oliveira Dal'molin C. G., Quek L. E., Palfreyman R. W., Brumbley S. M., Nielsen L. K. (2010). AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 152, 579–589. 10.1104/pp.109.148817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt S., Wuyts N., Inze D. (2013). Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 18, 428–439. 10.1016/j.tplants.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Espinoza C., Degenkolbe T., Caldana C., Zuther E., Leisse A., Willmitzer L., et al. (2010). Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 5:e14101. 10.1371/journal.pone.0014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A. M., Palsson B. O. (2010). The biomass objective function. Curr. Opin. Microbiol. 13, 344–349. 10.1016/j.mib.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell D. A., Poolman M. G., Gevorgyan A. (2010). Building and analysing genome-scale metabolic models. Biochem. Soc. Trans. 38, 1197–1201. 10.1042/BST0381197 [DOI] [PubMed] [Google Scholar]
- Folger O., Jerby L., Frezza C., Gottlieb E., Ruppin E., Shlomi T. (2011). Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7:501. 10.1038/msb.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Redestig H., Arita M., Saito K. (2009). Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13, 532–538. 10.1016/j.cbpa.2009.09.022 [DOI] [PubMed] [Google Scholar]
- Funahashi A., Matsuoka Y., Jouraku A., Morohashi M., Kikuchi N., Kitano H. (2008). CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE 96, 1254–1265. 10.1109/JPROC.2008.925458 [DOI] [Google Scholar]
- Gehlenborg N., O'donoghue S. I., Baliga N. S., Goesmann A., Hibbs M. A., Kitano H., et al. (2010). Visualization of omics data for systems biology. Nat. Methods 7, S56–S68. 10.1038/nmeth.1436 [DOI] [PubMed] [Google Scholar]
- Grafahrend-Belau E., Junker A., Eschenroder A., Muller J., Schreiber F., Junker B. H. (2013). Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiol. 163, 637–647. 10.1104/pp.113.224006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafahrend-Belau E., Schreiber F., Koschutzki D., Junker B. H. (2009). Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol. 149, 585–598. 10.1104/pp.108.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräßler J., Koschützki D., Schreiber F. (2012). CentiLib: comprehensive analysis and exploration of network centralities. Bioinformatics 28, 1178–1179. 10.1093/bioinformatics/bts106 [DOI] [PubMed] [Google Scholar]
- Hanumappa M., Preece J., Elser J., Nemeth D., Bono G., Wu K., et al. (2013). WikiPathways for plants: a community pathway curation portal and a case study in rice and arabidopsis seed development networks. Rice (N. Y). 6:14. 10.1186/1939-8433-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. S., Dejongh M., Best A. A., Frybarger P. M., Linsay B., Stevens R. L. (2010). High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982. 10.1038/nbt.1672 [DOI] [PubMed] [Google Scholar]
- Hucka M., Finney A., Sauro H. M., Bolouri H., Doyle J. C., Kitano H., et al. (2003). The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531. 10.1093/bioinformatics/btg015 [DOI] [PubMed] [Google Scholar]
- Hung J. H., Yang T. H., Hu Z., Weng Z., Delisi C. (2012). Gene set enrichment analysis: performance evaluation and usage guidelines. Brief. Bioinform. 13, 281–291. 10.1093/bib/bbr049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduke D. R., Lewis N. E., Palsson B. O. (2013). Analysis of omics data with genome-scale models of metabolism. Mol. Biosyst. 9, 167–174. 10.1039/c2mb25453k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. A., Papin J. A. (2011). Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics 27, 541–547. 10.1093/bioinformatics/btq702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby L., Shlomi T., Ruppin E. (2010). Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6:401. 10.1038/msb.2010.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker B. H. (2014). Flux analysis in plant metabolic networks: increasing throughput and coverage. Curr. Opin. Biotechnol. 26, 183–188. 10.1016/j.copbio.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Jupp S., Malone J., Bolleman J., Brandizi M., Davies M., Garcia L., et al. (2014). The EBI RDF platform: linked open data for the life sciences. Bioinformatics 30, 1338–1339. 10.1093/bioinformatics/btt765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr J. R., Sanghvi J. C., Macklin D. N., Gutschow M. V., Jacobs J. M., Bolival B. Jr., et al. (2012). A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401. 10.1016/j.cell.2012.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T., Wilkinson M. D., Aoki-Kinoshita K. F., Kawashima S., Yamamoto Y., Yamaguchi A., et al. (2014). BioHackathon series in 2011 and 2012: penetration of ontology and linked data in life science domains. J. Biomed. Semantics 5:5. 10.1186/2041-1480-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B. (2002). Genotype-phenotype mapping: genes as computer programs. Trends Genet. 18, 555–559. 10.1016/S0168-9525(02)02765-8 [DOI] [PubMed] [Google Scholar]
- Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., et al. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40, D841–D846. 10.1093/nar/gkr1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P., Sirota M., Butte A. J. (2012). Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8:e1002375. 10.1371/journal.pcbi.1002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. Y., Sohn S. B., Kim Y. B., Kim W. J., Lee S. Y. (2012). Recent advances in reconstruction and applications of genome-scale metabolic models. Curr. Opin. Biotechnol. 23, 617–623. 10.1016/j.copbio.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Krouk G., Crawford N. M., Coruzzi G. M., Tsay Y. F. (2010). Nitrate signaling: adaptation to fluctuating environments. Curr. Opin. Plant Biol. 13, 266–273. 10.1016/j.pbi.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Kruger N. J., Ratcliffe R. G. (2012). Pathways and fluxes: exploring the plant metabolic network. J. Exp. Bot. 63, 2243–2246. 10.1093/jxb/ers073 [DOI] [PubMed] [Google Scholar]
- Kusano M., Fukushima A. (2013). Current challenges and future potential of tomato breeding using omics approaches. Breed. Sci. 63, 31–41. 10.1270/jsbbs.63.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Huhman D. V., Sumner L. W. (2011). Mass spectrometry strategies in metabolomics. J. Biol. Chem. 286, 25435–25442. 10.1074/jbc.R111.238691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman J. A., Hyduke D. R., Latif H., Portnoy V. A., Lewis N. E., Orth J. D., et al. (2012). In silico method for modelling metabolism and gene product expression at genome scale. Nat. Commun. 3:929. 10.1038/ncomms1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N. E., Nagarajan H., Palsson B. O. (2012). Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 10, 291–305. 10.1038/nrmicro2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libourel I. G., Shachar-Hill Y. (2008). Metabolic flux analysis in plants: from intelligent design to rational engineering. Annu. Rev. Plant Biol. 59, 625–650. 10.1146/annurev.arplant.58.032806.103822 [DOI] [PubMed] [Google Scholar]
- Lucas M., Laplaze L., Bennett M. J. (2011). Plant systems biology: network matters. Plant Cell Environ. 34, 535–553. 10.1111/j.1365-3040.2010.02273.x [DOI] [PubMed] [Google Scholar]
- Luo W., Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Herrgard M. (2014). Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput. Biol. 10:e1003580. 10.1371/journal.pcbi.1003580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G. D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5:e13984. 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S., Meir S., Malitsky S., Ruppin E., Aharoni A., Shlomi T. (2012). Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 339–344. 10.1073/pnas.1100358109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., Kazi F., Wright G., Donaldson S. L., et al. (2010). GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928. 10.1093/bioinformatics/btq562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., Bader G. D., Morris Q. (2014). GeneMANIA: fast gene network construction and function prediction for Cytoscape. F1000Res 3:153. 10.12688/f1000research.4572.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele T., Weckwerth W. (2013). A workflow for mathematical modeling of subcellular metabolic pathways in leaf metabolism of Arabidopsis thaliana. Front. Plant Sci. 4:541. 10.3389/fpls.2013.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K., Ono K., Kanaya S., Takahashi K. (2014). KEGGscape: a cytoscape app for pathway data integration [v1; ref status: approved with reservations 2, http://f1000r.es/3qe]. F1000 Research 3:144. 10.12688/f1000research.4524.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J. D., Thiele I., Palsson B. O. (2010). What is flux balance analysis? Nat. Biotechnol. 28, 245–248. 10.1038/nbt.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman M. G., Miguet L., Sweetlove L. J., Fell D. A. (2009). A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 151, 1570–1581. 10.1104/pp.109.141267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N. (2012). Correlation networks visualization. Front. Plant Sci. 3:240. 10.3389/fpls.2012.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radrich K., Tsuruoka Y., Dobson P., Gevorgyan A., Swainston N., Baart G., et al. (2010). Integration of metabolic databases for the reconstruction of genome-scale metabolic networks. BMC Syst. Biol. 4:114. 10.1186/1752-0509-4-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V. K., Shen L., Moore J. H., Saykin A. J. (2012). Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 28, 323–332. 10.1016/j.tig.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redestig H., Kusano M., Fukushima A., Matsuda F., Saito K., Arita M. (2010). Consolidating metabolite identifiers to enable contextual and multi-platform metabolomics data analysis. BMC Bioinformatics 11:214. 10.1186/1471-2105-11-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn H., Hartmann A., Junker A., Junker B. H., Schreiber F. (2012a). FluxMap: a VANTED add-on for the visual exploration of flux distributions in biological networks. BMC Syst. Biol. 6:33. 10.1186/1752-0509-6-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn H., Junker A., Hartmann A., Grafahrend-Belau E., Treutler H., Klapperstuck M., et al. (2012b). VANTED v2: a framework for systems biology applications. BMC Syst. Biol. 6:139. 10.1186/1752-0509-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Chowdhury A., Maranas C. D. (2014). Recent advances in the reconstruction of metabolic models and integration of omics data. Curr. Opin. Biotechnol. 29C, 39–45. 10.1016/j.copbio.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Saha R., Suthers P. F., Maranas C. D. (2011). Zea mays iRS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6:e21784. 10.1371/journal.pone.0021784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Matsuda F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489. 10.1146/annurev.arplant.043008.092035 [DOI] [PubMed] [Google Scholar]
- Sakurai N., Ara T., Ogata Y., Sano R., Ohno T., Sugiyama K., et al. (2011). KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res. 39, D677–D684. 10.1093/nar/gkq989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardoni G., Tosadori G., Faizan M., Spoto F., Fabbri F., Laudanna C. (2014). Biological network analysis with CentiScaPe: centralities and experimental dataset integration [v1; ref status: approved with reservations 2, http://f1000r.es/3p0]. F1000Res 3:139. 10.12688/f1000research.4477.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. J., Ebrahim A., Metz T. O., Adkins J. N., Palsson B. O., Hyduke D. R. (2013). GIM3E: condition-specific models of cellular metabolism developed from metabolomics and expression data. Bioinformatics 29, 2900–2908. 10.1093/bioinformatics/btt493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver S. M., Gerdes S., Frelin O., Lerma-Ortiz C., Bradbury L. M., Zallot R., et al. (2014). High-throughput comparison, functional annotation, and metabolic modeling of plant genomes using the PlantSEED resource. Proc. Natl. Acad. Sci. U.S.A. 111, 9645–9650. 10.1073/pnas.1401329111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver S. M., Henry C. S., Hanson A. D. (2012). Frontiers in metabolic reconstruction and modeling of plant genomes. J. Exp. Bot. 63, 2247–2258. 10.1093/jxb/err371 [DOI] [PubMed] [Google Scholar]
- Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J. (2004). Mathematical models in microbial systems biology. Curr. Opin. Microbiol. 7, 513–518. 10.1016/j.mib.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Stitt M. (2013). Systems-integration of plant metabolism: means, motive and opportunity. Curr. Opin. Plant Biol. 16, 381–388. 10.1016/j.pbi.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Strömbäck L., Lambrix P. (2005). Representations of molecular pathways: an evaluation of SBML, PSI MI and BioPAX. Bioinformatics 21, 4401–4407. 10.1093/bioinformatics/bti718 [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Obata T., Fernie A. R. (2014). Systems analysis of metabolic phenotypes: what have we learnt? Trends Plant Sci. 19, 222–230. 10.1016/j.tplants.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Ratcliffe R. G. (2011). Flux-balance modeling of plant metabolism. Front. Plant Sci. 2:38. 10.3389/fpls.2011.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I., Palsson B. O. (2010). A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121. 10.1038/nprot.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topfer N., Caldana C., Grimbs S., Willmitzer L., Fernie A. R., Nikoloski Z. (2013). Integration of genome-scale modeling and transcript profiling reveals metabolic pathways underlying light and temperature acclimation in Arabidopsis. Plant Cell 25, 1197–1211. 10.1105/tpc.112.108852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D., Fernie A. R., Nikoloski Z., Fait A. (2013). Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol. 31, 29–36. 10.1016/j.tibtech.2012.10.011 [DOI] [PubMed] [Google Scholar]
- Tsesmetzis N., Couchman M., Higgins J., Smith A., Doonan J. H., Seifert G. J., et al. (2008). Arabidopsis reactome: a foundation knowledgebase for plant systems biology. Plant Cell 20, 1426–1436. 10.1105/tpc.108.057976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138, 1195–1204. 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Iersel M. P., Kelder T., Pico A. R., Hanspers K., Coort S., Conklin B. R., et al. (2008). Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics 9:399. 10.1186/1471-2105-9-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roey K., Orchard S., Kerrien S., Dumousseau M., Ricard-Blum S., Hermjakob H., et al. (2013). Capturing cooperative interactions with the PSI-MI format. Database (Oxford) 2013:bat066. 10.1093/database/bat066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W. (2011). Green systems biology - From single genomes, proteomes and metabolomes to ecosystems research and biotechnology. J. Proteomics 75, 284–305. 10.1016/j.jprot.2011.07.010 [DOI] [PubMed] [Google Scholar]
- Xia J., Wishart D. S. (2010). MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 38, W71–W77. 10.1093/nar/gkq329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M. A., Goh K. I., Cusick M. E., Barabasi A. L., Vidal M. (2007). Drug-target network. Nat. Biotechnol. 25, 1119–1126. 10.1038/nbt1338 [DOI] [PubMed] [Google Scholar]
- Yizhak K., Benyamini T., Liebermeister W., Ruppin E., Shlomi T. (2010). Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics 26, i255–i260. 10.1093/bioinformatics/btq183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zheng G., Shan L., Meng G., Vingron M., Liu Q., et al. (2014). Reconstruction of gene regulatory network related to photosynthesis in Arabidopsis thaliana. Front. Plant Sci. 5:273. 10.3389/fpls.2014.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Foerster H., Tissier C. P., Mueller L., Paley S., Karp P. D., et al. (2005). MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 138, 27–37. 10.1104/pp.105.060376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi K., Franz M., Rodriguez H., Montojo J., Lopes C. T., Bader G. D., et al. (2013). GeneMANIA prediction server 2013 update. Nucleic Acids Res. 41, W115–W122. 10.1093/nar/gkt533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.