Abstract
Rationale
Transverse tubules (TTs) couple electric surface signals to remote intracellular Ca2+ release units (CRUs). Diffraction-limited imaging studies have proposed loss of TT components as disease mechanism in heart failure (HF).
Objectives
Objectives were to develop quantitative super-resolution strategies for live-cell imaging of TT membranes in intact cardiomyocytes and to show that TT structures are progressively remodeled during HF development, causing early CRU dysfunction.
Methods and Results
Using stimulated emission depletion (STED) microscopy, we characterized individual TTs with nanometric resolution as direct readout of local membrane morphology 4 and 8 weeks after myocardial infarction (4pMI and 8pMI). Both individual and network TT properties were investigated by quantitative image analysis. The mean area of TT cross sections increased progressively from 4pMI to 8pMI. Unexpectedly, intact TT networks showed differential changes. Longitudinal and oblique TTs were significantly increased at 4pMI, whereas transversal components appeared decreased. Expression of TT-associated proteins junctophilin-2 and caveolin-3 was significantly changed, correlating with network component remodeling. Computational modeling of spatial changes in HF through heterogeneous TT reorganization and RyR2 orphaning (5000 of 20 000 CRUs) uncovered a local mechanism of delayed subcellular Ca2+ release and action potential prolongation.
Conclusions
This study introduces STED nanoscopy for live mapping of TT membrane structures. During early HF development, the local TT morphology and associated proteins were significantly altered, leading to differential network remodeling and Ca2+ release dyssynchrony. Our data suggest that TT remodeling during HF development involves proliferative membrane changes, early excitation-contraction uncoupling, and network fracturing.
Keywords: Ca2+ sparks, excitation-contraction coupling, heart failure, T-tubule, super-resolution imaging, calcium signaling
Due to large volumes of muscle cells, excitation-contraction (E-C) coupling depends critically on fast signaling mechanisms to overcome spatial diffusion barriers. Electron microscopy studies have suggested that transverse tubules (TTs) at sarcomeric Z-line striations enable action potential (AP) propagation.1 Intracellular voltage imaging provided evidence that TTs propagate APs inside cardiomyocytes.2 AP propagation by TTs is further supported by synchronization of local Ca2+ release signals (sparks) during E-C coupling in adult ventricular cardiomyocytes.3 As an E-C coupling mechanism, electron microscopy (EM) studies characterized ≈12-nm-wide nanodomains containing junctional TT segments and terminal sarcoplasmic reticulum (SR) Ca2+ release sites.4,5 Ca2+ release unit (CRU) nanodomains were defined as couplons,4,6 protein-membrane complexes that locally control the Ca2+ release (spark) function of CRU compartments.7 Recent models assume that TTs propagate APs from the surface to an estimated 20 000 CRUs throughout an adult ventricular cardiomyocyte during E-C coupling.8
In contrast, embryonic and early postnatal cardiomyocytes are largely devoid of TTs exhibiting slow intracellular Ca2+ transients limited by surface diffusion.9 Furthermore, regional loss of TTs was associated with delayed, dyssynchronous Ca2+ transients in heart failure (HF) in adult ventricular cardiomyocytes from animal models and patient samples (see Online Table IV for overview of reported TT changes).3,10,11 Lederer et al proposed abnormal spatial organization of the TT network as a mechanism of intracellular Ca2+ dyssynchrony through “orphaning” of ryanodine receptor (RyR2) Ca2+ release channels.3 Furthermore, TTs contain L-type Ca2+ channels (LCCs) in immediate proximity to RyR2 channels, and TT remodeling may disrupt functional colocalization.3 Since the morphology of individual TT structures appears typically as blurred fluorescence signal in conventional light microscopy, this may preclude understanding of the local nature of membrane changes in diseased cells, particularly of subtle remodeling processes during HF development.
Two-photon microscopy of healthy rat cardiomyocytes has shown a continuous cell-wide TT network composed mainly of transversal components at striations and longitudinal components.12 Magnified optical sections of individual TT components appear blurred in 2-photon images.12 Furthermore, confocal microscopy is limited by diffraction, which may hinder analysis of small subresolution TT structures or subtle changes in HF.3 Despite nanometric resolution, electron microscopy requires invasive histochemical protocols incompatible with live-cell imaging of intact membrane structures as discussed earlier.12 Therefore, live-cell imaging at the nanometer scale represents an important yet unattained goal to improve understanding of individual TT structures in E-C coupling, TT associated nanodomains such as caveolae, and changes in HF.
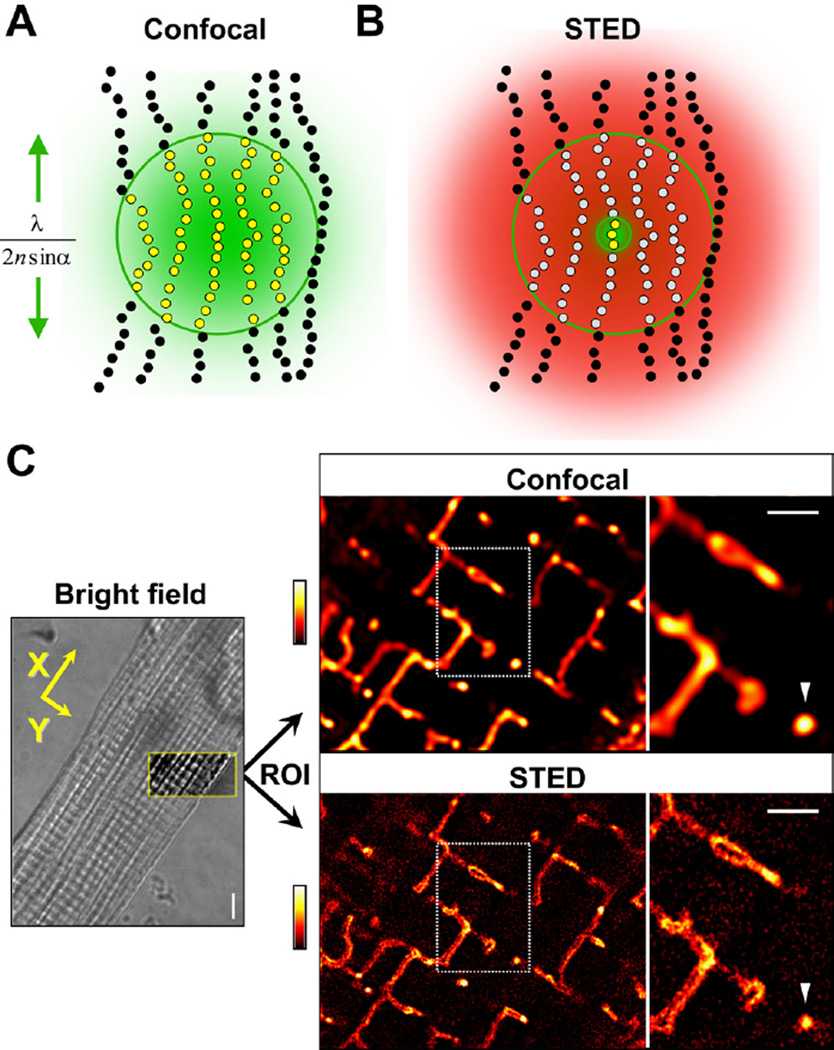
To overcome resolution limitations in imaging of TT structures in intact cells, we used stimulated emission depletion (STED) microscopy.13 A major advantage for live-cell imaging, STED was implemented as laser scanning microscope using a fast beam scanner (Online Figure IA).14 STED overcomes the diffraction barrier by switching (turning OFF) fluorescent molecules through a transient dark state.13 As in conventional confocal microscopy a focused laser excites all fluorescently labeled structures in the focal spot simultaneously therefore subresolution structures are not optically separated (Figure 1A). The diffraction barrier is overcome by switching peripheral fluorescent markers transiently into a dark state, confining the signal only to molecules at the focal center, by a second red-shifted laser beam with a toroidal (doughnut) profile with a central zero intensity (Figure 1B). Increasing the intensity of the STED beam further confines the central signal spot leading to higher, potentially infinite resolution, even resolving molecular scales.14,15 Practical limitations concern properties of the fluorescent STED dye in the sample and diffractive behaviors of large muscle cells.
Figure 1. Imaging of intracellular TT membrane structures by confocal versus STED microscopy in living cardiomyocytes.
A, Confocal microscopy: all fluorescent molecules within the focal spot region are excited and fluoresce (yellow dots); neighboring molecules are indistinguishable and biological structures appear blurred as indicated by the green area. The size (lateral extent) of the focal region indicated by the green line is a function of optical diffraction and determines resolution: λ/(2nsinα). Outside the focal region, molecules are not excited (black dots). B, STED: all peripheral fluorescing molecules are transiently deexcited into a dark state (gray dots) by light of sufficient intensity (red distribution) while exposed to the same focal excitation light. To realize the central region of zero intensity, the red STED laser beam is modified by a phase plate (inner green circle), confining fluorescence signals only to the zero center region (yellow dots) and a significantly smaller detection volume, resulting in higher resolution. Sufficiently high STED laser intensities confine arbitrarily small volumes of the zero center region. C, left, Transmitted light image (bright field) of the central cardiomyocyte section with the region of interest (ROI; framed yellow box) used for fluorescence imaging; scale, 5 µm. Yellow arrows indicate longitudinal (X) and transversal (Y) reference cell orientations for image analysis, corresponding with sarcomere orientations (striations). Right, Confocal and STED images of the same cardiomyocyte ROI showing TT membranes stained with di-8-ANEPPS. Dotted rectangles mark regions further magnified rightward. Arrowhead highlights exemplar “free” transversal TT cross section at a given striation; scale bars, 1 µm; fluorescence intensity coding as shown by look-up table.
Using STED microscopy with customized optics and calibration routines, we investigated TT membrane structures in living cardiomyocytes with nanometric resolution, confirmed by nanobead measurements (Online Figure IB). We show that STED provides a direct quantitative readout of the local TT membrane morphology and its changes during HF development. Progressive TT changes were analyzed side-by-side in samples from sham versus 4-week (4pMI) and 8-week (8pMI) post-MI hearts and through colocalization and expression analysis of TT associated proteins. We unravel a previously not recognized mechanism of proliferative spatial TT remodeling early during HF development resulting in a highly heterogeneous network phenotype. Furthermore, computational modeling of spatial TT and RyR2 cluster reorganization (orphaning) and Ca2+ release dyssynchrony in HF confirmed an aggravating mechanism of delayed subcellular Ca2+ release and AP prolongation, which may contribute to electric and contractile dysfunction.
Methods
For the myocardial infarction model, phenotyping, cardiomyocyte isolation, confocal microscopy protocols, and statistics, an expanded Methods section is available in the Online Data Supplement.
STED Microscopy Image Acquisition
A custom STED setup was modified for intracellular TT membrane imaging in isolated cardiomyocytes based on an inverted confocal microscope (Online Figure IA). For live-cell STED imaging of TT membranes we tested several dyes, of which di-8-ANEPPS provided (1) bright membrane signals and (2) quiescent cardiomyocytes (see Online Methods). The STED microscopy laser configuration is summarized in Online Figure IA. Excitation at 490 nm wavelength by a pulsed diode laser (Pico TA 490, Toptica, Munich, Germany) was focused through a 1.4 NA oil objective (PL APO oil 100×, Leica, Wetzlar, Germany). The collected fluorescence was passed through 2 dichroic mirrors, filtered via a 675/60 band pass filter, and recorded by an avalanche photodiode detector (PerkinElmer, Waltham, MA). STED pulses were delivered by a Ti:Sapphire laser (MaiTai, Spectra-Physics, Darmstadt, Germany) operating at 80 MHz emitting at 750 nm. The STED beam was passed through a Vortex phase plate producing a focal doughnut (RPC Photonics, Rochester, NY). Excitation pulses were temporally overlaid with STED pulses by external triggering. Imaging planes were recorded by resonant mirror scanning along the first lateral axis (15.8 kHz, SC-30, Electro-Optical Products Corp, Glendale, NY) and by piezostage scanner along the second lateral axis (P-733–3DD, Physik Instrumente GmbH, Karlsruhe, Germany). If optical aberrations are negligible, the focal plane resolution is estimated to be ≈60 nm, derived from measurements of 20-nm fluorescent beads (Online Figure IB).
Quantitative STED Image Analysis of TT Cross-Sections
A 2D Gauss function was fitted to the fluorescence signal distribution of individual transversal TT cross-sections. Full width at half-maximum (FWHM) was determined for longitudinal (X) and transversal (Y) cell directions as indicated in Figure 1B. TT cross-section area was calculated by formula: A=π (0.5·X)·(0.5·Y). (2) For the same TT cross sections, contour lines were computed with a resolution of 120 points using a 50% threshold of the maximal pixel intensity. Using the center of mass, contour lines of a given TT population were superimposed in space (X, Y), presented as 2D probability histogram. Furthermore, the mean circumference of individual TT cross-section contours are reported (Tables). (3) The radius size of individual TT cross sections was determined as the median of 120 contour radii and independent of cell orientation. Radius size distributions were plotted. Using a nonparametric test (Mann-Whitney), significant changes in median distribution are reported, and 2-peak Gaussian fitting was used to document deviations (increased heterogeneity) from symmetrical distributions. (4) A separate group of superenlarged highly irregular TT cross sections (Figure 3C) was characterized by manual contour analysis and not included in the automated contour data as detailed in the corresponding Results section. For further details, please refer to the Online Methods.
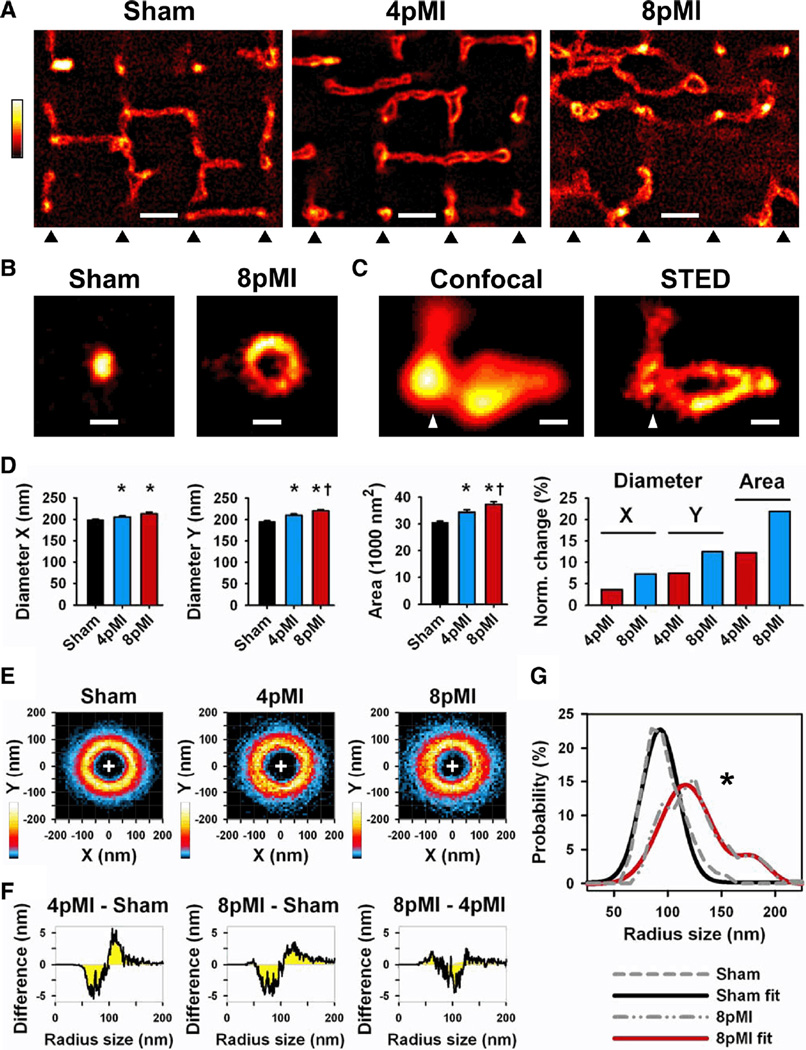
Figure 3. Transversal TT cross sections in intact cardiomyocytes are progressively enlarged through heterogeneous remodeling during HF development.
A, Comparison of striation-aligned STED images showing intracellular TT structures from sham, 4pMI, and 8pMI cells (bottom triangles indicate position of striations). TTs appear enlarged and misaligned in 4pMI and 8pMI cells. Scale, 1 µm. B, STED examples of TT cross sections from sham and 8pMI cells. Scale, 200 nm. C, Confocal versus STED images of a superenlarged TT cross section complex at 4pMI (triangle: position of striation). Scale, 200 nm. D, Longitudinal (X) and transversal (Y) diameters of TT cross sections were determined (see Methods). Bar graphs summarize mean TT diameters X and Y and cross-section area; right, change in TT cross-section dimensions normalized to sham. *P<0.05 versus sham; †P<0.05 versus 4pMI. E, Two-dimensional probability histograms of contours from TT cross-sections of indicated treatment groups (same cross-sections as in D). TT cross-sections were analyzed by automated contour algorithm (see Methods). Colors represent high (white) versus low (black) contour pixel probabilities as indicated. F, Difference integrals were calculated for radius size distributions between the indicated treatment groups. Radius sizes were determined from individual TT contours (see Methods); for example, at 4pMI-sham, a decrease of small versus an increase of large TT radius sizes occurred during HF development. G, Median radius size distributions were calculated from individual TT contours of sham and 8pMI cells (gray dashed lines). The 8pMI distribution is significantly right-shifted (*P<0.05). Furthermore, 2-peak Gaussian fitting of the 8pMI data (red line) confirmed a rightward shift and an additional second peak documenting heterogeneous changes of TT cross-sections. Mean data are summarized in Table 2.
Skeleton and Spatial Orientation Analysis
Two-dimensional skeletons of intact TT networks were extracted from STED images by ImageJ (imagej.nih.gov) and Fiji (pacific.mpi-cbg.de plugins). The following analysis steps were applied: local contrast enhancement (CLAHE), median filtering, automated thresholding, and “skeletonizing” of the binary image. TT skeletons were analyzed by Fiji plug-in algorithm (“directionality”) to quantify spatial orientations of network components. Histogram analysis of orientation probability of TT network components relative to principal cell directions is based on local gradient orientations detected by a 5×5 Sobel filter. Network complexity is analyzed through the amount of triple junctions; STED and confocal imaging data are compared in Online Figure VIII.
Computational Modeling of Local Ca2+ Release Function
Local CRU function was investigated through Ca2+ sparks, Ca2+ transients, [Ca2+]SR, and AP behavior using the fully stochastic mathematical model of Williams et al,8 which includes spatial determinants of individual CRU nanodomain organization and a realistic number of 20 000 independent release sites. The whole-cell model examines changes in local subspace [Ca2+]i signaling in HF by implementing previously established changes in ion transport proteins (eg, NCX and SERCA; for details see Online Methods). Orphaning of RyR2 clusters during HF due to heterogeneous TT and/or RyR2 cluster remodeling was implemented for only 25% of CRUs through a 30-fold increase in subspace volume, whereas 75% of CRUs remained unchanged.
Results
Nanometric Mapping of TT Membranes in Living Cells
After staining with the membrane probe di-8-ANEPPS (for selection of STED dyes see Online Methods), the central cardiomyocyte region of interest was sequentially imaged by confocal and STED mode and sarcomere orientation (striations) were documented by transmitted light (Figure 1C). Compared with confocal imaging (Figure 1C), STED immediately revealed sharper TT structures including membrane structures deep inside living cardiomyocytes and individual cross-sections of transversal TTs relevant during E-C coupling (Figure 1C; arrowhead).
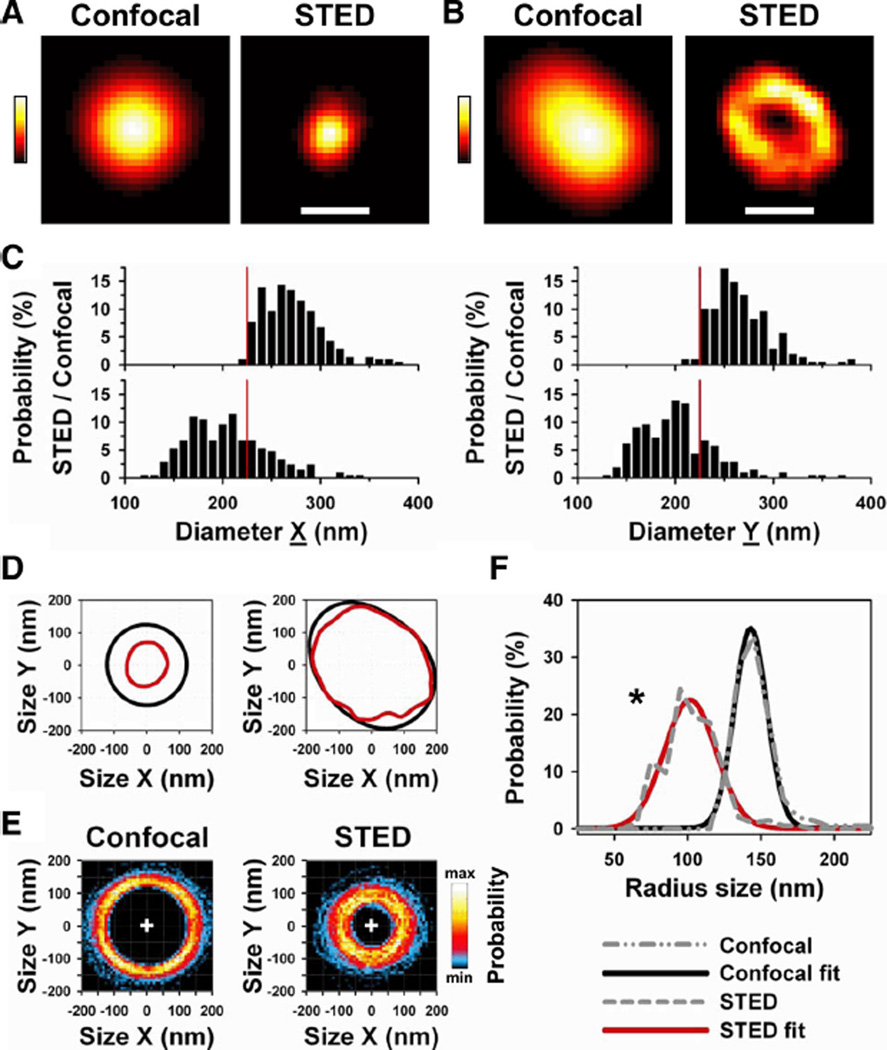
Figure 2A shows a representative TT cross-section where STED produced a sharper image of the membrane morphology where the lumen was not resolved due to object limitations, whereas Figure 2B shows a TT cross-section where the interior was resolved confirming the underlying hollow membrane structure. To assess the TT dimensions in situ, we analyzed perpendicular TT diameters corresponding to the longitudinal (X) and transverse (Y) cell orientations as full width at half maximum (FWHM; Figure 1C defines X and Y orientations applied henceforth; see Methods). On average, longitudinal TT diameters (n=205) from healthy control cells measured 198.7±2.8 nm by STED mode at 3- to 8-µm imaging depth (Table 1). In contrast, the same TTs measured by confocal mode appeared significantly larger (265.9±2.1 nm; P<0.001). Consistent with sharper STED images, the TT cross-section area appeared 44% smaller when imaged by STED as compared with confocal mode (Table 1).
Figure 2. STED nanoscopy shows transversal TT cross sections with subresolution dimensions in living cells.
A, The same TT cross section was imaged by confocal and STED mode. STED shows the morphology of the TT cross section at 3.9 µm imaging z depth. Note: images are rotated according to X (horizontal) and Y (vertical) cell orientations as given by Figure 1B. Scale, 200 nm; colors indicate fluorescence intensity. B, Example of a TT cross section where STED resolved the underlying hollow membrane structure at 5.9 µm z depth. Scale, 200 nm. C, Histograms of diameter dimensions X and Y of the same 205 TT cross sections measured by confocal (upper) versus STED (lower) mode. Diameters represent full-width at half maximum (FWHM) determined by 2D Gauss fitting (see Methods). Vertical red line marks 220 nm, the confocal resolution limit. D, Same TT cross-section pairs as in A and B analyzed by 50% intensity threshold contour algorithm (see Methods). Graphs represent paired contour data each for confocal (black) and STED (red) mode. Note: only STED detected the underlying TT membrane morphology. E, Two-dimensional probability histograms of 205 contours each for confocal and STED mode. Color bar (right) indicates high (max) versus low (min) pixel probability; plus sign (+) is point of origin used for overlay of contour data. F, Median radius distribution from 205 individual TT contours (see Methods) each for confocal and STED mode (gray dashed lines; see legend in figure). STED imaging resulted in a significant leftward shift toward smaller radius sizes confirming faithful detection of subresolution structures (*P<0.05). Furthermore, Gaussian fitting confirmed that STED (red line) detected a wider distribution of smaller TT radius sizes as compared with confocal imaging (black line). Mean data are presented in Table 1.
Table 1.
Intracellular TT Dimensions of Untreated Control Cardiomyocytes
| Imaging Mode |
Diameter X, nm |
Diameter Y, nm |
Area, 103 nm2 |
Circumference, nm |
|---|---|---|---|---|
| Confocal | 265.9±2.1 | 261.1±2.0 | 54.6±0.6 | 918.9±5.9 |
| STED | 198.7±2.8* | 195.2±2.6* | 30.8±0.7* | 672.9±8.7* |
TT cross sections were imaged by confocal and STED modes (at 3 to 8 µm depth). TT diameters represent FWHM measured by automated 2D Gauss fitting (see Methods) for X (longitudinal) and Y (transverse) directions as indicated in Figure 1A. Circumference was determined by automated contour analysis (see Methods). Data represent paired measurements.
P<0.001 versus confocal imaging; mean±SEM from 205 randomly selected TT cross sections of 43 cells.
We wondered if the dimensions of TT cross sections are differently distributed between confocal and STED measurements. Figure 2C shows that the majority of longitudinal (X) and transversal (Y) TT diameters appeared smaller by STED (lower histograms) as compared with confocal imaging (upper histograms), consistent with smaller mean cross-section dimensions (Table 1). Furthermore, similar distributions of TT dimensions were confirmed for STED less than 3 µm near the surface membrane (Online Figure IIA). Notably, for confocal measurements a brisk cutoff in TT diameter sizes occurred at 220 nm (Figure 2C), the confocal resolution limit.
To characterize the local morphology of TT cross sections, we developed an automated 2D contour analysis (see Methods). Figure 2D shows paired contour examples from the same TT cross-sections each for Figure 2A and 2B. Apparently, STED imaging resulted in realistic, detailed contour shapes (red) as compared with confocal mode (black). Figure 2E shows contour overlay plots of the same 205 cross sections (analyzed in Figure 2C) as 2D probability histogram each for confocal and STED imaging. Compared with confocal imaging, STED resulted in a dramatically different probability distribution of TT contours enclosing a smaller area, consistent with a significantly decreased mean circumference (Table 1). Next, we calculated the radius size from individual contours (see Methods) and analyzed the distribution of all median radius sizes for confocal and STED imaging (Figure 2F, gray dashed lines): the radius size distribution shows a significant leftward shift for STED imaging (P<0.05). In addition, Gaussian fitting confirmed a significantly left-shifted peak distribution (Figure 2F, red line). Furthermore, we found similar contour and radius distributions for subsurface TT cross sections (Online Figure IIB and C, and Online Table I). In summary, STED captures TT cross-sections with greater accuracy providing sharper images of individual TT cross sections, which can be directly quantified and characterized for representative cell populations.
Myocardial Infarction Leads to Enlarged TT Cross-Sections
In chronic HF, global changes of the TT network have been documented in living cells by confocal microscopy showing a loss of the transverse component at Z-line striations.3 So far, neither individual TT structures in living cells, nor quantitative side-by-side comparison of TT changes during different time points of HF development have been quantitatively characterized. Therefore, we investigated transversal TTs each 4 weeks (4pMI) and 8 weeks (8pMI) after myocardial infarction, using a previously established model of progressive HF in mice.16 To induce HF, the left anterior descending (LAD) coronary artery was proximally ligated; hearts were analyzed at 4pMI and 8pMI and compared with sham-treated control hearts. Normalized heart weight (HW/BW) and left ventricular cross-section area in diastole (Area,d) were progressively increased, whereas ejection fraction was significantly decreased (Online Figure III), confirming the expected phenotype of progressive cardiac dilation, hypertrophy, and pump failure. Furthermore, cardiomyocytes from 4pMI and 8pMI hearts showed significantly increased cell dimensions consistent with progressive hypertrophic cardiomyocyte remodeling (Online Figure IV).
Whereas regular sarcomere striations were documented by transmitted light in living cardiomyocytes (data not shown), STED nanoscopy revealed profound changes of TT cross-sections and networks after MI. Cardiomyocytes from sham hearts showed regularly aligned TT components with transverse and longitudinal orientations consistent with a rectilinear network (Figure 3A, sham). In contrast, cardiomyocytes at 4pMI showed enlarged TT cross-sections and minor changes in sarcomere alignment (Figure 3A, 4pMI). Furthermore, 8pMI cardiomyocytes showed severe, heterogeneous TT network misalignment and enlarged TT cross-sections (Figure 3A, 8pMI). Enlarged TT cross-sections showed circular morphologies (Figure 3B and Online Figure VA) or complex morphologies connected with other network components both at intracellular (Figure 3C) and subsurface locations (Online Figure VB). Two-dimensional Gauss analysis confirmed progressively increased dimensions of TT cross-sections as evidenced by significant changes of longitudinal (X) and transversal (Y) diameters and cross-section area (Figure 3D and Table 2). Notably, the average TT cross-section area was enlarged by 12% at 4pMI and by 22% at 8pMI, consistent with progressive remodeling (Figure 3D, right). Subsurface TT cross-sections showed significant increases in TT dimensions only at 8pMI (Online Figure VC; and Online Table II).
Table 2.
Intracellular TT Cross-Section Dimensions 4 Weeks or 8 Weeks After Myocardial Infarction Compared With Sham Control Treatment
| Treatment Group |
Diameter X, nm |
Diameter Y, nm |
Area, 103 nm2 |
Circumference, nm |
|---|---|---|---|---|
| Sham | 198.4±1.9 | 195.5±1.9 | 30.6±0.4 | 656.5±5.6 |
| 4pMI | 205.7±2.9* | 210.1±3.2* | 34.3±0.8* | 692.6±9.5* |
| 8pMI | 213.0±3.4* | 219.9±3.3*† | 37.2±0.9*† | 748.9±11.2*† |
TT cross sections were imaged by STED deep inside cardiomyocytes each for sham, 4pMI, and 8pMI treatment groups. TT diameters represent FWHM measurements by automated 2D Gauss fitting corresponding to X (longitudinal) and Y (transversal). Circumference was determined by automated contour analysis. Data represent unpaired measurements.
P<0.05 versus sham;
P<0.05 versus 4pMI; mean±SEM from the following numbers of TT cross sections (cells) for each treatment group: sham, 627 (90); 4pMI, 237 (44); and 8pMI, 290 (46).
We analyzed TT cross-sections by contour method and summarized the data for each treatment group by 2D histogram. TT cross-sections from sham hearts showed a symmetrical, concentric probability distribution characterized by a circular, uniform crest (Figure 3E, sham). However, at 4pMI and 8pMI, both the high probability crest distribution and the periphery of the 2D probability histograms were increasingly altered as reflected by a less uniform distribution and increasing heterogeneity (Figure 3E). We further analyzed contour changes through the difference in radius size distribution for the different treatment groups. Difference integrals revealed a prominent loss of smaller and an increase of larger cross-section radius sizes each at 4pMI and 8pMI versus sham (Figure 3F). Furthermore, a loss of intermediate radius sizes occurred at 8pMI versus 4pMI (Figure 3F). In addition, we determined the median radius size distribution of TT cross-sections for sham and 8pMI. As compared with sham, at 8pMI the median distribution was blunted and significantly right-shifted toward larger radius sizes (Figure 3G, gray dashed lines; P<0.05, Mann-Whitney test). Furthermore, 2-peak Gaussian fitting confirmed heterogeneous changes through a second peak at ≈180 nm, indicating a population of TT cross-sections with dramatically increased radius sizes at 8pMI (Figure 3G).
We summarize that the physiological in situ morphology of TT cross-sections is progressively altered during HF development and replaced by an increasingly heterogeneous population of TT components with increased sizes. Furthermore, due to limitations with the automated analysis of complex superenlarged TT morphologies (eg, Figure 3C), we performed a manual contour analysis to estimate cross-section dimensions. At 8pMI, the mean circumference of superenlarged TT complexes was 1458±51 nm (n=40 from 24 cells), a 2-fold increase compared with sham (Table 2; P<10−10). In addition, we observed superenlarged TT cross sections at subsurface locations at 8pMI, confirming a cell-wide phenotype of heterogeneous proliferative remodeling (Online Figure VA and B). Although many superenlarged TTs approached micrometer dimensions, only STED imaging was able to resolve the underlying membrane morphology of complex luminal configurations (Figure 3C) and supported further quantitative analysis of superenlarged TT cross-sections within intact networks.
STED Reveals Proliferative Network Remodeling After Myocardial Infarction
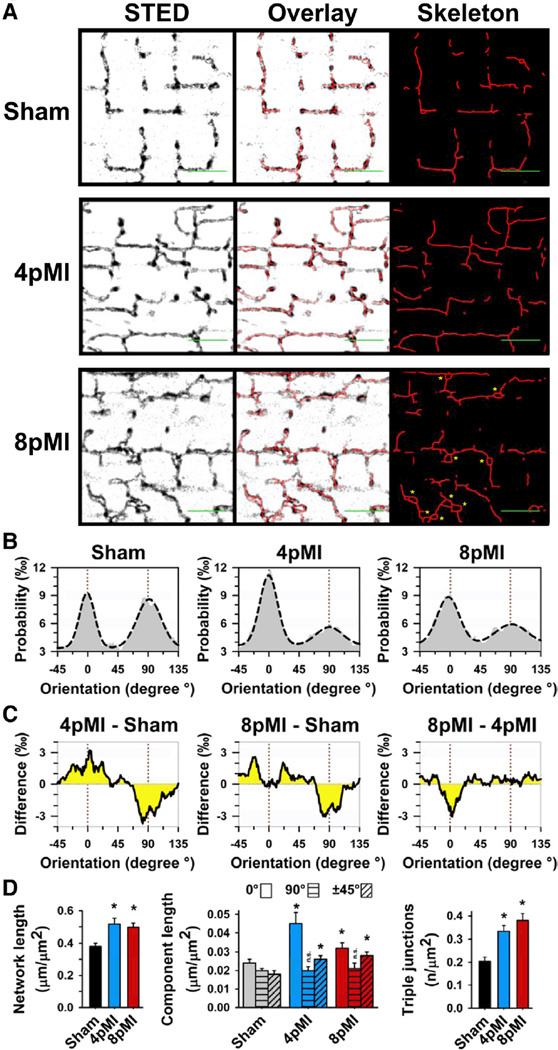
Originally, a regional loss of TTs has been described in different HF models (see Online Table IV for overview). Confocal imaging of cardiomyocytes from hypertensive rats with congestive HF showed a decrease in transversal TT network components at ≈19 months of age.3 However, TT network properties were evaluated through the striation-associated signal periodicity,3 an indirect strategy not addressing individual or irregular structures. Using STED imaging, we directly characterized intact TT networks comprehensively through all local component orientations and segment lengths (see Methods). Cardiomyocytes from sham control hearts showed rectilinear network architectures composed of transversal and longitudinal components (Figure 4A, sham). Accordingly, all-component orientation analysis from sham cells resulted in a probability distribution with 2 peaks corresponding to longitudinal (0°) and transversal (90°) network orientations (Figure 4B, left). In contrast, at 4pMI the TT network showed differential changes of the major network components (Figure 4B), a relative increase in longitudinal (0°) versus a decrease in transversal (90°) components (Figure 4C, 4pMI-sham). In addition, at 8pMI a relative increase in oblique and a reduction of longitudinal components occurred (Figure 4C, 8pMI-4pMI). Gauss fitting of the longitudinal (0°) and transversal (90°) histogram peaks (Figure 4B, dashed line) confirmed replacement of physiological rectilinear by oblique components at 8pMI, documented by wider 0° and 90° peaks (Figure 4B). Thus, early remodeling of the TT network during HF development at 4pMI is characterized by relative changes of longitudinal versus transversal components, whereas late HF changes at 8pMI are characterized by relative changes in longitudinal versus oblique components. Overall, the TT network appears increasingly fractured and misaligned at striations.
Figure 4. Individual components of the TT network are profoundly remodeled during HF development.
A, Representative TT network grayscale images from sham, 4pMI, and 8pMI cardiomyocytes show STED raw data, overlay of skeleton data, and extracted intact skeleton network data after automated thresholding. In addition, superenlarged TT cross sections were manually evaluated (8pMI: yellow asterisks; see also Results section). Scale bars (green) indicate 2 µm and longitudinal (0°) orientation. B, Probability histograms of individual (relative) orientations of network components for sham, 4pMI, and 8pMI groups. Dashed lines represent Gauss fits. The majority of network components correspond either to longitudinal (0°) or transversal (90°) orientations (marked by red dotted lines). C, Difference integrals showing changes between indicated treatment groups. For instance, the difference integral between 4pMI and sham (left, 4pMI-sham) shows a relative increase in longitudinal (0°) and a decrease in transversal (90°) component orientations during early HF development. D, Bar graphs summarizing average amount of TT network components for sham, 4pMI, and 8pMI data. Left, Total network length normalized to cell area. Center, Total component lengths normalized to cell area, each for longitudinal (0°), transversal (90°), and oblique (±45°) orientations. Right, Number of triple junctions composed of 3 individual components normalized to cell area. *P<0.05 versus sham; n.s. indicates not significant.
To further explore the prevailing hypothesis of a regional loss of network components (Online Table IV), we analyzed the total length and complexity of intact network skeletons. Surprisingly, we found a significantly increased total network length both at 4pMI and 8pMI (Figure 4D, left), seemingly contrasting with earlier reports and with our own STED observation of sarcomere regions devoid of TT components (Figure 4A, 8pMI). Notably, the total amount of longitudinal and oblique components if directly measured through individual segment lengths was significantly increased, but the total amount of transverse components was unchanged (Figure 4D, center). Furthermore, we found a nearly 2-fold increase in triple junctions consistent with increased post-MI network branching and complexity (Figure 4D, right). Manual contour analysis confirmed an increase in superenlarged TT cross sections at 8pMI (Figure 4A, 8pMI; asterisks). Consistent with Figure 3C, the underlying membrane morphology of superenlarged TT structures was only resolved by STED (Figure 4A and 8pMI), including a significant increase of triple junctions only detected by STED (Online Figure VIII). Therefore, our data suggest increases in TT network length, complexity, and TT dimensions through additive, proliferative membrane remodeling processes early during HF development. Furthermore, network fracturing and sarcomere misalignment at 8pMI indicate severe disorganization with loss of local TT associations with CRUs at striations, consistent with earlier studies showing a loss of striation associated TTs at relatively late stages of HF (Online Table IV).
Remodeling of TT-Associated Nanodomains
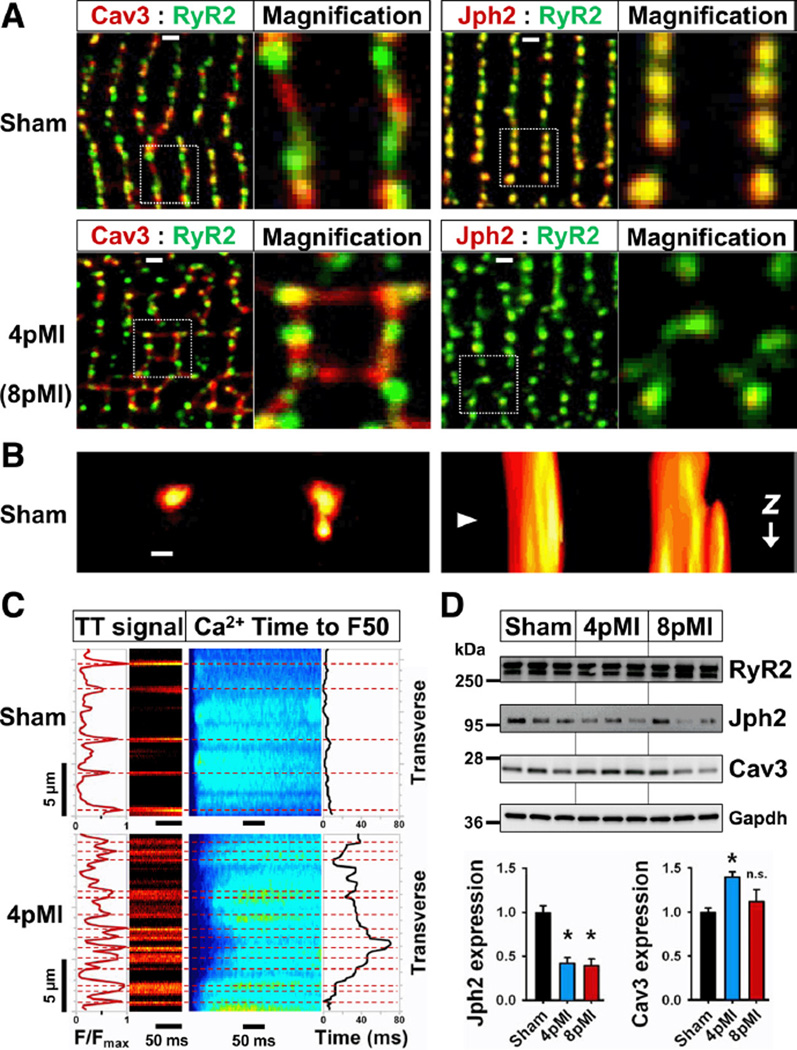
During muscle development, TTs and caveolae exhibit membrane interactions, depending on the muscle-specific caveolin-3 (Cav3) isoform.17 Both Cav3 and junctophilin-2 (Jph2) proteins showed punctate, yet different colocalization behaviors with RyR2 clusters at striations (Figure 5A, sham), consistent with reported nanodomain structures.18 To directly confirm caveolae like TT nanodomains in living adult cardiomyocytes, we used 3D STED imaging (z-stack) of transversal TTs and identified local membrane evaginations in cross-sections and 3D reconstructions reminiscent of caveolae (Figure 5B and Online Figure VIA through D). Furthermore, Cav3-positive longitudinal structures between striations were increased after MI (Figure 5A, 4pMI; and Online Figure VII), consistent with the documented increase in longitudinal TT components (Figure 4D). The protein expression of Cav3 was significantly increased in ventricular cardiomyocytes at 4pMI (Figure 5D). Although the lumen of TT evaginations was not resolved at the given resolution of ≈60 nm (Online Figure IB), the membrane structures are consistent with the dimensions of caveolae. Whereas 3D reconstructions from sham cardiomyocytes showed spatially separated transversal TT structures at neighboring striations (Online Figure VIE), 8pMI cells often showed membranous connections between transversal TTs through longitudinal components and abnormally branched, superenlarged cross-section structures (Online Figure VIF). The complex TT network morphology at 8pMI together with the observed increase of Cav3-positive structures suggests a mechanism of proliferative remodeling through longitudinal and oblique TT growth as documented by component analysis during HF development (Figure 4D).
Figure 5. Remodeling of TT-associated nanodomains, Ca2+ release dyssynchrony, and protein expression in early HF (4pMI).
A, top, Sham cardiomyocytes show differences in Cav3 versus Jph2 colocalization with RyR2 clusters as evidenced by color separation of punctate Cav3 and RyR2 signals at striations (para-localization), whereas Jph2 and RyR2 are fully colocalized (yellow puncta). Note Cav3 signal (red) rarely occurs between striations. Dotted rectangles correspond to magnification. Lower left, 4pMI cardiomyocytes show changes both of Cav3 and RyR2 signals, including an increase of longitudinal Cav3-positive structures. Lower right, 8pMI cardiomyocytes showed decreased Jph2 signals compared to RyR2. An increased spatial variability with a loss of striation associated signals after MI compared with sham was confirmed by Fourier analysis, summarized in Online Figure VII. B, left, STED image of neighboring TT cross sections, only the right TT shows a membrane evagination; right, rotated 3D reconstruction of the same TTs reveals abrupt change in morphology of the right TT. Triangle indicates position of leftward imaging plane; scale, 200 nm; see Online Figure VI for extended data. C, Simultaneous imaging of Ca2+ and TT signals by transversal line scans of sham and 4pMI cardiomyocytes. Ca2+ release synchrony was quantified by half-maximal thresholding (F50) and temporal variability analysis (leading edge behavior) similar to Louch et al.10 The dyssynchrony index is 2.2 ms for sham and 15.6 ms for 4pMI. For average dyssynchrony indexes of transversal and longitudinal scans, see Results section. D, RyR2, Jph2, and Cav3 immunoblots from cardiomyocytes isolated from sham, 4pMI, and 8pMI hearts as indicated. Bar graphs summarize change in protein expression normalized to sham from at least 3 independent measurements; *P<0.05 versus sham; n.s. indicates not significant.
Notably, the localization of RyR2 clusters was significantly changed at 4pMI (Figure 5A, lower left), as documented by a significant decrease of the striation associated power function of the first periodic peak (Online Figure VII; sham n=6, 4pMI n=5 cells), indicating a loss of RyR2 clusters at striations. Furthermore, higher power values between periodic peaks indicated an increase of RyR2 clusters at atypical locations (Online Figure VII), consistent with post-MI spatial reorganization and orphaning. Because post-MI RyR2 protein expression was not changed (Figure 5D and data not shown), this suggests spatial reorganization of RyR2 clusters as possible orphaning mechanism. Furthermore, Jph2 signals appeared altered compared to RyR2 at 8pMI (Figure 5, lower right), and the striation associated peak power was significantly decreased (Online Figure VII). In agreement, heart-restricted inducible shRNA knock-down of Jph2 resulted in significantly increased variability of CRU spacing at TT junctions,19 and Jph2 knock-out showed that the SR transmembrane protein stabilizes CRU nanodomains through a cytosolic phospholipid binding domain and colocalization with LCC and RyR2 membrane complexes.20
We confirmed colocalization of Jph2 and RyR2 clusters (Figure 5A, top right), which was altered at 8pMI (Figure 5A, bottom right). Post-MI expression of Jph2 was significantly decreased (Figure 5D), to a similar degree as in conditional Jph2 shRNA knockdown hearts with increased RyR2 cluster/TT spacing and abnormal Ca2+ release.19 Therefore, we investigated changes in Ca2+ release both by combinatory (TT and Ca2+) and orientation differentiating strategies (transversal versus longitudinal confocal line scanning). As expected, sham cells showed widely spaced longitudinal TT signals between striations in transverse line scans with synchronous onset of Ca2+ release, analyzed by time to F50 threshold, resulting in a low dyssynchrony index (Figure 5C, sham). In contrast, transverse line scans of 4pMI cells showed an increase of longitudinal/oblique TT signals between striations and heterogeneously delayed Ca2+ release, resulting in a high dyssynchrony index (Figure 5C, 4pMI). On average, sham cells showed the same low dyssynchrony index of 2.2±0.1 ms for transversal and longitudinal line scans, whereas 4pMI cells showed delayed Ca2+ release and increased dyssynchrony indexes both in transverse (6.7±1.2; n=11) and longitudinal (5.0±1.5; n=9) directions. As the most profound change in dyssynchronous Ca2+ release delay occurred between transversal striations with increased longitudinal/oblique TT components (Figure 4) and we found reorganization of RyR2 clusters at striations, we next investigated RyR2 orphaning as a spatial mechanism of Ca2+ release dysfunction using a recent CRU nanodomain model.8
Computational Modeling of Spatial CRU Changes
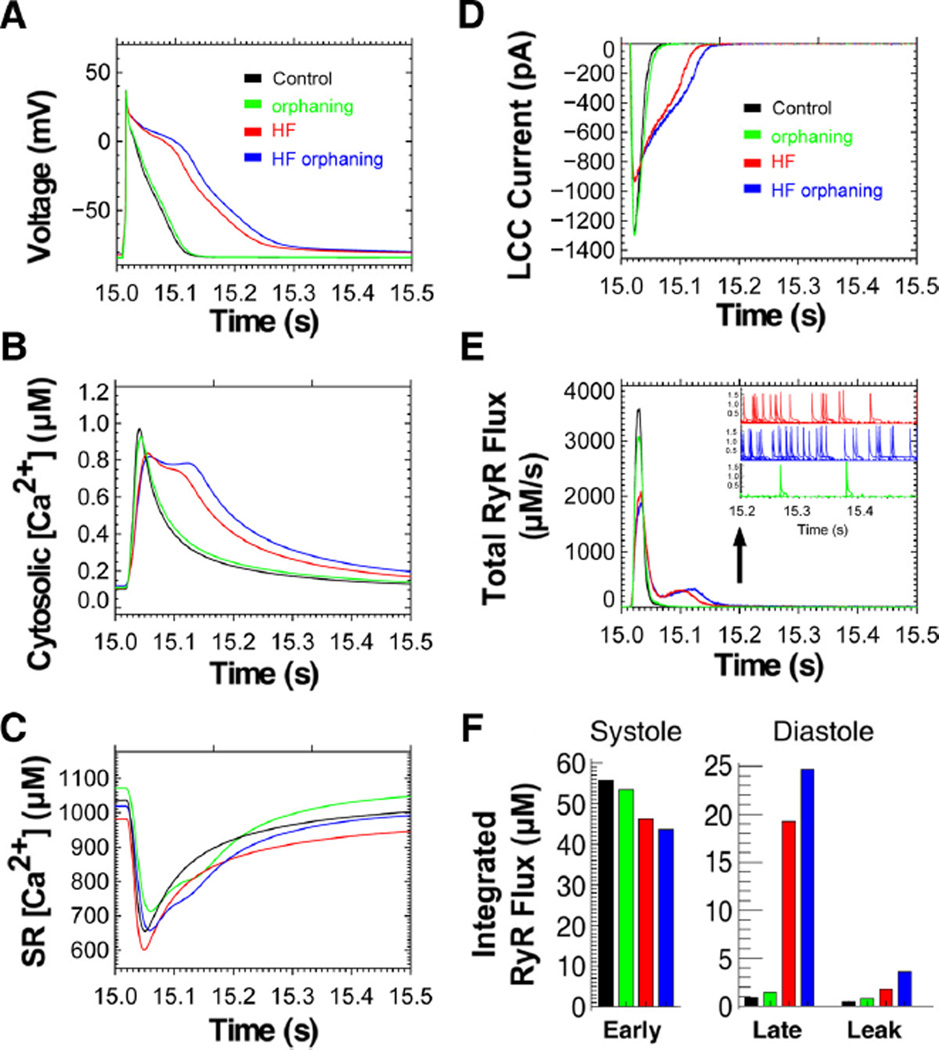
Our data suggest heterogeneous changes of individual TT components as well as RyR2 clusters and CRUs. The findings are consistent with and extend the previously described RyR2 “orphaning”3 and further show decreased Jph2 levels previously established as mechanism of increased CRU spacing at TT junctions.19 To assess the impact of increased, yet heterogeneous spatial CRU changes in HF, we used an advanced computational model based on 20 000 individual Ca2+ release sites, a realistic estimate for ventricular cardiomyocytes.8 Heterogeneous TT changes were investigated through increased spacing between TTs and RyR2 clusters at CRUs of only 25% of TT junctions (whereas 75% of CRU nanodomains remained unchanged), consistent with our data and earlier studies.3 HF was investigated by implementing previously established changes in ion transport (NCX, SERCA; see Methods). Under steady-state pacing conditions (1 Hz), the CRU model readily reproduced AP prolongation in HF, which was further worsened by additional RyR2 orphaning, whereas isolated RyR2 orphaning (without any HF changes) at 25% of CRUs caused only minimal AP prolongation (Figure 6A). In HF, AP prolongation was paralleled by a delayed cytosolic [Ca2+]i transient, and additional RyR2 orphaning clearly aggravated the [Ca2+]i transient delay (Figure 6B). Importantly, the CRU model reproduced decreased SR Ca2+ load, a hallmark of E-C coupling pathophysiology in HF, which was blunted by RyR2 orphaning (Figure 6C). Unexpectedly, combined RyR2 orphaning with HF normalized SR [Ca2+] load in diastole (Figure 6C). Furthermore, in HF depletion of diastolic SR [Ca2+] load resulted in an increased late L-type Ca2+ current (LCC) due to decreased Ca2+ dependent inactivation, further increased by RyR2 orphaning in HF (Figure 6D).
Figure 6. Computational modeling of local CRU Ca2+ release in HF and during increased TT spacing of RyR2 clusters.
Local [Ca2+]i signals from 20 000 CRUs were modeled for 15 seconds starting from the same initial conditions to reach steady-state Ca2+ transients at 1 Hz pacing. A, Action potential (AP) under normal control conditions (black); HF (red); HF with 25% orphaned RyR2 clusters (blue) resulting in AP prolongation; and orphaning without HF (green). B, The cell-wide cytosolic [Ca2+]i transient is delayed in HF (red), and RyR2 orphaning causes further, even biphasic prolongation (blue). C, In HF, SR Ca2+ content is decreased throughout E-C coupling (red). However, HF with RyR2 orphaning restores diastolic SR Ca2+ load (blue); orphaning without HF (green). D, L-type Ca2+ current (LCC) inactivation is delayed in HF (red) and further delayed by RyR2 orphaning (blue). E, Total peak RyR2 Ca2+ flux is decreased in orphaning without HF (green), HF without (red) and with RyR2 orphaning (blue) including abnormally delayed RyR2 Ca2+ flux causing a second Ca2+ flux peak in diastole. E, inset, During late diastole (after 15.2 seconds) RyR2 Ca2+ flux (leak) from Ca2+ sparks is increased in HF (red) compared with control (black). RyR2 orphaning further increases leak through Ca2+ sparks (blue). F, Integrated RyR2 Ca2+ flux during systole and diastole. Early RyR2 Ca2+ flux during systole (15.01–15.06 seconds) is decreased in HF and further decreased with RyR2 orphaning. During diastolic AP repolarization (15.06 –15.21 seconds) late RyR2 Ca2+ flux is abnormally increased in HF and further increased by RyR2 orphaning, corresponding with AP prolongation in A. During the diastolic AP phase 4 (15.21–15.50 seconds), RyR2 Ca2+ leak is increased in HF (red) and worsened by additional RyR2 orphaning (blue). Note: diastolic RyR2 Ca2+ leak corresponds to increased Ca2+ spark frequency and duration in E, inset.
We investigated the local CRU function through total peak RyR2 Ca2+ release flux, which was strongly decreased in HF, contributing to a delayed [Ca2+]i transient and an abnormal second peak in diastole (Figure 6E). Both decreased peak RyR2 Ca2+ release flux and delayed [Ca2+]i release in diastole corresponded with prolonged AP repolarization, which was further delayed by RyR2 orphaning (Figure 6A and 6E). Moreover, the early systolic integrated RyR2 Ca2+ flux was blunted in HF, which was further decreased by RyR2 orphaning (Figure 6F). In diastole, a dramatic increase in late RyR2 Ca2+ release flux occurred during AP repolarization and was further increased by RyR2 orphaning (Figure 6F). Furthermore, after completion of AP repolarization, increased diastolic RyR2 Ca2+ leak persisted as evidenced by an increased spark rate and spark duration in HF (Figure 6E; inset). Whereas RyR2 orphaning by itself did not increase spark dependent Ca2+ leak, in HF additional orphaning of RyR2 clusters further increased diastolic Ca2+ sparks (Figure 6E; inset) and Ca2+ leak (Figure 6F). Thus, RyR2 orphaning as occurs during heterogeneous spatial TT and CRU reorganization in HF resulted in macroscopic AP prolongation explained by a pronounced delay of SR Ca2+ release late in diastole and in a blunted systolic Ca2+ transient. Therefore, in addition to E-C uncoupling in HF, heterogeneous spatial reorganization of TTs and RyR2 clusters may aggravate the experimentally observed delayed and dyssynchronous SR Ca2+ release, promote severe diastolic SR Ca2+ leak, and potentially contribute to afterdepolarizations and Ca2+-triggered arrhythmias.
Discussion
Earlier live-cell studies in models of late stage HF showed either a regional loss of TT density or spatial network reorganization.3 Because the dimensions of individual TTs were below the resolution limit of conventional light microscopy,4 we have established noninvasive super-resolution imaging of intact TT membrane structures deep inside living cardiomyocytes, based on the STED concept.13 In addition, we developed quantitative image analysis strategies which showed circular morphologies of intact TT cross-sections in situ with diameters of ≈200 nm. Based on these strategies, we investigated the hypothesis that TT structures undergo early, progressive remodeling during HF development. Using STED, we provide direct in situ evidence of TT remodeling through heterogeneous enlargement of cross-sections and an increase of TT network length, establishing (1) an early time point (4pMI) of TT remodeling during HF development and (2) a proliferative mechanism of membrane reorganization. In particular, we show by quantitative analysis that the rectilinear TT network was differentially remodeled during HF development at 4pMI through significantly increased longitudinal and oblique components. The TT changes deteriorated rapidly during HF development resulting in a heterogeneous, fractured and misaligned TT network at 8pMI. In addition, Jph2 and Cav3 protein levels were significantly changed during HF development, each indicating altered TT associated CRU and caveolae nanodomain functions, respectively. Finally, Ca2+ imaging, analysis of RyR2 clusters, and computational modeling of heterogeneous TT and RyR2 orphaning changes uncovered spatial mechanisms of Ca2+ release delay and AP prolongation in HF. To our best knowledge, this is the first super-resolution study of membrane structures in cardiomyocytes establishing live-cell STED imaging as direct quantitative strategy to investigate TT membranes in healthy and diseased cells.
This study overcomes the diffraction barrier deep inside large excitable cells, using STED nanoscopy and the membrane probe di-8-ANEPPS. We have carefully evaluated different STED-compatible membrane dyes to ensure effective staining and cardiomyocyte integrity. This information is essential to achieve sufficient fluorescence membrane signals from the typical attoliter-sized spot volumes used by STED nanoscopy (Figure 1B). STED has previously shown intracellular antibody tagged vesicles in living neurons14 and has resolved YFP-tagged ER structures in PtK2 cells.21 However, YFP expression strategies require prolonged cell culture, which promotes TT loss in cardiomyocytes, and neither are antibody-based strategies advantageous to delineate TT membranes. In addition, we have shown that STED is not limited by intracellular imaging depth by comparing subsurface and deep intracellular imaging data and through 3D reconstruction of TT membrane invaginations. Nanoscale imaging of even smaller TT substructures such as caveolae is limited by the given lateral and axial resolutions of ≈60 nm and ≈500 nm, respectively, which could potentially be improved by additional instrumentation such as combining STED with 4Pi microscopy22 or by additional STED beams for further resolution enhancement. Other super-resolution methods such as STORM and (f)PALM depend on time-multiplexed localization of random signals from single molecules and image reconstruction.23 In comparison, for freely diffusing membrane dyes, STED nanoscopy might be advantageous as local membrane signals are directly characterized. Compared with STORM or STED, structured illumination is limited by a maximal 2-fold resolution increase.23 We conclude that STED nanoscopy has clear advantages for studies of subresolution membrane structure and disease changes (Online Figure VIII), even for deep intracellular imaging of individual TT structures in living cells and under challenging imaging conditions in large muscle cells with densely packed diffractive myofilaments and mitochondria. STED nanoscopy together with the presented analytic framework of quantitative TT nanopathology may therefore open avenues for future translational studies of human HF samples and pathophysiological interpretation through temporally controlled studies of progressive heart disease development.
Considering existing concepts of TT changes (for overview see Online Table IV), regional loss of TTs remains an important model. Furthermore, spatial network reorganization has been proposed as E-C uncoupling mechanism.3 Electron microscopy studies of cat hearts have estimated TT cross-section dimensions at 150 to 200 nm, depending on fixation methods,24 which is clearly below the resolution limit of conventional light microscopy as confirmed by our confocal imaging studies of TT cross-sections (Figure 2). In contrast, using noninvasive super-resolution techniques, we have directly determined the in situ dimensions of hundreds of TT cross-sections from healthy cells with mean dimensions approximating 200 nm (Table 1 and Online Table I). Furthermore, contour analysis and optical sectioning established cylindrical TT membrane structures in healthy cells (Figures 2 and 5 and Online Figure VI). Based on typical in situ TT dimensions and circular cross-section morphologies, we propose that TTs function as “nanowires” during intracellular AP propagation in muscle cells. If intracellular AP propagation depends on local TT properties, then maintenance of membrane integrity and physiological network structures is essential to ensure synchrony of E-C coupling at an estimated 20 000 CRUs. Indeed, combined Ca2+ and TT imaging of 4pMI myocytes showed increased spatial heterogeneity of intracellular Ca2+ release between striations with increased longitudinal/oblique TT components. This suggests that dyssynchronous Ca2+ release, reported earlier in HF,3,10,11,25 might be caused by TT proliferation and membrane reorganization of CRU nanodomains. Further, we find significant reorganization of RyR2, Jph2, and Cav3 clusters after MI, representing distinct TT associated nanodomains and potential RyR2 orphaning mechanisms.
Consistent with earlier observations in late-stage HF,3 we found a relative decrease of transversal network components at 8pMI (Figure 4). In addition, STED imaging established an early network phenotype at 4pMI, characterized by significantly increased longitudinal components reminiscent of immature cells.9 Although an increase of longitudinal network components has been documented during intermediate26 and late3 HF stages by confocal imaging, STED identified a previously unknown significant increase in longitudinal TT components already 4 weeks after infarction, based on sharper images and direct component analysis (Online Figure VIII). To characterize local TT signals from STED images, we used a novel strategy of quantitative analysis and found significant increases of the total amount of longitudinal and oblique TT components at 4pMI (Figure 4D). This raises the possibility that additive membrane processes and/or reactivation of fetal genes contribute to early TT remodeling during HF development. Previous work established an immature embryonic TT system not connected to the surface membrane and SR terminals until completion of postnatal maturation through transverse TT invaginations.9 Our data showing a total increase in TT network length through longitudinal and oblique components during HF development (Figure 4D) are consistent with a recent confocal study showing less uniform and often oblique TTs with increased dimensions in human HF27 and with earlier electron microscopy examples of failing and hypertrophied human hearts describing TT “proliferation” and increased dimensions.28 Electron microscopy studies showed that themajority of longitudinal tubules in healthy rat myocytes form SR junctions with ultrastructural features similar to couplons, suggesting the potential for E-C coupling and Ca2+ release.29
Junctional complexes stabilize CRUs between terminal SR release sites and TTs through lipid-protein interactions. Jph2 bridges the junctional cleft through interactions with phospholipids at TT membranes.20 We found significant Jph2 downregulation early in HF comparable in extent with heartspecific shRNA targeted Jph2 knockdown, which resulted in TT remodeling with increased CRU spacing and Ca2+ leak.19 After MI, we showed significant reorganization of Jph2 and RyR2 clusters. We note that Ca2+ release dysfunction can by itself accelerate cardiac remodeling as evidenced earlier under comparable HF conditions.16 Furthermore, we found a significant early increase in Cav3 expression together with a significant increase of longitudinal TT components early during HF development. Cav3 overexpression has been shown to induce cardiomyopathy through a dystrophin-dependent mechanism.30 We note that cardiomyocytes are exposed to abnormal stretch conditions during dilative HF (Online Figure III) and have previously shown that abnormal load conditions lead to differential activation of embryonic gene programs.31 Since Cav3 is trafficked through vesicle carriers to TT membranes via the Golgi complex, it is strategically positioned to directly participate in TT membrane remodeling. We documented para-localization of Cav3 with RyR2 clusters at transversal striations, a pattern recently shown at the cardiomyocyte surface during revision of this manuscript.18 Therefore, post-MI changes in Cav3 expression may influence para-localized RyR2 clusters, for example, through recruitment and stabilization of cholesterol-enriched glycolipid rafts, repeated caveolae formation, proliferation of TTs, and possibly altered aggregate behaviors of RyR2 cluster proteins.32 We speculate that the observed increase in Cav3 expression during TT network remodeling at 4pMI represents an adaptive response during hypertrophic remodeling (Online Figure IV). Therefore, it will be particularly important to extend our observations to molecular Cav3 mechanisms of TT remodeling in future studies.
We used computational modeling to investigate a realistic, massive number of functionally independent CRUs,8 a prerequisite to characterize CRU changes, which we found during heterogeneous TT remodeling, and earlier studies during orphaning of RyR2 clusters in HF3 or during HF induced by Jph2 knockdown.19 Our data show that heterogeneous CRU spacing and RyR2 orphaning on top of global HF changes aggravated the phenotype through delayed SR Ca2+ release and AP prolongation. Unexpectedly, HF with RyR2 orphaning resulted in normalization of diastolic SR [Ca2+] load explained by reduced LCC current inactivation due to increased spacing of RyR2 release sites (Figure 6D and Online Figure VII). Normalization of SR load is in agreement with a previous study in a pig model of ischemic heart disease that found even increased SR Ca2+ load based on NCX current measurements.11 Normalized SR load in HF with RyR2 orphaning contributed to amore pronounced RyR2 Ca2+ flux delay and to increased diastolic SR Ca2+ leak through abnormally increased spark activity. Different from Williams et al,8 an earlier modeling study of LCC gating changes showed delayed Ca2+ signaling.33 However, we found reduced LCC current inactivation in HF based on individual CRU nanodomain properties due to increased local heterogeneity of release sites, supporting complex subcellular changes in Ca2+ signaling. Therefore, future super-resolution studies will need to assess local mechanisms of SR Ca2+ leak directly at CRUs, potentially through novel STED dyes or combined imaging techniques.
In summary, we demonstrate that STED imaging provides fundamentally new insight through local information of the TT morphology in living cardiomyocytes that was previously unattainable and show how critical super-resolution studies are to understand the integrity of complex intracellular membrane systems. In addition, STED nanoscopy provided reproducible membrane localization information, which is not subject to potential artifacts from postprocessing or indirect strategies. We have developed novel quantitative analysis strategies which allow unbiased, automated data processing and characterization of individual TTs and network components in living cells. Through STED imaging and molecular analysis, we have uncovered a mechanism of additive TT remodeling consistent with an early HF phenotype of TT proliferation previously not recognized. Our studies further implicate caveolae as TT substructures that may contribute to TT remodeling through membrane trafficking. In addition, significantly decreased Jph2 expression indicates increased CRU spacing in the context of heterogeneous TT membrane changes, a phenotype investigated through computational modeling of heterogeneous CRU changes aggravating delayed Ca2+ release and AP prolongation. STED nanoscopy of living cardiomyocytes will be particularly useful to gain mechanistic insight about the function of TT changes in different heart disease models, to explore early and potentially reversible disease mechanisms with high accuracy, to test interventions which may prevent TT remodeling, and to translate basic mechanisms and concepts through nanopathology studies of patient samples.
Supplementary Material
Novelty and Significance.
What Is Known?
Post–myocardial infarction heart failure (HF) is a leading cause of morbidity and mortality that is increasing worldwide.
Remodeling of the T-tubule membrane system, which has been shown in patients and animals with HF, may represent an important mechanism of pathological remodeling.
Due to small subresolution dimensions, the imaging of subcellular T-tubule membrane structures may require super-resolution imaging. Recently developed light microscopy techniques may allow for quantitative investigation of intact subresolution membrane structures in cardiac cells or their changes under pathological conditions.
What New Information Does This Article Contribute?
A super-resolution technique called stimulated emission depletion (STED) microscopy shows T-tubule membrane structures with nanometric resolution deep inside living cardiac myocytes for the first time.
In cardiac myocytes, T-tubule remodeling occurs through an additive, proliferative remodeling process that results in profound membrane network reorganization and Ca2+ signaling dysfunction as early as 4 weeks after myocardial infarction in mice.
Previous studies suggest that the T-tubule system contributes to abnormal Ca2+ signaling in animals and patients with HF. We developed novel strategies for super-resolution imaging and quantitative analysis of T-tubules in living cardiac myocytes using STED microscopy. We investigated the nature of T-tubule remodeling based on significantly improved nanometric resolution and show that membrane remodeling underlies subcellular Ca2+ cycling defects in HF. We found that individual T-tubule cross sections increase progressively in dimensions over several weeks after myocardial infarction, suggesting additive membrane remodeling processes. The physiological rectilinear T-tubule network organization in mice was found to undergo dramatic reorganization with an increase in length and the addition of new components. The time course and increase of T-tubule membrane components correlated well with an increase in the expression of muscle-specific caveolin-3 proteins that are known to contribute to T-tubule organization in the developing heart. The spatial reorganization of the T-tubule network correlated with increased dyssynchrony of intracellular Ca2+ release in mice after myocardial infarction. The Ca2+ release dyssynchrony is explained by orphaning of ryanodine receptor (RyR2) Ca2+ release channels due to disruption of junctional signaling microdomains during T-tubule remodeling. Thus, our data suggest that T-tubule remodeling and orphaning of RyR2 clusters might play important roles in the development of HF by contributing directly to electric and contractile dysfunction.
Acknowledgments
We thank K. Koschel and P. Mannheim for excellent technical assistance.
Sources of Funding
This work received support through Deutsche Forschungsgemeinschaft (KFO 155 subprojects KO 1872/2-1, LE 1313/2-1, and SFB 1002, to G.H., S.E.L., S.L., and S.W.H.); a Halbach Foundation award to S.E.L. supporting E.W.; and a DAAD exchange program supporting T.K. as visitor at the University of Maryland. The research leading to these results has received funding from the European Community’s Seventh Framework Program FP7/2007-2013 under grant agreement No. HEALTH-F2-2009-241526, EUTrigTreat (S.E.L., S.L., and W.J.L.). M.S.J. was supported by NIH 1RO1 HL1105239-01. G.H., S.W.H., and S.E.L. are investigators of the German Center for Cardiovascular Research (DZHK).
Non-standard Abbreviations and Acronyms
- AP
action potential
- CRU
calcium release unit
- E-C
excitation-contraction coupling
- FWHM
full width at half-maximum
- HF
heart failure
- LCC
L-type Ca2+ channel (cardiac isoform Cav1.2)
- RyR2
ryanodine receptor type 2 (cardiac isoform)
- SR
sarcoplasmic reticulum
- STED
stimulated emission depletion nanoscopy
- TT
transverse tubule; T-tubule
- 4pMI and 8pMI
4 and 8 weeks after myocardial infarction
Footnotes
The online-only Data Supplement is available with this article at http://circres.ahajournals.org/lookup/suppl/doi:10.1161/CIRCRESAHA.112274530/-/DC1.
Disclosures
None.
References
- 1.Endo M. Entry of a dye into the sarcotubular system of muscle. Nature. 1964;202:1115–1116. doi: 10.1038/2021115b0. [DOI] [PubMed] [Google Scholar]
- 2.Bu G, Adams H, Berbari EJ, Rubart M. Uniform action potential repolarization within the sarcolemma of in situ ventricular cardiomyocytes. Biophys J. 2009;96:2532–2546. doi: 10.1016/j.bpj.2008.12.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song LS, Sobie EA, McCulle S, Lederer WJ, Balke CW, Cheng H. Orphaned ryanodine receptors in the failing heart. Proc Natl Acad Sci U S A. 2006;103:4305–4310. doi: 10.1073/pnas.0509324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzini-Armstrong C, Protasi F, Ramesh V. Shape, size, and distribution of Ca(2+) release units and couplons in skeletal and cardiac muscles. Biophys J. 1999;77:1528–1539. doi: 10.1016/S0006-3495(99)77000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi T, Martone ME, Yu Z, Thor A, Doi M, Holst MJ, Ellisman MH, Hoshijima M. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J Cell Sci. 2009;122(Pt 7):1005–1013. doi: 10.1242/jcs.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern MD, Pizarro G, Rios E. Local control model of excitation-contraction coupling in skeletal muscle. J Gen Physiol. 1997;110:415–440. doi: 10.1085/jgp.110.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannell MB, Cheng H, Lederer WJ. The control of calcium release in heart muscle. Science. 1995;268:1045–1049. doi: 10.1126/science.7754384. [DOI] [PubMed] [Google Scholar]
- 8.Williams GSB, Chikando AC, Tuan HTM, Sobie EA, Lederer WJ, Jafri MS. Dynamics of calcium sparks and calcium leak in the heart. Biophys J. 2011;101:1287–1296. doi: 10.1016/j.bpj.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddock PS, Coetzee WA, Cho E, Porter L, Katoh H, Bers DM, Jafri MS, Artman M. Subcellular [Ca2+]i gradients during excitation-contraction coupling in newborn rabbit ventricular myocytes. Circ Res. 1999;85:415–427. doi: 10.1161/01.res.85.5.415. [DOI] [PubMed] [Google Scholar]
- 10.Louch WE, Mork HK, Sexton J, Stromme TA, Laake P, Sjaastad I, Sejersted OM. T-tubule disorganization and reduced synchrony of Ca2+ release in murine cardiomyocytes following myocardial infarction. J Physiol. 2006;574(Pt 2):519–533. doi: 10.1113/jphysiol.2006.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzel FR, Bito V, Biesmans L, Wu M, Detre E, von Wegner F, Claus P, Dymarkowski S, Maes F, Bogaert J, Rademakers F, D’Hooge J, Sipido K. Remodeling of T-tubules and reduced synchrony of Ca2+ release in myocytes from chronically ischemic myocardium. Circ Res. 2008;102:338–346. doi: 10.1161/CIRCRESAHA.107.160085. [DOI] [PubMed] [Google Scholar]
- 12.Soeller C, Cannell MB. Examination of the transverse tubular system in living cardiac rat myocytes by 2-photon microscopy and digital imageprocessing techniques. Circ Res. 1999;84:266–275. doi: 10.1161/01.res.84.3.266. [DOI] [PubMed] [Google Scholar]
- 13.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 14.Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 15.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 16.Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell. 2005;123:25–35. doi: 10.1016/j.cell.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parton RG, Way M, Zorzi N, Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baddeley D, Crossman D, Rossberger S, Cheyne JE, Montgomery JM, Jayasinghe ID, Cremer C, Cannell MB, Soeller C. 4D super-resolution microscopy with conventional fluorophores and single wavelength excitation in optically thick cells and tissues. PLoS One. 2011;6:e20645. doi: 10.1371/journal.pone.0020645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Oort RJ, Garbino A, Wang W, Dixit SS, Landstrom AP, Gaur N, De Almeida AC, Skapura DG, Rudy Y, Burns AR, Ackerman MJ, Wehrens XH. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 21.Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci U S A. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, Hell SW. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 23.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 24.Fawcett DW, McNutt NS. The ultrastructure of the cat myocardium. I. Ventricular papillary muscle. J Cell Biol. 1969;42:1–45. doi: 10.1083/jcb.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litwin SE, Zhang D, Bridge JH. Dyssynchronous Ca(2+) sparks in myocytes from infarcted hearts. Circ Res. 2000;87:1040–1047. doi: 10.1161/01.res.87.11.1040. [DOI] [PubMed] [Google Scholar]
- 26.Sachse FB, Torres NS, Savio-Galimberti E, Aiba T, Kass DA, Tomaselli GF, Bridge JH. Subcellular structures and function of myocytes impaired during heart failure are restored by cardiac resynchronization therapy. Circ Res. 2012;110:588–597. doi: 10.1161/CIRCRESAHA.111.257428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crossman DJ, Ruygrok PN, Soeller C, Cannell MB. Changes in the organization of excitation-contraction coupling structures in failing human heart. PLoS One. 2011;6:e17901. doi: 10.1371/journal.pone.0017901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaper J, Froede R, Hein S, Buck A, Hashizume H, Speiser B, Friedl A, Bleese N. Impairment of the myocardial ultrastructure and changes of the cytoskeleton in dilated cardiomyopathy. Circulation. 1991;83:504–514. doi: 10.1161/01.cir.83.2.504. [DOI] [PubMed] [Google Scholar]
- 29.Asghari P, Schulson M, Scriven DR, Martens G, Moore ED. Axial tubules of rat ventricular myocytes form multiple junctions with the sarcoplasmic reticulum. Biophys J. 2009;96:4651–4660. doi: 10.1016/j.bpj.2009.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravamudan B, Volonte D, Ramani R, Gursoy E, Lisanti MP, London B, Galbiati F. Transgenic overexpression of caveolin-3 in the heart induces a cardiomyopathic phenotype. Hum Mol Genet. 2003;12:2777–2788. doi: 10.1093/hmg/ddg313. [DOI] [PubMed] [Google Scholar]
- 31.Toischer K, Rokita AG, Unsold B, et al. Differential cardiac remodeling in preload versus afterload. Circulation. 2010;122:993–1003. doi: 10.1161/CIRCULATIONAHA.110.943431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carozzi AJ, Ikonen E, Lindsay MR, Parton RG. Role of cholesterol in developing T-tubules: analogous mechanisms for T-tubule and caveolae biogenesis. Traffic. 2000;1:326–341. doi: 10.1034/j.1600-0854.2000.010406.x. [DOI] [PubMed] [Google Scholar]
- 33.Cannell MB, Crossman DJ, Soeller C. Effect of changes in action potential spike configuration, junctional sarcoplasmic reticulum microarchitecture and altered t-tubule structure in human heart failure. J Muscle Res Cell Motil. 2006;27:297–306. doi: 10.1007/s10974-006-9089-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.