Abstract
Ribonuclease inhibitor (RI) is a conserved protein of the mammalian cytosol. RI binds with high affinity to diverse secretory ribonucleases (RNases) and inhibits their enzymatic activity. Although secretory RNases are found in all vertebrates, the existence of a non-mammalian RI has been uncertain. Here, we report on the identification and characterization of RI homologs from chicken and anole lizard. These proteins bind to RNases from multiple species, but exhibit much greater affinity for their cognate RNases than for mammalian RNases. To reveal the basis for this differential affinity, we determined the crystal structure of mouse, bovine, and chicken RI·RNase complexes to a resolution of 2.20, 2.21, and 1.92 Å, respectively. A combination of structural, computational, and bioinformatic analyses enabled the identification of two residues that appear to contribute to the differential affinity for RNases. We also found marked differences in oxidative instability between mammalian and non-mammalian RIs, indicating evolution toward greater oxygen-sensitivity in RIs from mammalian species. Taken together, our results illuminate the structural and functional evolution of RI, along with its dynamic role in vertebrate biology.
Keywords: leucine-rich repeat (LRR), protein–protein interaction, reactive oxygen species (ROS), redox homeostasis, ribonuclease
Introduction
Understanding the sequence–structure–function relationships of proteins, as well as how evolution has guided and shaped these relationships, is a central aim of biology. A protein that is especially worthy of study—due to its unique structure, fascinating biology, and emerging evolution—is ribonuclease inhibitor (RI).
RI is a highly conserved, 50-kDa protein present in the cytosol of all mammalian cells. Its name originates from its ability to inhibit the ribonucleolytic activity of a large variety of secretory ribonucleases (RNases) [1]. The structure of RI is composed entirely of leucine-rich repeats (LRR), a domain specifically associated with protein–protein and protein–ligand interactions [2]. Crystal structures of both free [3] and RNase-bound [4–7] RI have yielded a wealth of information about the LRR fold and its interaction with ligands. Beyond its unique shape, RI also possesses a large number of conserved cysteine residues, which must be reduced to maintain form and function [8,9]. Indeed, oxidation of even a single cysteine leads to a cooperative “all-or-none” cascade of disulfide-bond formation, resulting in the complete inactivation of RI [10]. Tellingly, treatment of cultured cells with oxidants is sufficient to cause the rapid disappearance of RI [8].
Despite vast knowledge about its structure, the biological function of RI remains enigmatic. Based on its extremely tight affinity for diverse secretory RNases[11], RI could serve to regulate the localization and function of RNases in vivo. Engineering RNases to evade RI binding imbues them with latent cytotoxicity for human cells [12], and overproduction of RI makes cells less susceptible to cytotoxic RNases[13]. Recent studies indicate that RI might dynamically regulate the function of the secretory RNases angiogenin [14,15] and RNase 7 [16].
In addition to controlling the activity of RNases, RI could play a role in maintaining intracellular redox homeostasis. The cytosolic localization of RI, coupled with its many free cysteine residues, suggests that RI might scavenge reactive oxygen species (ROS)[17–19]. ROSen compass a variety of highly reactive chemical species including superoxide anion, hydroxyl radical, and hydrogen peroxide [20]. The role of ROS and oxidative stress in ageing, cancer, and other diseases is now well known [21]. Knockdown of RI in various human cell lines leads to enhanced susceptibility to oxidant-induced DNA damage [18]. Similarly, overproduction of RI can protect cells against the effects of oxidative stress[22]. In vivo, oxidation of RI has been linked to the progression of pancreatitis [23], as well as to the effectiveness of certain cancer treatments [24]. Intriguingly, RI is present in red blood cells, which contain neither a nucleus nor an RNA. RI might play a role in protecting red blood cells from oxidative-stress-related ageing and turnover[25,26].
An overarching mystery in RI biology has been its apparent absence from non-mammalian species. Secretory ribonucleases are known to be present in all vertebrates [27,28]. Inhibition of ribonucleolytic activity had been detected in cellular lysates from non-mammalian hosts [29]. However, the source of this inhibition was never characterized, and no non-mammalian RI homologs have been isolated.
We have identified and characterized homologous RIs from two non-mammalian species: chicken and anole lizard. Our efforts provide much insight into the evolution of RI structure and function, as well as on its biological role. We show pronounced differences in oxidation-sensitivity across homologs, suggesting a dynamic evolutionary shift between mammals and non-mammals. Our observation that RI occurs in a wide range of animals indicates an essential role for this protein.
Results
Production of RI from mouse, chicken, and anole
Prior to our work, the presence of a homologous ribonuclease inhibitor (RI) in a non-mammalian species had never been confirmed. We located genes encoding avian and reptile homologs of RI, and we produced these proteins heterologously in Escherichia coli. In addition, we produced the mouse homolog of RI, which had never been characterized. To enable comparisons, we also produced the previously characterized human RI and bovine RI [30]. All RI homologs have similar molecular mass, unusually high cysteine and leucine content, and a strong overall anionic charge (Table 1). Mammalian RI homologs have relatively high aminoacid sequence identity and similarity. Avian and reptilian RI homologs are more similar to each other than to any of the mammalian RIs (Table S3). Our initial characterization determined that RI from each species bound tightly to its cognate ribonuclease in a 1:1 ratio and completely inhibited ribonucleolytic activity (Fig. S1a and S1b).
Table 1.
Properties of homologous ribonuclease inhibitors
| Species | MW (Da) | No. residues | No. leucine residues (%) | No. cysteine residues (%) | Za | Tmb | GenBank Accession No. |
|---|---|---|---|---|---|---|---|
| Human (H. sapiens) | 49973 | 461 | 92 (20%) | 32 (6.9%) | −22 | 51.7 ± 1.1 | NP_976323 |
| Mouse (M. musculus) | 49816 | 456 | 92 (20%) | 30 (6.6%) | −20 | 48.8 ± 0.5 | NP_001165571 |
| Bovine (B. taurus) | 48850 | 456 | 98 (22%) | 29 (6.4%) | −22 | 52.5 ± 0.9 | NP_001030396 |
| Chicken (G. gallus) | 49846 | 456 | 81 (18%) | 30 (6.6%) | −20 | 52.2 ± 0.4 | NP_001006473 |
| Lizard (A. carolinensis) | 49581 | 456 | 78 (17%) | 29 (6.4%) | −10 | 50.0 ± 0.7 | XP_003214831 |
Values of Z refer to the net molecular charge: Arg + Lys − Asp − Glu.
Values of Tm are the temperature at the midpoint of thermal denaturation, as determined by incorporation of a hydrophobic dye and quantitation by DSF.
Contrasts between intra- and inter-species RI·RNase binding affinity
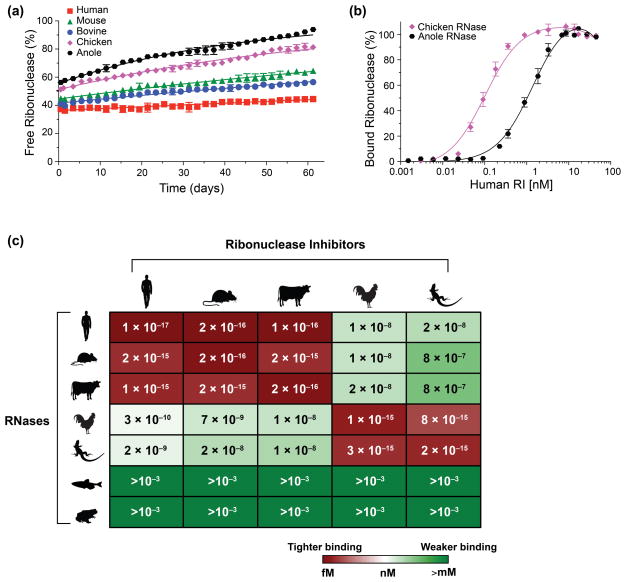
To quantify the stability of both endogenous RI·RNase complexes and inter-species complexes, we used binding assays that employ a fluorescently labeled RNase (Fig. 1). From these data, we determined equilibrium dissociation constants for each RNase paired with each RI in our study (Fig. 1c; Table S2). We found that each endogenous RI·RNase complex was extremely tight (Kd ≤ 1 fM). Indeed, these RI–RNase interactions are the tightest known amongst biomolecules. In addition, mammalian RIs bind tightly to mammalian RNases, and avian and reptilian RIs bind tightly to avian and reptilian RNases. Interestingly, complexes formed between evolutionarily distant classes (i.e., mammalia versus aves or reptilia) were ~7–8 orders of magnitude weaker than endogenous complexes (Fig. 1c; Table S2). Surprisingly, none of the RIs in our study exhibited detectable binding to RNases from either frog or fish.
Fig. 1.
Stability of endogenous and inter-species RI·RNase complexes. (a) Representative normalized fluorescence data showing the gradual dissociation of fluorescently labeled RNases from endogenous RI·RNase complexes over time. Data were fitted to derive kd values for each RI·RNase complex. (b) Representative normalized fluorescence data showing inter-species RI·RNase complex formation with increasing concentration of RI. Data were fitted to derive Kd values for each RI·RNase pair. (c) Heat map of the Kd values for 35 RI·RNase complexes. Red indicates lower Kd values; green indicates higher Kd values.
Increased thermostability of RI complexes correlates to binding strength
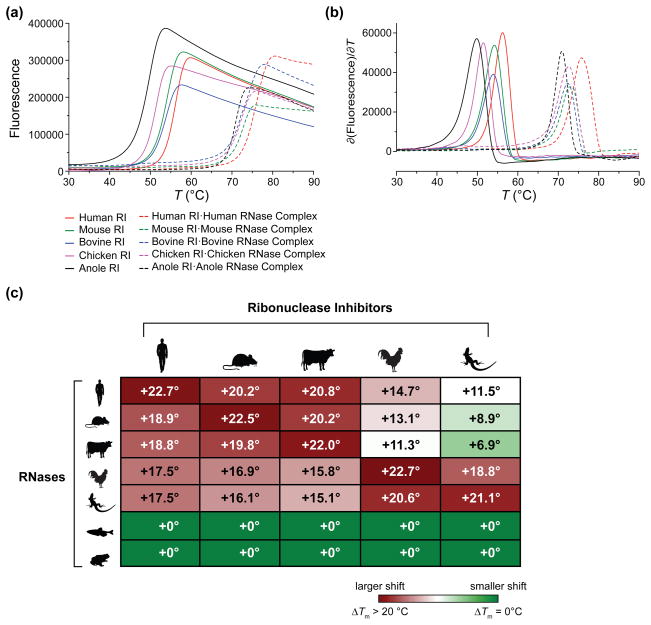
We next determined if differences in the affinity of RI for an RNase correlated to differences in the thermostability of an RI·RNase complex. To do so, we measured the thermal denaturation of RI in both an unbound state and an RNase-bound state. For each species, the thermostability of RI increased dramatically (>21 °C) when bound to its cognate RNase (Fig. 2a and 2b). We also determined the shift in Tm for each RI bound to every RNase in our study. We found that changes in RI thermostability upon RNase binding correlated well with the measured Kd for that RNase (Fig. 2c). Similarly, there was no change in RI thermostability when incubated with either frog or fish RNase.
Fig. 2.
Effect of bound RNase on the thermostability of RI. (a) Temperature-dependence of the fluorescence of SYPRO Orange dye at 578 nm in the presence of RI alone and in the presence of RI plus an RNase. (a) Thermal denaturation curves. (b) Derivatives of the data in panel (a). Values of Tm are listed in Table 1. (c) Heat map summarizing the change in the thermostability of RI conferred upon its binding various RNases. Numbers represent ΔTm from the unbound to the bound state.
Structural characterization of endogenous RI·RNase complexes
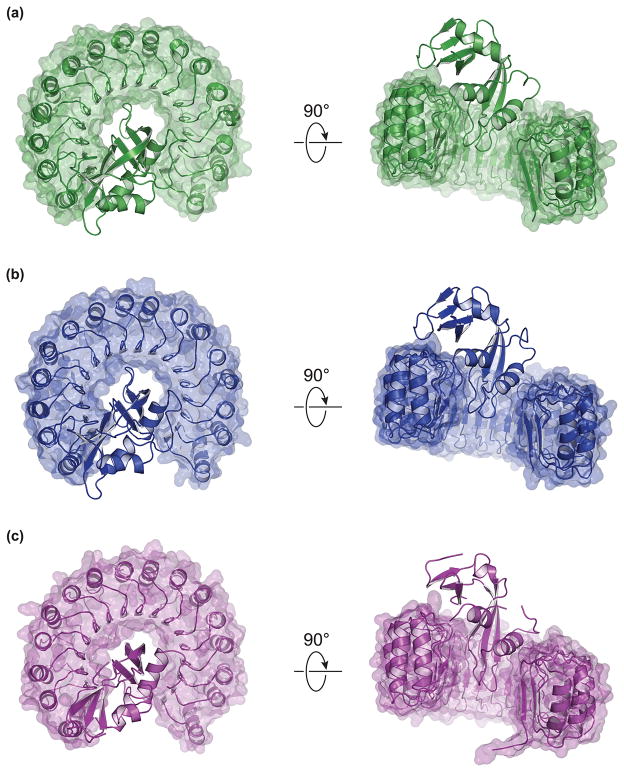
Intrigued by the large differences in binding affinity between mammals and non-mammals, we sought structural explanations to account for the change in Kd. Accordingly, we determined high-resolution crystal structures for three complexes: mouse RI·mouse RNase, bovine RI·bovine RNase, and chicken RI·chicken RNase (Table 2; Fig. 3). We were unable to generate diffraction-quality crystals for the anole RI·anole RNase complex.
Table 2.
Computational analysis of the interface in RI·RNase complexes
| No. of Contacts Residuesc | Character of Interface Residuesc [No. (%)] | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| RI·RNase Complex | Buried ASAa (Å2) | Scb | From RI | From RNase | Non-polar | Uncharged Polar | Charged | Hydrogen Bondsd (Å) | Non-bonded contactse |
| Humanf | 2801 | 0.688 | 28 | 23 | 17 (33%) | 14 (27%) | 20 (39%) | 19 (2.79) | 177 |
| Mouseg | 2650 | 0.645 | 23 | 25 | 15 (30%) | 18 (36%) | 16 (33%) | 13 (2.92) | 126 |
| Cowh | 2793 | 0.605 | 28 | 25 | 15 (28%) | 17 (32%) | 21 (40%) | 15 (2.90) | 150 |
| Chicken | 2757 | 0.599 | 26 | 20 | 21 (46%) | 7 (15%) | 18 (39%) | 12 (2.90) | 118 |
| Pig·Cow | 2582 | 0.590 | 26 | 23 | 14 (29%) | 13 (27%) | 22 (45%) | 8 (3.01) | 90 |
Buried accessible surface area (ASA) was calculated with the program PDBsum.
The value of Sc reports on geometrical shape complementarity, where Sc = 1.0 for two perfectly complementary surfaces and Sc = 0 for two completely dissimilar surfaces [74]. Sc values were calculated with SC v6.4.
Contact residues were identified by PDBsum as non-polar (A,F,G,I,L,M,P,V,W,Y), uncharged polar (C,N,Q,S,T), or charged (D,E,H,K,R).
Hydrogen bonds were calculated by the HBPLUS[66] algorithm of PDBsum (rX···X<3.3 Å).
Non-bonded contacts were calculated by HBPLUS [66] and defined as any contacts between proteins involving either a carbon or a sulfur atom, where the interaction distance is ≤3.9 Å.
Calculations were performed with chain Y (hRI) and chain Z (RNase 1) from PDB entry 1z7x due to the presence of bound citrate in the active site of RNase 1 in the other complex in the asymmetric unit.
Calculations represent the average values from four complexes in the asymmetric unit.
Calculations represent the average values from two complexes in the asymmetric unit.
Fig. 3.
Crystal structures of homologous RI·RNase complexes. (a) Mouse RI with mouse RNase 1 (PDB entry 3tsr). (b) Bovine RI with bovine RNase 1 (PDB entry 4peq). (c) Chicken RI with chicken RNase A-1 (PDB entry 4per).
In general, the structures of the RI homologs bear striking resesmblance to each other and to the previously characterized structures of human and porcine RI (Fig. 3) [3,5]. The structures are repetitive and symmetrical, and they have a vast surface area that is largely concave. The conserved LRR units are arranged in a horseshoe shape, and correspond to structural units consisting of a β-strand and an α-helix. Each RI molecule binds to its cognate ribonuclease in a similar position and orientation.
Analyses of binding interface regions highlight key differences across classes
Beyond the outward similarities of each RI·RNase complex, we probed for subtle differences at the interface region between the two bound proteins. We found each interface to contain a similarly large amount of buried surface area (Table 2). The number and character of interface residues were similar across the complexes, with the exception of that in the chicken complex, which has more non-polar residues and fewer uncharged residues than do the mammalian complexes (Table 2). Shape complementarity (Sc) calculations appeared to correlate with buried surface area and followed a general trend, with the human interface having the greatest complementarity, followed by mouse, bovine, and chicken. The human complex has the greatest number of both hydrogen bonds and non-bonded interactions, whereas the chicken complex has the fewest. As a comparison, we also analyzed the inter-species porcine RI·bovine RNase complex [4]. Interestingly, this non-endogenous complex displays less buried surface area, the lowest Sc value, and fewer hydrogen bonds and non-bonded interactions than do any of the endogenous complexes (Table 2).
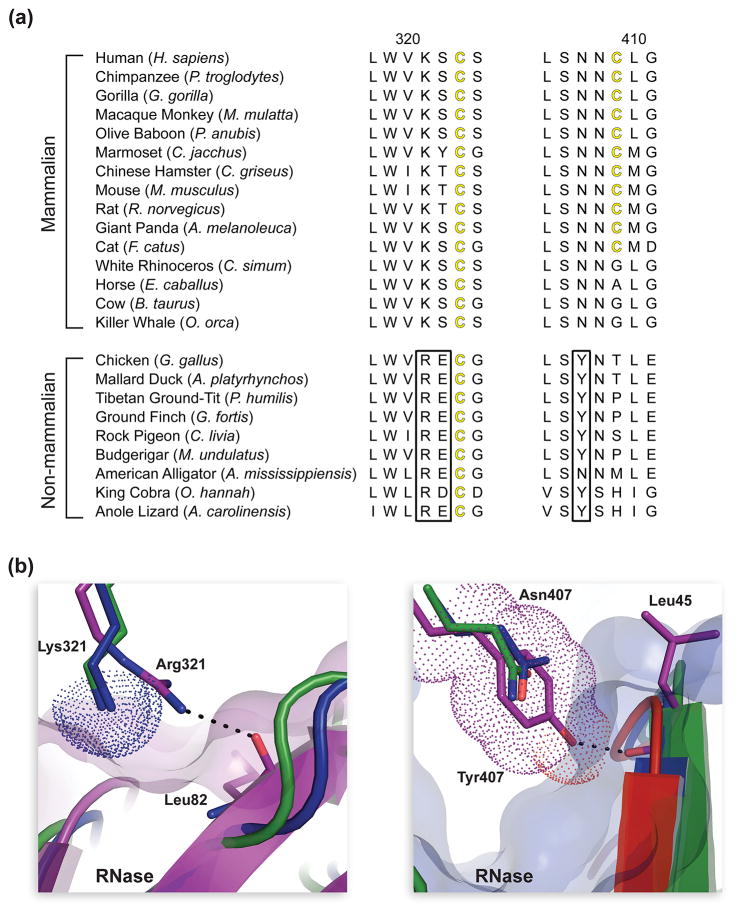
Upon mapping the interface residues of each complex onto protein sequence alignments, we discovered that the interface residues contributed by both RIs and RNases were conserved across homologs (Fig. 4 and Fig. S2). We analyzed each RI·RNase interface for the presence of predicted “hot spots”—residues predicted to have a large contribution to binding energy [31,32]. We found two hotspot regions in chicken RI that are particularly divergent from those in mammalian RIs: Arg321 and Tyr407 (human RI nomenclature) (Fig. 5). Analysis of these regions at the atomic level indicated that Arg321 and Tyr407 might play a role in the differential RI binding described above. These two residues were highly conserved across 9 non-mammalian species, but were completely absent from 15 mammalian species (Fig. 5a). Arg321 in chicken RI forms a hydrogen bond with the main-chain oxygen of Leu86 in chicken RNase (human RNase nomenclature). Due to the replacement of Arg321 with a lysine residue in mammalian RIs, this interaction is lost. In addition, the side chain of Lys321, which is conserved in human, cow, and mouse RI, could sterically hinder the binding of chicken RNase (Fig. 5b). Similar to Arg321, Tyr407 in chicken RI forms a hydrogen bond to the main-chain oxygen of Leu43 of its cognate RNase, an interaction that is not observed in the cow and mouse RI·RNase structures. The larger Tyr residue, which is conserved in non-mammalian RIs, could lead to a significant steric clash in a cow RI·chicken RNase or mouse RI·chicken RNase complex (Fig. 5b). Thus, these two substitutions result in the loss of two direct RI–RNase interactions in the chicken complexes and generate potential steric clashes during the formation of non-endogenous RI·RNase complexes. Interestingly, these two residues are present in anole RI as well.
Fig. 4.
Amino-acid sequence alignment of homologous RIs. Residues participating in binding to endogenous RNases (as identified in crystal structures) are shaded. Black boxes indicate predicted “hotspots” for binding affinity [32]. Gray coils represent α-helices, and black arrows represent β-sheets.
Fig. 5.
Evolution of residues at the interface of mammalian and non-mammalian RI·RNase complexes. (a)Two sections of amino-acid sequence alignment for homologous mammalian and non-mammalian RIs. Black boxes highlight residues conserved in non-mammalian RIs that are absent from mammalian homologs. (b) Two sections of tertiary structural alignment of the mouse (green), bovine (blue), and chicken (purple) RI·RNase complexes to illustrate the affect of amino-acid substitutions at position 321 and 407 of RI. The hydrogen bond of Arg321 in chicken RI with the main-chain oxygen of Leu86 in chicken RNase is not present in mammalian RI·RNase complexes, which contain a lysine residue at this position in RI. Lys321 (dotted surfaces) of cow and mouse RI could clash with bound chicken RNase (purple transparent surface). The hydrogen bond of Tyr407 in chicken RI with the main-chain oxygen of Leu43 in chicken RNase is not present in mammalian RI·RNase complexes, which contain an asparagine residue at this position in RI. Tyr407 (dotted surface) of chicken RI could clash with bound bovine RNase (blue transparent surface).
RI·RNase complexes are differentially sensitive to oxidation
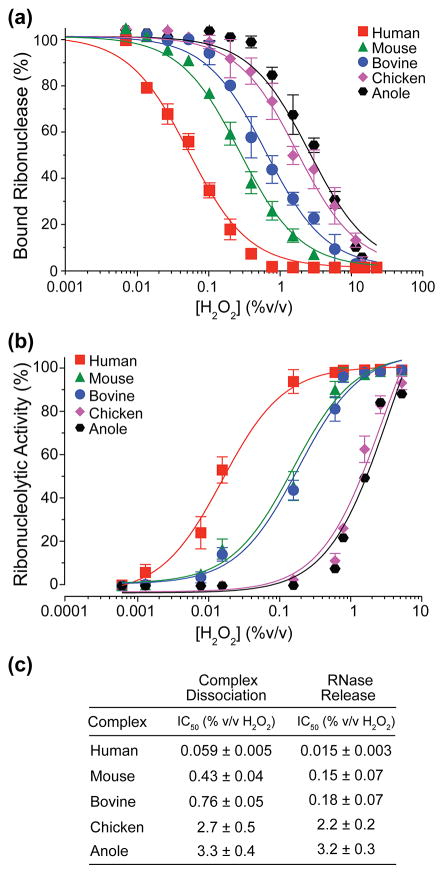
Upon oxidation, RI undergoes rapid unfolding and inactivation, subsequently releasing bound ribonuclease [1]. To determine the oxidation-sensitivity of each RI complex, we assessed the ability of hydrogen peroxide to disrupt RI·RNase complexes using two distinct assays. Upon measuring the dissociation of a fluorescently labeled RNase, we found that human RI was the most sensitive to oxidation, with H2O2 IC50 values 7-, 13-, 46-, and 56-fold lower than mouse, bovine, chicken, or anole RI, respectively (Fig. 6a). Oxidation of each endogenous RI·RNase complex yielded a catalytically active RNase. Upon measuring the release of fully active RNase, we found that human RI was again most sensitive to oxidation, with H2O2 IC50 values 10-, 12-, 147-, and 213-fold lower than mouse, bovine, chicken, or anole RI, respectively (Fig. 6b).
Fig. 6.
Comparison of the oxidation sensitivity of homologous RI·RNase complexes. (a) The dissociation of fluorescently-labeled RNases upon treatment of RI·RNase complexes with increasing concentrations of H2O2. (b) The release of active ribonucleases from RI·RNase complexes upon treatment with increasing concentrations of H2O2. (c) H2O2 IC50 values derived from the data in panels (a) and (b).
Cysteine solvation correlates to RI oxidation-sensitivity
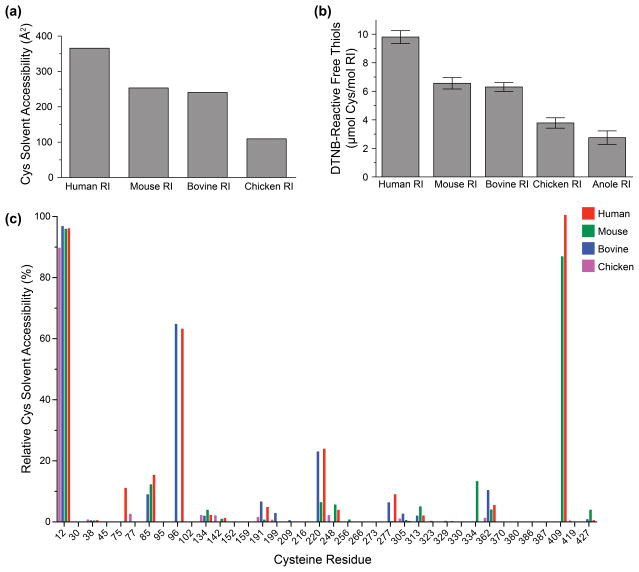
To explain the extreme differences in oxidation-sensitivity measured for RI homologs, we calculated the solvent-exposed surface area of each cysteine residue in human, mouse, bovine, and chicken RI. We found that human RI contained the highest overall cysteine solvent accessibility, followed by mouse, bovine, and chicken RI (Fig. 7a). Next, we empirically measured the amount of reactive thiol content for each RI protein using an assay based on the reduction of dithionitrobenzoic acid. Our experimental results matched closely with the computational data: human RI had the highest reactive thiol content, followed by mouse, cow, chicken, and anole RI (Fig. 7b).
Fig. 7.
Comparison of cysteine residues of homologous RIs. (a) Combined solvent-exposed surface area for cysteine residues, as calculated from crystal structures with PyMOL. (b) Quantitation of solvent-exposed thiol groups in recombinant proteins, based on reaction with dithionitrobenzoic acid. (c) Relative solvent accessibility calculations for each cysteine residue in human, mouse, bovine, and chicken RIs.
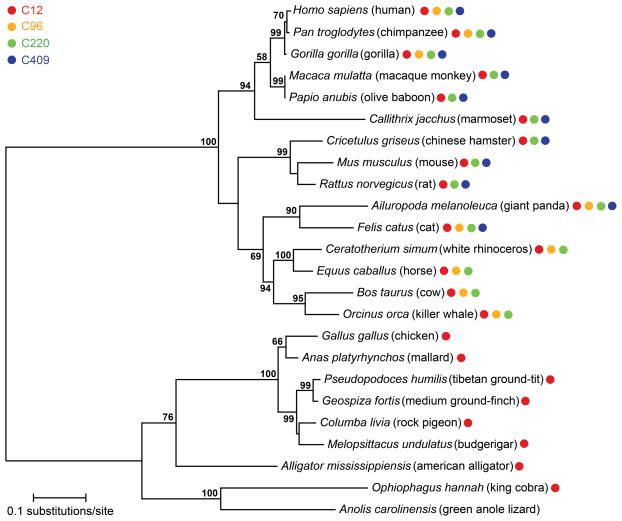
Finally, we mapped the relative solvent accessibility of the cysteine residues for each RI homolog (Fig. 7c). We determined that there were four cysteines with the highest overall solvent-exposed surface area: Cys12, Cys96, Cys220, and Cys409. Of these four cysteines, human RI contains all four, mouse RI contains three, bovine RI contains three, chicken RI contains one, and anole RI contains zero (Fig. 4 and 6c). We expanded our analysis to include 15 mammalian and 9 non-mammalian RI homologs. We determined that although all mammalian RI homologs possessed at least three highly solvated cysteine residues, non-mammalian RI homologs only contained one or none (Fig. 8).
Fig. 8.
Evolution of solvated cysteine residues in homologous RIs. A phylogenetic tree depicts the evolutionary relationship among homologous RIs, and colored circles indicate the presence of each of the four most highly solvated cysteine residues (Fig. 7c). Bootstrap values >50 are shown.
Discussion
Secretory ribonucleases have been characterized from every class of vertebrate [33,34]. Typically, these proteins have high, non-specific activity against RNA substrates, circulate freely in extracellular fluids, and can enter cells spontaneously [35,36]. A potent, cytosolic inhibitor for such RNases is critical. Indeed, mammalian ribonuclease inhibitor (RI) was discovered and characterized over 50 years ago [37,38]. Still, multiple early studies proclaimed the total absence of RI in avian and reptilian tissues (for reviews, see refs. [29,39]). Our data nullify this proclamation, as we have identified RIs from both chicken and anole lizard. We find many similarities between these proteins and their more characterized mammalian counterparts, along with key differences.
Importantly, we determined that non-mammalian ribonuclease inhibitors bind their cognate ribonucleases with tight affinity, similar to that of mammalian inhibitors. This observation implies that a critical role for non-mammalian RIs—like mammalian RIs— is to regulate the biological activity of secretory ribonucleases. Further evidence for this hypothesis is the apparent co-evolution of RIs from different species to bind to their endogenous RNases. We find that proteins bind as tightly or tighter to their cognate partner than to any inter-species partner (regardless of pI or overall charge), suggesting the presence of subtle changes in the binding interface to promote better molecular recognition.
Our observation that avian and reptilian RI binds ~108-fold more weakly to mammalian RNases (and vice versa) has other implications. These data explain the previous difficulties in detecting and purifying non-mammalian RIs, which do not bind tightly to the bovine RNase used in early detection assays and affinity chromatography. Whereas nanomolar binding affinities are seemingly tight, in the RI·RNase system they are not especially relevant. For example, mammalian RNases engineered to evade mammalian RI possess nanomolar affinity for RI but are highly toxic to human cells[11]. For many of these cytotoxic variants, substituting a single interface residue results in enormous decreases in affinity for RI [12,40].
Accounting for the specific changes that have led to the diversity between species, as well as demonstrating co-evolution between intraspecies binding partners, is difficult. The similarity of the various RI·RNase binding interfaces suggests that the changes driving the divergent binding are subtle. This notion corresponds well with the hypothesis of interface “hot spots”, or the small subset of residues that are predicted to account for most of the binding affinity between two proteins [31,41]. Tellingly, detailed dissection of the binding interface between human RI and human angiogenin revealed that, although the binding affinity relied upon relatively few key contacts, multiple residues function cooperatively, suggesting a complicated landscape and highlighting the difficultly of assigning the sources of binding energy rigorously [42]. Still, as difficult as they are to study, co-evolutionary changes in protein–protein interactions do occur, and are an important driver of speciation [43,44].
Surprisingly, we were unable to detect binding between fish or frog RNase and any of the RI molecules in our study. An exhaustive search of amphibian and fish genomes did not yield any viable RI-homolog candidates. RI could be quite divergent in these classes. Fish and frog RNases share a low level of sequence identity and similarity to other secretory RNases (Table S4). Early studies in bullfrogs indicated the presence of a cytosolic protein that could inhibit the activity of bullfrog RNase (but not bovine RNase), and was sensitive to thiol-reactive agents. The estimated size of the complex between this molecule and RNase was, however, ~130–140 kDa, which is much larger than the ~65 kDa size noted for mammalian RI·RNase complexes [37,45,46]. This dissimilarity could reflect intrinsic differences in the amphibian RI homolog, such as in molecular mass or binding stoichiometry. Methods such as affinity chromatography using frog or fish RNase could be necessary to identify these more divergent RI homologs.
The apparent evolving oxidation-sensitivity of mammalian RI homologs implies the emergence of new functionality. Indeed, RI has been identified as a potential keystone in the maintenance of cellular redox homeostasis [18,22]. The ability of oxidized RI to release functional, active ribonuclease is particularly fascinating. Potentially, the intracellular redox state could serve as a trigger to release a caged ribonuclease. Previous studies have shown that partially oxidized RI can allow partial RNase activity [47]. Thus, cells might have a “redox switch” that regulates RNases. Under oxidative stress, the manifestation of ribonucleolytic activity could induce apoptosis. This hypothesis has important implications, given the well-characterized association of oxidative stress with ageing, cancer, and other diseases.
In conclusion, we have confirmed the existence of avian and reptile homologs of RI that display characteristics unique from mammalian homologs. Our discovery that non-mammalian RIs exhibit extremely tight binding to their endogenous RNases but remarkably lower sensitivity to oxidation suggests that the primary role of non-mammalian RI is to regulate the biological activities of secretory ribonucleases. Intriguingly, these data also imply that mammalian RIs have not only retained and even improved upon their avid RNase binding, but also evolved greater sensitivity to oxidation. This redox reactivity might be driving new biological roles—such as scavenging intracellular free radicals—or might be adding complexity to existing roles, such as triggering the release of active RNases as a cellular stress-response mechanism. Further in vivo characterizations are necessary to continue probing the dynamic biology of RI.
Materials and Methods
Materials and Instrumentation
E. coli BL21(DE3) cells and the plasmid pET22b(+) were from EMD Millipore. 6-FAM–dArU(dA)2–6-TAMRA, a fluorogenic ribonuclease substrate, as well as DNA oligonucleotides for PCR, sequencing, and mutagenesis were from Integrated DNA Technologies. Protein purification columns were from GE Healthcare. Costar 96-well NBS microtiter plates were from Corning Life Sciences. Restriction and PCR enzymes were from Promega. All other chemicals were of commercial grade or better and were used without further purification.
The molecular mass of each RI and ribonuclease was determined by matrix-assisted laser desorption/ionization-time-of-flight (MALDI–TOF) mass spectrometry using a Voyager-DE-PRO Biospectrometry Workstation (Applied Biosystems). MALDI–TOF mass spectrometry experiments were performed at the campus Biophysics Instrumentation Facility. All fluorescence and absorbance measurements were made using a Tecan M1000 fluorimeter plate reader, unless stated otherwise. All data were fit and analyzed using the graphing software package Prism 5 (GraphPad), unless stated otherwise.
RI cDNA cloning and protein purification
Human RI[5] and bovine RI[30] constructs were inserted previously into the pET22b expression vector for tagless expression in BL21(DE3) E. coli. The gene encoding mouse RI (Gene ID: 107702) was amplified from Musmusculus liver cDNA and inserted in the pET22b vector. The sequences of chicken RI (Gene ID: 423111) and anole RI (Gene ID:100553617) were identified by their hypothetical annotation in the GenBank database. The genes encoding chicken RI and anole RI were amplified from Gallus gallus liver cDNA and Anolis carolinensis liver cDNA (Reptile Rapture, Madison, WI), respectively, and inserted into pET22b with an N-terminal, protease-cleavable 6× His tag. All primers used for cloning are listed in Table S1.
Human, bovine, and mouse RIs were purified via RNase A–affinity chromatography and ion–exchange chromatography as described previously [5,30]. Chicken RI and anole RI were produced as described previously [30], with the following modifications. In lieu of RNase A–affinity chromatography, chicken and anole RIs were purified over a nickel column and eluted over a linear gradient of imidazole. They were then purified again over an anion–exchange column to yield nearly pure protein. The N-terminal 6× His purification tag was cleaved by incubation with TEV (tobacco etch virus) protease [48], yielding native RI proteins with a single N-terminal glycine residue. Molecular masses of RI proteins were confirmed by MALDI–TOF mass spectrometry. Protein concentration was determined by using a Bradford assay kit (Pierce) with bovine serum albumin as a standard.
Ribonuclease cDNA cloning and protein purification
Human RNase 1[30], bovine RNase A[30] and frog RNase (Ranapipiens)[49] constructs were inserted previously into the pET22b expression vector for tagless expression in BL21(DE3) E. coli. The gene encoding mouse RNase 1 (Gene ID: 19752) was amplified from M.musculus pancreas cDNA and inserted into pET22b. The gene encoding chicken RNase A-1[50] (Gene ID: 396194) was amplified from G.gallus liver cDNA and inserted into pET22b. The novel anole RNase used in this study, referred to as “anole RNase 1”, was identified by BLAST analysis using human RNase 1 as an input. This RNase was the most evolutionarily similar to human RNase 1 from all returned BLAST hits, as determined by phylogram analysis (data not shown), and possessed the identifier “LOC100555482 ribonuclease-like”. The gene encoding anole RNase 1 (Gene ID: 100555482) was amplified from Anoliscarolinensis liver cDNA and inserted into pET22b. The gene encoding zebrafish RNase 3/4[51,52] (Gene ID: 100101462) was amplified from Danio rerio cDNA and inserted into pET22b. The program Signal P was used to predict and exclude peptide leader sequences for all proteins. All primers used for cloning are listed in Table S1.
To enable site-specific fluorescent labeling of ribonucleases, we introduced cysteine residues by site-directed mutagenesis into loop regions that are distal to both the enzymic active site and the RI-binding interface. The ensuing variants were P19C human RNase 1, S19C mouse RNase 1, A19C bovine RNase 1, T17C chicken RNase A-1, S20C anole RNase 1, A14C zebrafish RNase 3/4, and S61C frog RNase. RNases were purified from inclusion bodies by using cation-exchange chromatography, and free-cysteine variants were labeled with diethylfluorescein (DEFIA)[53] as described previously [30,54,55]. Molecular masses of RNase conjugates were confirmed by MALDI–TOF mass spectrometry. Protein concentration was determined by using a bicinchoninic acid (BCA) assay kit (Pierce) with wild-type RNase A as the standard.
Dissociation rate of RI·RNase complexes
For the tightest-binding RI·RNase complexes (Kd ≤ 10–15 M), the dissociation rate constant (kd)was determined by following the release of diethylfluorescein (DEFIA)-labeled ribonuclease from the RI·RNase complex over time, as described previously [54]. Briefly, RI and a DEFIA-labeled RNase were mixed in equimolar ratios, and the resulting solution was incubated at 25°C for 5 min. A 50-fold molar excess of human RNase 1 was added to scavenge dissociated RI. Complex dissociation was measured by monitoring the increasing fluorescence of dissociated RNase over time (≥60 days). A value of Kd for each complex was determined as described previously [54]. These values represent the mean from at least three independent experiments.
For weaker-binding complexes (Kd ≥ 10–9 M), an RI-saturation binding assay was used, as described previously [56]. Briefly, fluorescence spectroscopy was used to monitor the binding of an RI to a DEFIA-labeled ribonuclease, availing the decrease in fluorescence upon binding to RI. Data were normalized to unbound DEFIA-RNase and fitted with nonlinear regression analysis to obtain a value of Kd for each complex. These values are the mean from at least three independent experiments.
Determination of Tm values
Thermal unfolding of RIs (unbound and bound to an RNase) was monitored in the presence of a fluorescent dye by using differential scanning fluorimetry (DSF). DSF was performed using a Vii A 7 Real-Time PCR machine (Applied Biosystems) as previously described[57,58]. Briefly, a solution of protein (10 μg) was placed in the wells of a MicroAmp optical 96-well plate, and SYPRO Orange dye (Sigma Chemical) was added to a final dilution of 1:333 in relation to the stock solution of the manufacturer. The temperature was increased from 20 to 96°C at 1°C/min in steps of 1°C. Fluorescence intensity was measured at 578 nm, and the resulting data were analyzed with Protein Thermal Shift software (Applied Biosystems). A solution with no protein was used for background correction. Values of Tm were calculated from curves of ∂fluorescence/∂T and are the mean from three independent experiments.
Purification of RI·RNase complexes
Mouse, bovine, and chicken RI·RNase complexes were purified for crystallization as described previously [5]. Briefly, purified RNase (~50 mg/ml) and RI (~10 mg/ml) were mixed at a 1.2:1.0 molar ratio, and this solution was incubated at 25 °C for 20 min to allow for complex formation. The solution was then purified using anion-exchange chromatography to remove any unbound RNase. Purified complex was dialyzed for 16 h at 4 °C against 20 mM Hepes–NaOH buffer (pH 7.5) containing DTT (10 mM) and glycerol (2% v/v), and was concentrated to ~10 mg/ml. Aliquots were flash frozen and stored at −80 °C.
Crystallization of RI·RNase complexes
All RI·RNase complexes were screened for initial crystallization conditions using a Mosquito nanoliter liquid handling robot (TTP LabTech), and the resulting crystals were optimized using the hanging-drop vapor diffusion method at 20 °C. Crystals for bovine RI·RNase were observed in the PACT premier HT screen (Molecular Dimensions) and grew to maximum size within a week[59]. Optimized bovine RI·RNase crystals that were used for structure determination were grown by mixing 1 μL of protein solution with 1 μL of 25% w/v polyethylene glycol (PEG) 1500 and 100mM malic acid/MES/Tris (MMT) buffer(pH 4.0). Initial chicken RI·RNase crystals were observed in the PEGRx HT screen (Hampton Research) and grew to maximum size within 24 h.
Crystals used to determine the chicken RI·RNase structure were grown by mixing 1 μL of protein solution with 1 μL of 21% w/v PEG 1500 and 100mM sodium citrate buffer (pH 3.5). Mouse RI·RNase crystals were observed in the Index HT screen (Hampton research) and were optimized further. The crystals that were used to determine the mouse RI·RNase structure were grown by mixing 1 μL of protein solution with 1 μL of 25% w/v PEG 3350 and 100 mM sodium citrate buffer (pH 3.5). All RI·RNase crystals were frozen directly in liquid N2 before data collection. The bovine, chicken, and mouse crystals were cryoprotected by the addition of ethylene glycol to 15%, 15%, and 20% v/v, respectively, to the solutions described above.
Structure Determination of RI·RNase complexes
Diffraction images for bovine RI·RNase, chicken RI·RNase, and mouse RI·RNase were collected at the Life Sciences Collaborative Access Team 21-ID-G, 21-ID-G, and 21-ID-D beamlines, respectively, at the Advanced Photon Source, Argonne National Laboratory. All the RI·RNase structures presented here were solved by molecular replacement with Phaser[60]using PDB entries 1dfj [61], 1z7x [5], and 1z7x as a starting model for bovine, chicken, and mouse, respectively. All RI·RNase structures were completed with altering rounds of model building in Coot[62] and refinement in Phenix[63]. Model quality was assessed with Molprobity[64] before deposition to the PDB. Structural images were generated with PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). Data collection, refinement, and model statistics are presented in Table 3. All structures used in this study were analyzed with the program PDBsum[65] to identify intermolecular hydrogen bonds and van der Waals contacts. PDBsum uses the algorithm HBPLUS[66] to identify hydrogen bonds (rX···X<3.3 Å). Structures were also analyzed using the Knowledge-based FADE and Contacts (KFC2) server [32,67].
Table 3.
Summary of crystal parameters, data collection, and refinement statistics. Values in parentheses are for the highest-resolution shell.
| Mouse RI·Mouse RNase | Bovine RI·Bovine RNase | Chicken RI·Chicken RNase | |
|---|---|---|---|
| Crystal parameters | |||
| Space group | P21 | I222 | P212121 |
|
a = 72.40 b = 125.34 |
a = 117.79 b = 123.55 |
a = 52.66 b = 84.54 |
|
| Unit-cell parameters (Å) | |||
|
c = 123.06 β = 94.72° |
c = 179.30 | c = 121.66 | |
|
| |||
| Data collection statistics | |||
| Wavelength (Å) | 0.9794 | 0.97857 | 0.97857 |
| Resolution range (Å) | 50.00–2.20 (2.25–2.20) | 50.00–2.21 (2.25–2.21) | 50.00–1.82 (1.85–1.82) |
| Completeness (%) | 97.9 (88.2) | 100.0 (99.4) | 99.4 (99.8) |
| Rmerge* | 0.145 (0.478) | 0.084 (0.747) | 0.134 (0.687) |
| Redundancy | 4.2 (2.6) | 7.2 (5.6) | 4.0 (3.7) |
| Mean I / sigma (I) | 9.9 (2.1) | 22.72 (2.31) | 9.16 (1.69) |
|
| |||
| Refinement and model statistics | |||
| Resolution range (Å) | 34.38–2.20 | 49.24–2.21 | 39.83–1.92 |
| No. of reflections (work / test) | 102210 / 1897 | 57584 / 1930 | 39152 / 2064 |
| Rcryst§ | 0.183 (0.234) | 0.176 (0.218) | 0.207 (0.234) |
| Rfree¶ | 0.233 (0.338) | 0.226 (0.298) | 0.254 (0.263) |
| RMSD bonds (Å) | 0.003 | 0.008 | 0.009 |
| RMSD angles (°) | 0.679 | 1.192 | 1.158 |
| Average B-factor (Å2) | 25.5 | 20.5 | 30.7 |
| No. of protein atoms | 17650 | 8698 | 4404 |
| No. of waters | 882 | 562 | 233 |
|
| |||
| Ramachandran plot (%) | |||
| Favorable | 97.20 | 97.65 | 96.83 |
| Allowed | 2.80 | 2.35 | 3.17 |
| Disallowed | 0.00 | 0.00 | 0.00 |
| PDB ID | |||
| 3tsr | 4peq | 4per | |
Rmerge= Σh Σi | Ii(h) − <I(h)>| / ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
Rcryst=Σh||Fobs| − |Fcalc|| / Σh |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using randomly selected unique reflections that were omitted from the structure refinement.
Oxidation sensitivity of RI·RNase complexes
The sensitivity of RI·RNase complexes to oxidation by hydrogen peroxide (H2O2) was assessed by following the release of DEFIA-labeled ribonuclease upon RI dissociation, as previously described [30]. Briefly, fresh H2O2 (30% v/v, Fisher Scientific) was diluted serially in reaction buffer (20 mM HEPES–HCl buffer, pH 7.0, containing 50 mMKCl) to produce a final range of 30–0.001% v/v H2O2. Desalted RI (100 nM) and ribonuclease (100 nM) were combined in 50 μL of reaction buffer across a 96-well plate and incubated for 20 min at 25 °C to allow for complex formation. Initial fluorescent readings were taken, and 50 μL of H2O2 serial dilutions was added to each well containing the RI·RNase complex. Plates were incubated at 37 °C for 1 h, and final fluorescent readings were taken. Data were normalized to control wells containing only labeled RNase at each H2O2 concentration and fitted using nonlinear regression to generate H2O2 IC50 values for complex dissociation. Values represent the mean from at least three independent experiments.
The release of active ribonuclease from the RI·RNase complex in response to H2O2 treatment was measured by assessing the ability of ribonucleases to cleave a fluorogenic RNA substrate, as described previously [68]. Briefly, RIs and RNases were incubated in equimolar ratios (50 nM for human, mouse, bovine, and chicken; 500 nM for anole) in 50 μL of reaction buffer and allowed to form RI·RNase complexes. Initial fluorescent readings were recorded, and 50 μL of H2O2 serial dilutions (see above) was added to each well containing a RI·RNase complex. Plates were incubated at 37 °C for 1 h, and final fluorescent readings were recorded. Data were normalized to control wells containing only labeled RNase at each H2O2 concentration and fitted with nonlinear regression analysis to generate values of IC50 for complex dissociation. These values represent the mean from at least three independent experiments.
Quantification of RI thiol groups and cysteine solvent-exposed surface area
Accessible protein sulfhydryl groups were quantified by UV spectroscopy using Ellman’s Reagent (Pierce) according to the manufacturer’s protocol. Briefly, 10μM RI was eluted from PD-10 columns (GE Healthcare) to remove all traces of reducing agents. A 250-μL aliquot of desalted RI (10 μM) was added to 2.5 mL of reaction buffer (0.10 M sodium phosphate buffer, pH 8.0, containing 1 mM ethylenediaminetetraacetic acid) and 50 μL of Ellman’s Reagent solution (4mg/mL in reaction buffer). The resulting solutions were incubated for 15 min at 25 °C, and their absorbance was recorded at 412 nm and converted to absolute values using N-acetylcysteine as the standard (0–0.1 mM). Samples were analyzed in triplicate and values represent the mean from three independent experiments.
The solvent-accessible surface area of cysteine residues in RI crystal structures was calculated with PyMOL[69].
Construction of a RI phylogenetic tree
Annotated RI protein sequences were obtained from the National Center for Biotechnology Information (NCBI) database. Only 100% complete sequences were used for analysis. RI protein sequence alignments were made using MUSCLE[70] with manual adjustments. A maximum–likelihood phylogenetic tree was generated in MEGA5.2[71] using the Jones–Taylor–Thornton (JTT)[72] substitution model and 1000 bootstrap replicates. Non-uniformity of evolutionary rates was modeled using a discrete Gamma distribution [73], assuming for the presence of invariable sites. Bootstrap values >50 are reported.
Supplementary Material
Acknowledgments
We are grateful to University of Wisconsin–Madison Professor Mark E. Cook for G. gallus tissue, Professor Yevgenya Grinblat for D. rerio tissue, and Dr. Craig A. Bingman for expertise on X-ray diffraction analysis. J.E.L. was supported by a National Science Foundation Graduate Research Fellowship. C.M.B. was supported by grant DE-FC02-07ER64494 (Department of Energy). A.C. and G.N.P. were supported by grant U01 GM098248 [National Institutes of Health (NIH)]. This work was supported by grant R01 CA073808 (NIH). Access to crystallization equipment was provided by the Center for Eukaryotic Structural Genomics, which was supported by grants U54 GM074901 and P50 GM064598 (NIH). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-ENG-38. The use of Life Science Collaborative Access Team at the Advanced Photon Source was supported by the College of Agricultural and Life Sciences, Department of Biochemistry, and Graduate School of the University of Wisconsin–Madison.
Abbreviations
- LRR
leucine-rich repeat
- RI
ribonuclease inhibitor
- RNase
ribonuclease
- ROS
reactive oxygen species
- MALDI–TOF
matrix-assisted laser desorption/ionization–time-of-flight
- DSF
differential scanning fluorimetry
- PEG
polyethylene glycol
- NIH
National Institutes of Health
Footnotes
Accession Numbers
Structures were deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under the following codes: mouse RI·mouse RNase complex, 3tsr; bovine RI·bovine RNase complex, 4peq; chicken RI·chicken RNase complex, 4per.
GenBank accession numbers for proteins used for phylogenetic analysis are listed in Table S5.
References
- 1.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: Structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–74. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–27. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 3.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–6. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 4.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996;264:1028–43. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007;367:434–49. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papageorgiou AC, Shapiro R, Acharya KR. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 Å resolution. EMBO J. 1997;16:5162–77. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karstens T, Kobe K. Rhodamine B and Rhodamine 101 as reference substances for fluorescence quantum yield measurements. J Phys Chem. 1980;84:1871–2. [Google Scholar]
- 8.Blázquez M, Fominaya JM, Hofsteenge J. Oxidation of sulfhydryl groups of ribonuclease inhibitor in epithelial cells is sufficient for its intracellular degradation. J Biol Chem. 1996;271:18638–42. doi: 10.1074/jbc.271.31.18638. [DOI] [PubMed] [Google Scholar]
- 9.Kim B-M, Schultz LW, Raines RT. Variants of ribonuclease inhibitor that resist oxidation. Protein Sci. 1999;8:430–4. doi: 10.1110/ps.8.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fominaya JM, Hofsteenge J. Inactivation of ribonuclease inhibitor by thiol–disulfide exchange. J Biol Chem. 1992;267:24655–60. [PubMed] [Google Scholar]
- 11.Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. 2008;9:185–9. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–32. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson KA, Kang DK, Kwon YS, Kim JC, Leland PA, Kim BM, et al. Ribonuclease inhibitor regulates neovascularization by human angiogenin. Biochemistry. 2009;48:3804–6. doi: 10.1021/bi9005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pizzo E, Sarcinelli C, Sheng J, Fusco S, Formiggini F, Netti P, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126:4308–19. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer JD, Schwaderer AL, Eichler T, Wang H, Kline J, Justice SS, et al. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int. 2013;85:1179–91. doi: 10.1038/ki.2013.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Li H. Radical scavenging activity of ribonuclease inhibitor from cow placenta. Biochemistry (Mosc) 2006;71:520–4. doi: 10.1134/s0006297906050087. [DOI] [PubMed] [Google Scholar]
- 18.Monti DM, Montesano Gesualdi N, Matoušek J, Esposito F, D’Alessio G. The cytosolic ribonuclease inhibitor contributes to intracellular redox homeostasis. FEBS Lett. 2007;581:930–4. doi: 10.1016/j.febslet.2007.01.072. [DOI] [PubMed] [Google Scholar]
- 19.Furia A, Moscato M, Cali G, Pizzo E, Confalone E, Amoroso MR, et al. The ribonuclease/angiogenin inhibitor is also present in mitochondria and nuclei. FEBS Lett. 2011;585:613–7. doi: 10.1016/j.febslet.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 21.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med. 2010;49:1603–16. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui XY, Fu PF, Pan DN, Zhao Y, Zhao J, Zhao BC. The antioxidant effects of ribonuclease inhibitor. Free Radic Res. 2003;37:1079–85. doi: 10.1080/10715760310001600408. [DOI] [PubMed] [Google Scholar]
- 23.Moreno ML, Escobar J, Izquierdo-Alvarez A, Gil A, Perez S, Pereda J, et al. Disulfide stress: A novel type of oxidative stress in acute pancreatitis. Free Radic Biol Med. 2014 doi: 10.1016/j.freeradbiomed.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Das K, Wu J, Lee MH, Tan P. RNH1 regulation of reactive oxygen species contributes to histone deacetylase inhibitor resistance in gastric cancer cells. Oncogene. 2014;33:1527–1537. doi: 10.1038/onc.2013.104. [DOI] [PubMed] [Google Scholar]
- 25.Moenner M, Vosoghi M, Ryazantsev S, Glitz D. Ribonuclease inhibitor protein of human erythrocytes: Characterization, loss of activity in response to oxidative stress, and association with Heinz bodies. Blood Cells Mol Dis. 1998;24:149–64. doi: 10.1006/bcmd.1998.0182. [DOI] [PubMed] [Google Scholar]
- 26.Nadano D, Yasuda T, Takeshita H, Kishi K. Ribonuclease inhibitors in human blood: Comparative studies on the inhibitors detected in erythrocytes, platelets, mononuclear leukocytes and granulocytes. Int J Biochem Cell Biol. 1995;27:971–9. doi: 10.1016/1357-2725(95)00063-u. [DOI] [PubMed] [Google Scholar]
- 27.Dyer KD, Rosenberg HF. The RNase A superfamily: Generation of diversity and innate host defense. Mol Divers. 2006;10:585–97. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 28.Beintema JJ, Breukelman HJ, Carsana A, Furia A. Evolution of vertebrate ribonucleases: Ribonuclease A superfamily. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. New York: Academic Press; 1997. pp. 245–69. [Google Scholar]
- 29.Hofsteenge J. Ribonuclease inhibitor. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. New York: Academic Press; 1997. pp. 621–58. [Google Scholar]
- 30.Johnson RJ, Lavis LD, Raines RT. Intraspecies regulation of ribonucleolytic activity. Biochemistry. 2007;46:13131–40. doi: 10.1021/bi701521q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–12. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Mitchell JC. KFC2: A knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins. 2011;79:2671–83. doi: 10.1002/prot.23094. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg HF. RNase A ribonucleases and host defense: An evolving story. J Leukoc Biol. 2008;83:1079–87. doi: 10.1189/jlb.1107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: Identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–20. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J Cell Sci. 2003;116:313–24. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao T-Y, Raines RT. Mechanism of ribonuclease A endocytosis: Analogies to cell-penetrating peptides. Biochemistry. 2011;50:8374–82. doi: 10.1021/bi2009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth JS. Ribonuclease. IX. Further studies on ribonuclease inhibitor. Biochim Biophys Acta. 1962;61:903–15. doi: 10.1016/0926-6550(62)90007-5. [DOI] [PubMed] [Google Scholar]
- 38.Girija NS, Sreenivasan A. Characterization of ribonucleases and ribonuclease inhibitor in subcellular fractions from rat adrenals. Biochem J. 1966;98:562–6. doi: 10.1042/bj0980562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee FS, Vallee BL. Structure and action of mammalian ribonuclease (angiogenin) inhibitor. Prog Nucleic Acid Res Mol Biol. 1993;44:1–30. doi: 10.1016/s0079-6603(08)60215-9. [DOI] [PubMed] [Google Scholar]
- 40.Leland PA, Schultz LW, Kim B-M, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;98:10407–12. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Sawyer N, Regan L. Protein–protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–5. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro R, Ruiz-Gutierrez M, Chen C-Z. Analysis of the interactions of human ribonuclease inhibitor with angiogenin and ribonuclease A by mutagenesis: Importance of inhibitor residues inside versus outside the C-terminal “Hot Spot”. J Mol Biol. 2000;302:497–519. doi: 10.1006/jmbi.2000.4075. [DOI] [PubMed] [Google Scholar]
- 43.Sandler I, Abu-Qarn M, Aharoni A. Protein co-evolution: How do we combine bioinformatics and experimental approaches? Mol Biosyst. 2013;9:175–81. doi: 10.1039/c2mb25317h. [DOI] [PubMed] [Google Scholar]
- 44.Lovell SC, Robertson DL. An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol. 2010;27:2567–75. doi: 10.1093/molbev/msq144. [DOI] [PubMed] [Google Scholar]
- 45.Nagano H, Kiuchi H, Abe Y, Shukuya R. Purification and properties of an alkaline ribonuclease from the hepatic cytosol fraction of bullfrog, Rana catesbeiana. J Biochem. 1976;80:19–26. doi: 10.1093/oxfordjournals.jbchem.a131251. [DOI] [PubMed] [Google Scholar]
- 46.Malicka-Blaszkiewicz M, Kubicz A. The RNAse–RNAse inhibitor system in the liver of the frog Rana esculenta: Subcellular distribution and differential binding of inhibitor with multiple RNAses. Acta Physiol Pol. 1981;32:317–26. [PubMed] [Google Scholar]
- 47.Ferreras M, Gavilanes JG, Lopéz-Otín C, García-Segura JM. Thiol–disulfide exchange of ribonuclease inhibitor bound to ribonuclease A. J Biol Chem. 1995;270:28570–8. doi: 10.1074/jbc.270.48.28570. [DOI] [PubMed] [Google Scholar]
- 48.Miladi B, Bouallagui H, Dridi C, El Marjou A, Boeuf G, Di Martino P, et al. A new tagged-TEV protease: Construction, optimisation of production, purification and test activity. Protein Expr Purif. 2011;75:75–82. doi: 10.1016/j.pep.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Chao T-Y, Lavis LD, Raines RT. Cellular uptake of ribonuclease A relies on anionic glycans. Biochemistry. 2010;49:10666–73. doi: 10.1021/bi1013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nitto T, Dyer KD, Czapiga M, Rosenberg HF. Evolution and function of leukocyte RNase A ribonucleases of the avian species, Gallus gallus. J Biol Chem. 2006;281:25622–34. doi: 10.1074/jbc.M604313200. [DOI] [PubMed] [Google Scholar]
- 51.Cho S, Zhang J. Zebrafish ribonucleases are bactericidal: Implications for the origin of the vertebrate RNase A superfamily. Mol Biol Evol. 2007;24:1259–68. doi: 10.1093/molbev/msm047. [DOI] [PubMed] [Google Scholar]
- 52.Pizzo E, Merlino A, Turano M, Russo Krauss I, Coscia F, Zanfardino A, et al. A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish. Biochem J. 2011;433:345–55. doi: 10.1042/BJ20100892. [DOI] [PubMed] [Google Scholar]
- 53.Lavis LD, Rutkoski TJ, Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal Chem. 2007;79:6775–82. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lomax JE, Eller CH, Raines RT. Rational design and evaluation of mammalian ribonuclease cytotoxins. Methods Enzymol. 2012;502:273–90. doi: 10.1016/B978-0-12-416039-2.00014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundlass NK, Eller CH, Cui Q, Raines RT. Contribution of electrostatics to the binding of pancreatic-type ribonucleases to membranes. Biochemistry. 2013;52:6304–12. doi: 10.1021/bi400619m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal Biochem. 2002;306:100–7. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 57.Menzen T, Friess W. High-throughput melting-temperature analysis of a monoclonal antibody by differential scanning fluorimetry in the presence of surfactants. J Pharm Sci. 2013;102:415–28. doi: 10.1002/jps.23405. [DOI] [PubMed] [Google Scholar]
- 58.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 59.Newman J, Egan D, Walter TS, Meged R, Berry I, Ben Jelloul M, et al. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: The PACT/JCSG+ strategy. Acta Crystallogr D Biol Crystallogr. 2005;61:1426–31. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 60.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–6. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laskowski RA. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–2. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–93. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 67.Darnell SJ, LeGault L, Mitchell JC. KFC Server: Interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:W265–9. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–6. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 70.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 73.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 74.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.