Abstract
The skin, the human body’s largest organ, is home to a diverse and complex variety of innate and adaptive immune functions. Despite this potent immune system present at the cutaneous barrier, the skin encourages colonization by microorganisms. Characterization these microbial communities has enhanced our knowledge of the ecology of organisms present in normal skin; furthermore, studies have begun to bring to light the intimate relationships shared between host and resident microbes. In particular, it is apparent that just as host immunological factors and behaviors shape the composition of these communities, microbes present on the skin greatly impact the functions of human immunity. Thus, today the skin immune system should be considered a collective mixture of elements from the host and microbes acting in a mutualistic relationship. In this article we will review recent findings of the interactions of skin microbial communities with host immunity, and discuss the role that dysbiosis of these communities plays in diseases of the skin.
Keywords: Keratinocyte, Staphylococcus, Propionibacterium acnes, Antimicrobial peptides, Dysbiosis
1. Introduction
The skin is our most exposed organ, responsible for providing a barrier to the external environment that can resist a wide range of challenges and respond appropriately to penetrating dangers. However, despite a potent cutaneous immune system, many different microbial communities thrive on the surface. The challenge for the skin’s immune system is that it is charged with resisting infections, but must do so under normal conditions in the absence of cell recruitment and inflammation. Failure to properly control and tolerate resident microbes results in skin disease. More recently, it has been hypothesized that the skin commensal microbial communities not only co-exist despite our immune defense network but actually modify immunity, therefore influencing normal skin health as well as participating in various dermatological conditions [1]. The microbiota of the skin have therefore become the subject of much recent interest from the perspective of better understanding cutaneous disease and as a source for developing novel therapies for skin disease.
Historically, detection and characterization of the skin microbes depended on their cultivation from swabs of the skin surface [2]. With the advent of DNA-based technologies for the detection and identification of microbial genes, it is now clear that the culturable microbes represent only a small fraction of the total organisms that interact at the surface. DNA sequencing techniques have sought to describe the diversity of microbes residing on and within our bodies, and as a shorthand for describing the ecology of the human body as a “biome,” the microbial communities inhabiting us have collectively been called the “human microbiome.” This field received a great boost in 2007 when the National Institutes of Health initiated the Human Microbiome Project (HMP) with the intent of surveying and characterizing the microbes that reside at different body sites [3]. The seminal HMP analysis of microbes from 18 body sites in over 240 healthy volunteers, completed in 2012, has begun to reveal the complex nature of the human microbial inhabitants and the incredible amount of both intra- and interpersonal variation in the communities residing throughout our bodies [4]. Today, with the groundwork prepared by descriptive studies, we are now poised to uncover the intimate relationships that microbes share with their hosts and the influences they have on human health.
In this article, we will review recent findings of the role skin microbial communities play in host immunity, as well as explore the topic of dysbiosis as a participant in various pathologies of the skin. One finding made clear through the Human Micro-biome Project is that the communities residing at different body sites are not at all uniform – the gastrointestinal, oral, nasal, vaginal, and skin-associated microbes vary greatly in their compositions [4]. Adding to this complexity is the observation that different areas of the skin, which vary in physiologic aspects such as moisture, oiliness, and exposure to the external environment, harbor distinct groups of microbes. While this review will briefly address the nature of this diversity, we point the reader to several excellent publications for a more comprehensive characterization of the communities residing across the surfaces of our skin [5–9]. Additionally, thorough discussions of the technologies currently used to analyze microbiome composition and how these methods have revolutionized the way the human microbiome is viewed can be found elsewhere [10,11]. Our goal in this review is to begin to show how our existing information can be translated into a deeper functional understanding of how these may act to influence health.
2. The physical and cellular immune barrier of the skin against microbes
To understand the ecology of the skin surface and the factors influencing the skin microbiome it is first necessary to understand the elements contributing to the cutaneous environment. The skin barrier is most frequently thought of in terms of the outermost layer of the epidermis. For the purpose of this discussion of skin microbial communities, the epidermis will be discussed in greatest detail. However, it is important to recognize that the skin barrier consists of several layers below the epidermis that profoundly affect function and also harbor microbes [12]. Additionally, it is important to recognize that an aqueous and lipid layer exists above the epidermis, also contributing to the ecology of the surface. Combined, all layers of the skin must prevent infection and the entry of harmful substances while controlling the loss of water and nutrients. At the forefront of this process to maintain homeostasis is the highly keratinized epidermis, the result of a specialized differentiation process of keratinocytes, the main cell type in the epidermal barrier [13].
2.1. Epidermal structure and composition control the microbiome
The surface of the skin is formed by a network of cross-linked cornified cell envelopes and specialized lipid molecules creating the “bricks and mortar” structure of the epidermis [14–16]. The ultrastructure of the skin surface is also riddled with invaginations, including sweat glands, hair follicles, and sebaceous glands. Two types of sweat glands exist: eccrine and apocrine. Eccrine sweat glands, which are distributed across nearly the entire skin surface, play a crucial role in thermoregulation. They secrete sweat that is almost entirely water directly onto the skin surface, the evaporation of which allows the body to cool [17]. Eccrine sweat also contains salt and electrolytes which work to acidify the skin. On the whole, the result of this process is a barrier that is cool, dry, and slightly acidic. This environment plays a major role in limiting the composition of microbes that can survive and proliferate. Furthermore, eccrine sweat glands constitutively express several antimicrobial peptides (AMPs), including cathelicidin and β-defensins [18–20]. Thus, the density of eccrine sweat glands impacts microbial colonization of the skin. Apocrine sweat glands, which exist at birth but do not become active until puberty, have a more limited distribution, found primarily in sites such as the axilla, genitalia and perianal regions. These glands secrete their contents – an oily, odorless mixture of proteins, lipids, and steroids – into the hair canal [17]. It is the degradation of these apocrine-derived compounds by resident bacteria that produces the characteristic odor of sweat [21,22]. Sebaceous glands are connected to hair follicles, forming the pilosebaceous unit. Sebaceous glands secrete the lipid-rich substance called sebum, which works to lubricate the hair and skin. Indeed, it has long been realized that the pilosebaceous unit harbors distinct microbial communities, dominated by bacteria capable of thriving in the anoxic, lipid-rich environment, such as Propioni-bacterium acnes [23,24]. The breakdown of sebum generates free fatty acids, which work to control microbial colonization along with sebocyte-derived cathelicidin, β-defensins, and antimicrobial histones [25–27].
The differences in the physical characteristics of skin from different body sites are easily observed macroscopically. In some locations, such as the palms of the hands or soles of the feet, the skin is thick and hairless; other sites are thin and delicate, such as the eyelids. Sites like the scalp or axilla may support dense hair growth, and other locations produce more oil, such as the face, back, and chest. As described above, these anatomic differences can strongly impact the microbial communities that reside on the skin. Fig. 1 illustrates some of the fundamental differences in skin anatomy from various body sites. However, these innate, genetically defined differences in the anatomy of the skin at different sites are only a partial explanation of skin microbial diversity. An additional major variable to consider are individual behavioral factors that alter surface conditions. For example, the amount of exposure versus occlusion of body sites, the degree of detergent use, the application of lotions or cosmetic products, occupation, and where one lives all dramatically alter surface environments. Thus, the micro-biome will be influenced by the structure and composition of the epidermis as well as individual behaviors that dictate the total nature of this environment. The skin’s location at the interface with the outside world therefore makes is most subject to environmental influences that will affect the microbiota.
Fig. 1.
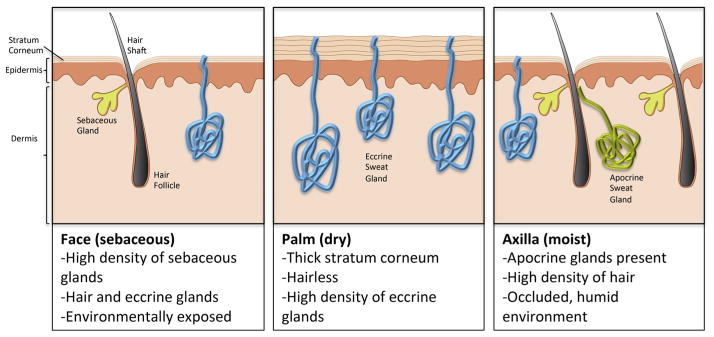
Diversity of the skin at different body sites. The physical and chemical features of the skin are not uniform across the body; rather, different anatomical locations show vast diversity in organization and the distribution of appendages and glandular structures. Shown are representative illustrations of the three major categories of skin environments.
2.2. Skin immunocytes control the microbiome
Within the skin, both innate and adaptive mechanisms contribute to immune function [28–30]. Keratinocytes are the first active participant in the skin immune response. These epithelial cells express a number of pattern recognition receptors (PRRs) that sense microbes through recognition of conserved molecular entities such as lipoproteins, nucleic acids, cell wall components, and flagella. While keratinocytes express a number of antimicrobial peptides, cytokines, and chemokines at steady state, activation of PRRs can rapidly increase the expression these molecules, resulting in direct antimicrobial effects as well as recruitment and education of additional immune cells [31]. Also found in the epidermis are Langerhans cells (LCs), a specific subset of dendritic cells. Historically viewed as constitutive immune-activating cells through their antigen-presenting roles, recent evidence supports the notion that LCs participate in promoting tolerance to self-antigens and commensal microbes through the induction of regulatory T cells at steady state [32]. The history and currently changing views of the role of LCs is reviewed in depth elsewhere [33,34]. Furthermore, within and below the epidermis reside many more cells types with functional roles in cutaneous immunity. Cells involved in both innate and adaptive immunity can be found here: dendritic cells, macrophages, mast cells, natural killer cells, and a variety of T cells including CD8+ memory T cells, CD4+ TH1, TH2, and TH17 cells, γδ T cells, NKT cells, and regulatory T cells (Treg) [28,29]. Combined, there is considerable specialized capacity in the skin cellular immune system to react and change in response to microbes.
Therefore, to understand the skin microbial flora it is essential to recognize that unlike all other commonly studied areas of the microbiome such as gut and oral mucosa, the skin has the greatest diversity of variables that influence its surface characteristics and a wide variety of cell types that are positioned to interact with microbes. One can imagine the skin as a melting pot of different microenvironments, constantly shifting with influences from the outside world and the host immune system. As we shall describe, this creates a potential problem in describing the “skin micro-biome.” Given the vast diversity of skin environments with different physical and chemical features it is logical to predict that each will support vastly different populations of microorganisms.
3. Characterization of the “skin microbiome” – does it really exist?
Several recent studies have set out to investigate the composition of microbial communities on the skin at various anatomical locations. One of the key take-home points arising from these studies is that just as the microbiome of the skin differs greatly from that of the gastrointestinal tract or oral cavity, so too do the populations from different areas of the skin. This finding makes sense when one considers the vast differences in features of the skin from different body sites as described above. In light of these findings, the term “skin microbiome” must be used carefully: such a broad term, while distinguishing microbes on the skin from those in the gut or oral cavity, fails to address the site-based variation in microbes residing on the skin. One must be especially critical when analyzing and comparing investigations of the “skin microbiome,” as the site studied and the environment the individual is exposed to can drastically impact the types of microbes identified. The definition of “biome” as being of a similar condition harboring a distinct community cannot be applied to the skin as a whole. However, as limited information has been assembled to accurately describe the composition and function of a “palm microbiome” or “axillary microbiome,” for now we must frequently lapse into the inherently incorrect term of the “skin microbiome.”
3.1. What is normal?
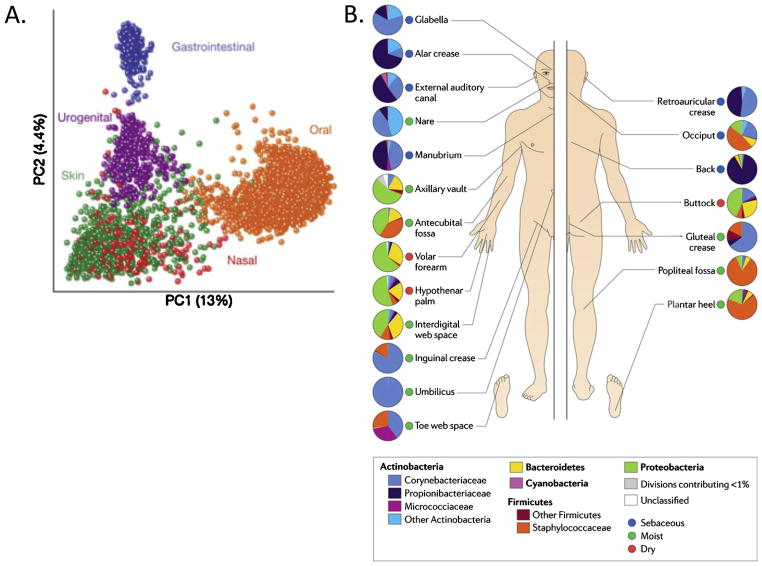
Based on 16S ribosomal RNA gene sequencing, the majority of “normal” bacterial inhabitants of the skin fall into four phyla: Actinobacteria, Bacteroidetes, Firmicutes, or Proteobacteria. While these are indeed the same four phyla that compose the majority of the bacteria present in the GI tract and oral cavity, these inhabitants are present at different ratios, and Actinobacteria are the most abundant inhabitants of many sites on the skin [4,35,36]. Commensal microbes on the skin have also been classified as resident of transient depending on their ability to populate the skin-resident microbes survive on the skin for long periods of time, whereas transient microbes may be detected during one sampling event and absent the next. However, as we have seen earlier, the most influential factor in regards to the microbes inhabiting the skin is the body site being studied, and as a result skin has the most diverse bacterial communities of any of the major epithelial surfaces studied to date (Fig. 2). Current information on these sites can be divided into three categories: moist, sebaceous, and dry [37]. Moist sites include areas such as the navel, the axilla, the inguinal crease (groin), the sole of the foot, antecubital fossa (inner elbow) and the popliteal fossa (back of the knee). The most abundant microbes colonizing these moist areas are Staphylococcus and Corynebacterium species, of the phyla Firmicutes and Actinobacteria, respectively [37,38]. Sebaceous sites, such as the forehead, the alar crease (side of the nostril), the retroauricular crease (behind the ear), and the back seem to harbor the least diverse populations of microbes. Propioni-bacterium species of the phyla Actinobacteria are the most common isolates from sebaceous areas, probably due to their ability to survive in these anaerobic, lipid-rich environments. Dry areas of the skin, including the forearm, various locations on the hand, and the buttocks, have been shown to have the most diversity in microbial inhabitants, with varying representations of the main four phyla (Fig. 2). At present is still remains unclear what proportion of these organisms can actually survive or replicate on the skin and which are simply frequent environmental encounters.
Fig. 2.
Diversity of the microbial communities populating the skin. (A) Microbial communities inhabiting the skin are extremely diverse, more so than those residing at other epithelial surfaces of the body. (B) In addition, communities residing on the skin at different anatomical locations differ greatly in their compositions. Reprinted from Refs.[4,7] with permission from Nature Publishing Group.
Currently, the vast majority of microbiome-focused studies have concentrated on bacterial species. However, one must not forget additional microorganisms that are residents of the skin, including fungi, parasites, and viruses. While the methods for classifying and differentiating these microbes based on sequencing data are not as well developed as those for bacteria, some progress has been made toward characterizing these members of the microbiome. In regards to the fungi present on normal human skin, the most commonly identified organisms are species of Malassezia [39–41]. In fact, one study estimated Malassezia to account for up to 80% of fungi present, depending on the anatomical location sampled [42]. Further investigation is warranted for identifying additional fungi that are components of the skin microbiome. Demodex mites, which are small, parasitic arthropods residing in pilosebaceous units, have been isolated and implicated in the pathogenesis of rosacea [43,44]. However, as these mites can also be found in normal skin, more investigation into their role as commensal microbes is needed [45]. Viruses are perhaps the least studied of all members of the skin microbiota, though recent studies have begun to probe this subject. Due to the great diversity already identified in the viral component of the microbiota, it is likely that these microbes play an important role in cutaneous immunity [46,47]. Unfortunately, at the present time few conclusions can be drawn for either the description or functions of commensal non-bacterial microbes on the skin.
3.2. Hyper-diversity of bacterial communities of the skin
The most common theme to emerge from in-depth studies of the bacterial communities residing on the skin is the large amount of temporal and spatial diversity seen both between different individuals and within the same person. As mentioned, the body site sampled is a key determinant in the makeup of these communities, as different microbes have evolved to thrive in different ecological niches present across our bodies [37,38]. While some body sites that share similar characteristics do show significant similarities across individuals, a tremendous amount of interpersonal variation has been identified.
The skin is colonized by bacteria starting at birth. This initial skin microbiome has a very low diversity across the body, and is largely shaped by the delivery mode of the child-babies born through the vaginal canal will be colonized by microbes present in the mother’s vagina, while babies born through caesarian section will acquire a skin flora more similar to the mother’s skin [48]. In the first years of one’s life, the microbiota at various body sites develop more diversity as children explore their environment, change their diets, and are exposed to more people and animals [49]. Studies have indicated that by 2.5 years of age, the intestinal microbiome of infants has developed to resemble that of adults [50]; similarly, age has been shown to impact the composition of microbial communities, as great diversity was observed when comparing children at different stages of development with adults [51].
In healthy adults, the amount of diversity seen in skin commensal bacteria is staggering. In analyzing the bacteria present on the forearms of six individuals, Gao et al. observed that less than 10% of identified genera were present across all individuals [52]. Similarly, in surveying the palms of 51 healthy volunteers, Fierer et al. report even greater diversity, with the average palm harboring over 150 species-level bacterial phylotypes [53]. Furthermore, individuals displayed on average only 17% similarity between their two hands, and between individuals, the common members dropped to 13% [53]. Much of this diversity correlates with gender, dominant hand, and time since last washing their hands. Additional studies have investigated the diversity between individuals, and in all cases, diversity was a unifying theme [37,54,55]. Interestingly, a recent study showed that family members residing in the same home show much more similarity in their skin commensal bacteria than strangers; furthermore, pet owners showed a striking similarity with the bacterial communities present on their pets [56]. Such observations further challenge our concept of the human biome and beg the question if we should actually include all the residents of our home and place of work in these discussions.
4. Functional associations of skin commensals with cutaneous immunity
The most interesting and important question to arise from studies of the human microbiome is to what extent these microbes impact our health. While a number of discoveries have been made regarding the importance of microbial communities in the gastrointestinal tract [57–59], much less is currently known about the role of microbes on the skin in the development and maintenance of our immune system. Conclusions drawn regarding the function of the gut microbiome in immunity are always complicated by the important role these microbes play in processing and absorption of nutrients. Thus, “germ-free” mice used in immunological studies will have dramatic, and often overlooked differences in nutrition compared to the control population. Studies of the skin micro-biome are therefore somewhat more straightforward as their role in nutritional homeostasis is less likely to be a confounding variable.
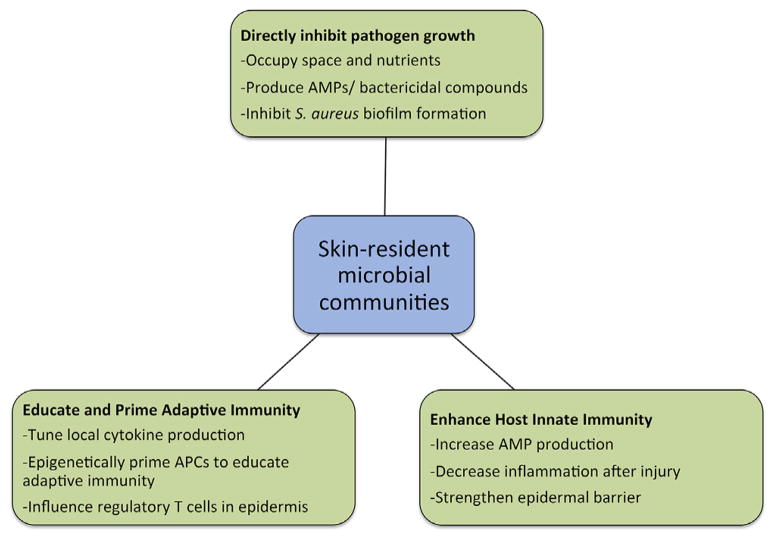
Fig. 3 summarizes the ways in which the commensal microbes of the skin are hypothesized to influence cutaneous immunity in the host. In the following sections, we will discuss recent experimental findings on the role of commensal microbes in cutaneous immunity.
Fig. 3.
Functional associations with microbial communities on the skin. Microbial communities on the skin contribute to host immune defense through a variety of mechanisms, as illustrated here.
4.1. Healthy competition from our commensal microbes
One method by which commensal microorganisms contribute to host immunity is through the inhibition of growth of pathogenic microbes. Indirectly, the presence of commensal microbes on the skin results in competition for nutrients and space, thus greatly impacting the potential for growth when pathogens are introduced on the skin surface. Additionally, a large number of bacteria are known to directly restrict the growth of competitors through the production of antimicrobial compounds [60]. These proteinaceous factors, called bacteriocins, are capable of inhibiting the growth of closely related species of bacteria while having no effect on the organisms which produce them. Recently, several lines of evidence have indicated that commensal bacteria from the skin produce molecules with antimicrobial properties that can function in vivo to restrict the growth of cutaneous pathogens [61].
One of the more abundant and frequently cultured members of the skin bacterial community is Staphylococcus epidermidis, a cousin of the frequent pathogen Staphylococcus aureus. While S. epidermidis does have the potential to cause serious infections, it has also been shown to produce several molecules that interfere with pathogen growth. Iwase et al. illustrated that clinical isolates of S. epidermidis are able to inhibit S. aureus biofilm formation through the production of a serine protease, Esp, which also boosts the antimicrobial effects of hBD2 [62]. Furthermore, the introduction of Esp-producing S. epidermidis into the nasal cavity of volunteers who were S. aureus carriers resulted in the clearance of S. aureus colonization, illustrating the clinical relevance of this commensal-produced protease [62]. The mechanism behind the biofilm-disrupting capabilities of Esp have been recently elucidated: it seems that Esp specifically degrades several S. aureus proteins involved in biofilm formation and many human receptor proteins important for S. aureus colonization and infection of host cells [63]. S. epidermidis produces a variety of additional molecules that influence the growth of pathogenic microbes. In particular, phenol-soluble modulins (PSMs) have potent antimicrobial functions, possessing the ability to strongly interact with and cause leakage of microbial lipid membranes [64]. These S. epidermidis-derived molecules selectively kill the skin pathogens Streptococcus pyogenes and S. aureus, cooperate with host-derived AMPs to increase bacterial killing, and can be incorporated into neutrophil extracellular traps (NETs), another innate host defense against infection [64,65]. Thus, S. epidermidis possesses several weapons that contribute to the innate immune defense arsenal present in human skin.
Another common skin commensal bacteria is P. acnes, and recent work indicates that this bacteria is capable of inhibiting the growth of MRSA [66]. Briefly, P. acnes ferments glycerol, a metabolite that naturally occurs in human skin, into a number of short-chain fatty acids that result in a decreased intracellular pH within S. aureus to inhibit its growth. These findings were recapitulated in vivo, as the application of P. acnes and glycerol to wounds on mouse skin greatly decreased the bacterial burden when challenged with CA-MRSA isolates [66]. These findings suggest that P. acnes may function to prevent pathogen growth in human skin, and could also be used to develop novel probiotic treatments for MRSA infections.
4.2. Commensal microbes control behavior of the host
Skin microbes also contribute to cutaneous immunity through their influence on the function of host cells. There is currently evidence for multiple mechanisms by which this can occur. Our group has shown that S. epidermidis, sensed by keratinocytes via Toll-like receptor 2, boosts host immunity to S. aureus infection through increased expression of antimicrobial peptides such as β-defensins 2 and 3 [67]. These results suggest a symbiotic relationship between hosts and commensal microbes that leaves the host more prepared to combat pathogenic infection. This is supported by findings from Wanke et al. which demonstrate that signaling from the commensal microbes through TLR2 results in increased antimicrobial peptide expression in keratinocytes, and blocks NF-KB inhibition induced by pathogenic S. aureus [68]. Indeed, the influence of commensal microbes extends to other cells types as well. For example, the role of commensals has been investigated in the setting of mast cell-mediated antiviral immunity. Wang et al. demonstrated that TLR2 activation, mediated by LTA from S. epidermidis, results in greater numbers of mast cells being recruited to sites of viral challenge in the skin [69]. Furthermore, the release of the AMP cathelicidin by these recruited mast cells was amplified by this TLR2 stimulus, resulting in increased antiviral immunity. Thus, signals from commensal microbes appear to be relevant to multiple cell types responding to a variety of microbial challenges in the skin. Together, these findings illustrate an important role for commensal bacteria in amplifying host immune defense against pathogens.
Commensal microbes like S. epidermidis also contribute to host immunity through maintenance of the epidermal barrier. TLR2-mediated recognition of LTA from S. epidermidis inhibited TLR3-driven inflammatory cytokine production in cultured keratinocytes and reduced levels of inflammation in vivo following wounding, a situation in which excessive inflammation would be detrimental to the host [70]. Furthermore, activation of TLR2 has been shown to increase the tight junction barrier in cultured keratinocytes, illustrating another role for commensal microbes in maintaining barrier homeostasis, a crucial aspect of host defense [71].
Other evidence that commensal skin microbes are necessary and sufficient for the generation of optimal skin immunity have come from germ-free mice that failed to mount an adequate immune response to Leishmania [72]. Recolonization of the gut was unable to restore cutaneous immune function, but exposure of the skin of these mice to S. epidermidis alone was sufficient to restore effect or T cell levels and rescue the immune deficiency. These observations were linked to IL-1 signaling, as germ-free mice showed significant decreases in cutaneous IL-1α production. This evidence suggests that communication between commensal microbes and skin-resident cells is important for proper tuning of the local inflammatory milieu. Future studies should continue to probe the effects of commensal microbiota on the development of an effective immune environment.
5. Consequences of dysbiosis in the microbiome
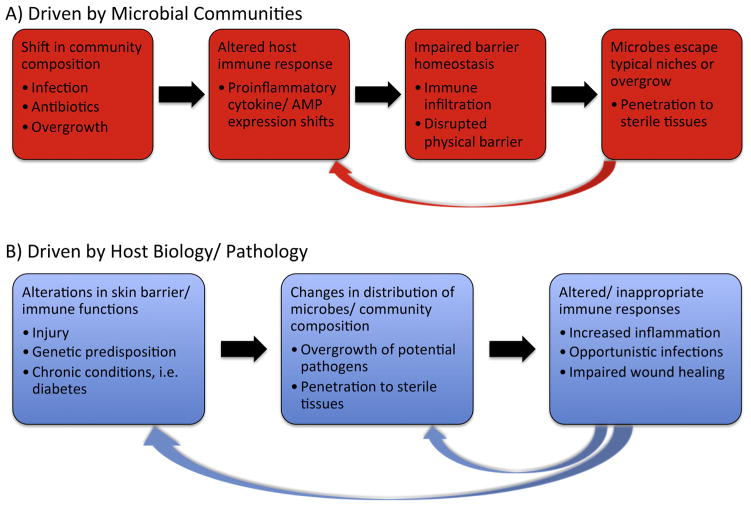
As it is becoming increasingly clear that the microbiota make important contributions to normal immune development and function, it is logical that disease can be correlated with alterations in microbial communities. Several descriptive studies have identified differences in the microbes present in diseased skin versus those present in healthy skin. While studies indicate that an imbalance of microorganisms, termed dysbiosis, exists in numerous pathologies, these results have presented a sort of “chicken-or-the-egg” type conundrum, as it is not entirely clear whether alterations in the microbiome lead to disease, or whether underlying conditions result in an imbalance in microbial communities. These models are displayed in Fig. 4.
Fig. 4.
Models of dysbiosis of skin-resident microbes. Dysbiosis of skin-resident microbes has been associated with a variety of dermatological pathologies; however, it remains unclear whether alterations in microbial communities or intrinsic features of the host initiate these processes.
Many microbes that are considered to be relatively harmless commensals can indeed cause serious infection in situations of immune suppression. This is supported by studies investigating the microbes present in chronic, non-healing ulcers that plague diabetic patients or the elderly, as well as the rates of coagulase-negative Staphylococcal infections seen in hospitals. Thus, one must keep in mind that even seemingly beneficial microorganisms can take on a pathogenic role when presented with the right opportunity.
5.1. Dysbiosis in atopic dermatitis
Atopic dermatitis (AD) is a chronic, relapsing, pruritic inflammatory skin condition that is non-contagious in nature, affecting approximately 15% of children in the United States. It has long been noted that AD flares are associated with colonization and infection by S. aureus, and that antibiotic treatments targeting S. aureus are sometimes successful in temporary improvement of disease [73]. However, it was not until very recently that a thorough comparison of the microbial communities present in lesional, non-lesional, and healthy skin was conducted [74]. In this study, disease flares were associated with a decrease in the overall diversity of microbial communities on the skin, due to an expansion of Staphylococcus species up to 90% of the microbes detected. Interestingly, both S. aureus and S. epidermidis were noted to increase in untreated lesions –given previous data indicating the ability of S. epidermidis to inhibit S. aureus, the relationship between these two microorganisms in lesional skin remains unclear. The observation that resolution of disease flares is preceded by a restoration of microbial diversity strengthens the case for a link between skin resident microbes and AD; however, more investigation is needed to definitively assign a causative role for the microbiota in AD.
Adding to the complexity of the AD-microbiome interaction is the number of host factors that have been implicated in the onset of AD [75]. To date, defects in several aspects of epidermal function have been implicated in AD: mutations in the filaggrin protein, an essential component of epidermal barrier formation [76]; mutations in receptors and signaling molecules that sense microbes, such as TLR2, CARD4, and CD14; and diminished expression or function of antimicrobial peptides such as defensins, cathelicidin, and dermicidin [77,78]. With so many compounding factors, it remains unclear whether changes in skin biology trigger alterations in microbial diversity or if overgrowth of Staphylococcus species occurs first and subsequently drives disease progression.
5.2. Dysbiosis in psoriasis
Psoriasis is an inflammatory skin condition, highlighted by erythematous, scaly plaques commonly occurring on the elbows, knees, scalp, and trunk [79]. Unlike AD, psoriasis is seldom complicated by infection and is usually not pruritic. In contrast, this disorder is complicated by auto-immune phenomena including arthritis and co-morbidities including coronary vascular disease. Detailed analysis of the microbes present in lesional and non-lesional skin of psoriasis has focused on both bacterial [80,81] and fungal [39,40] communities. While these studies have identified potential differences in the composition of the microbiota between psoriatic and normal skin, no consensus microorganisms have been directly identified and linked to disease pathogenesis. Additionally, the number of innate immune-related genetic factors identified in patients with psoriasis further complicates the issue, again leaving researchers without a definitive cause-and-effect relationship between disease and microbial diversity [79,82].
5.3. Opportunistic infections
Though commensal microorganisms typically inhabit our bodies peacefully, many do possess the ability to cause infection in the right setting. S. epidermidis, despite its many beneficial roles described previously, is a frequent cause of infections, particularly nosocomial deep tissue infections involving indwelling medical devices such as catheters [83,84]. Furthermore, numerous bacteria that are found in the normal skin microbiome frequently cause infection in chronic, non-healing wounds, which commonly occur in diabetic patients and the elderly [85,86]. While many factors of host biology contribute to the impaired healing of these wounds, it is noteworthy that the immune response to these commensal microbes in previously sterile tissues, resulting in prolonged inflammation, exacerbates the problem and creates a vicious cycle [86,87].
6. Conclusions
Recent efforts have greatly furthered our knowledge of the composition of microbial communities that inhabit the human body. With this descriptive information readily at hand, researchers are now poised to translate these findings into a deeper understanding of the complex relationships existing between commensals and their host. While several interesting discoveries have already been achieved, a number of important questions still remain to be answered. In particular, mechanistic details of the contributions of microbes to cutaneous immunity and the role of these commensal organisms in the initiation and progression of skin disease should be further pursued, as a deeper understanding of these processes will provide valuable information for the development of novel therapeutic strategies.
References
- 1.Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–64. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 2.Evans CA, Stevens RJ. Differential quantitation of surface and subsurface bacteria of normal skin by the combined use of the cotton swab and the scrub methods. J Clin Microbiol. 1976;3:576–81. doi: 10.1128/jcm.3.6.576-581.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong HH, Segre JA. Skin microbiome: looking back to move forward. J Invest Dermatol. 2012;132:933–9. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS ONE. 2012;7:e47075. doi: 10.1371/journal.pone.0047075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong HH. Skin microbiome: genomics-based insights into the diversity and role of skin microbes. Trends Mol Med. 2011;17:320–8. doi: 10.1016/j.molmed.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol. 2012;129:1204–8. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan XC, Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. 2012;8:e1002808. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55:856–66. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The micro-biome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elias PM. Stratum corneum defensive functions: an integrated view. J Invest Dermatol. 2005;125:183–200. doi: 10.1111/j.0022-202X.2005.23668.x. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 15.Proksch E, Brandner JM, Jensen J-M. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–72. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 16.Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–8. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilke K, Martin A, Terstegen L, Biel SS. A short history of sweat gland biology. Int J Cosmet Sci. 2007;29:169–79. doi: 10.1111/j.1467-2494.2007.00387.x. [DOI] [PubMed] [Google Scholar]
- 18.Rieg S, Seeber S, Steffen H, Humeny A, Kalbacher H, Stevanovic S, et al. Generation of multiple stable dermcidin-derived antimicrobial peptides in sweat of different body sites. J Invest Dermatol. 2006;126:354–65. doi: 10.1038/sj.jid.5700041. [DOI] [PubMed] [Google Scholar]
- 19.Rieg S, Garbe C, Sauer B, Kalbacher H, Schittek B. Dermcidin is constitutively produced by eccrine sweat glands and is not induced in epidermal cells under inflammatory skin conditions. Br J Dermatol. 2004;151:534–9. doi: 10.1111/j.1365-2133.2004.06081.x. [DOI] [PubMed] [Google Scholar]
- 20.Murakami M, Ohtake T, Dorschner RA, Schittek B, Garbe C, Gallo RL. Cathelicidin anti-microbial peptide expression in sweat, an innate defense system for skin. J Invest Dermatol. 2002;119:1090–5. doi: 10.1046/j.1523-1747.2002.19507.x. [DOI] [PubMed] [Google Scholar]
- 21.Natsch A, Schmid J, Flachsman F. Identification of odoriferous sulfanylalka-nols in human axilla secretions and their formation through cleavage of cysteine precursors by a C-S lyase isolated from axilla bacteria. Chem Biodivers. 2004;1:1058–72. doi: 10.1002/cbdv.200490079. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa Y, Yabuki M, Marsukane M. Identification of new odoriferous compounds in human axillary sweat. Chem Biodivers. 2004;1:2042–50. doi: 10.1002/cbdv.200490157. [DOI] [PubMed] [Google Scholar]
- 23.Leeming JPH, Cunliffe KTWJ. The microbial ecology of pilosebaceous units isolated from human skin. J Gen Microbiol. 1984;130:803–7. doi: 10.1099/00221287-130-4-803. [DOI] [PubMed] [Google Scholar]
- 24.Bruggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, et al. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–3. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- 25.Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, Huang CM. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upre-gulating beta-defensin-2 expression. J Invest Dermatol. 2010;130:985–94. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee DY, Huang CM, Nakatsuji T, Thiboutot D, Kang SA, Monestier M, et al. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol. 2009;129:2489–96. doi: 10.1038/jid.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy I, Pivarcsi A, Kis K, Koreck A, Bodai L, McDowell A, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006;8:2195–205. doi: 10.1016/j.micinf.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4:211–22. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–91. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afshar M, Gallo RL. Innate immune defense system of the skin. Vet Dermatol. 2013;24(32–38):e38–9. doi: 10.1111/j.1365-3164.2012.01082.x. [DOI] [PubMed] [Google Scholar]
- 31.Gallo RL, Nizet V. Innate barriers against infection and associated disorders. Drug Discov Today Dis Mech. 2008;5:145–52. doi: 10.1016/j.ddmec.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seneschal J, Clark RA, Gehad A, Baecher-Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity. 2012;36:873–84. doi: 10.1016/j.immuni.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igyarto BZ, Kaplan DH. Antigen presentation by Langerhans cells. Curr Opin Immunol. 2013;25:115–9. doi: 10.1016/j.coi.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romani N, Brunner PM, Stingl G. Changing views of the role of Langerhans cells. J Invest Dermatol. 2012;132:872–81. doi: 10.1038/jid.2011.437. [DOI] [PubMed] [Google Scholar]
- 35.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal micro-biota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–41. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulino LC, Tseng CH, Blaser MJ. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 2008;8:460–71. doi: 10.1111/j.1567-1364.2008.00359.x. [DOI] [PubMed] [Google Scholar]
- 41.Coelho MA, Sampaio JP, Goncalves P. Living and thriving on the skin: Malassezia genomes tell the story. mBio. 2013;4:e00113–7. doi: 10.1128/mBio.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol. 2010;48:3575–81. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacey N, Delaney S, Kavanagh K, Powell FC. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. Br J Dermatol. 2007;157:474–81. doi: 10.1111/j.1365-2133.2007.08028.x. [DOI] [PubMed] [Google Scholar]
- 44.Jarmuda S, O’Reilly N, Zaba R, Jakubowicz O, Szkaradkiewicz A, Kavanagh K. Potential role of Demodex mites and bacteria in the induction of rosacea. J Med Microbiol. 2012;61:1504–10. doi: 10.1099/jmm.0.048090-0. [DOI] [PubMed] [Google Scholar]
- 45.Lacey N, Ni Raghallaigh S, Powell FC. Demodex mites – commensals, parasites or mutualistic organisms? Dermatology. 2011;222:128–30. doi: 10.1159/000323009. [DOI] [PubMed] [Google Scholar]
- 46.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–15. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial micro-biota across multiple body habitats in newborns. Proceed Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131:2026–32. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceed Natl Acad Sci U S A. 2011;108:4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm super-ficial skin bacterial biota. Proceed Natl Acad Sci U S A. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proceed Natl Acad Sci U S A. 2008;105:17994–9. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hulcr J, Latimer AM, Henley JB, Rountree NR, Fierer N, Lucky A, et al. A jungle in there: bacteria in belly buttons are highly diverse, but predictable. PLoS ONE. 2012;7:e47712. doi: 10.1371/journal.pone.0047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blaser MJ, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Estrada I, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013;7:85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song SJ, Lauber CL, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–73. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceed Natl Acad Sci U S A. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of pep-tidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bastos MCF, Ceotto H, Coelho MLV, Nascimento JS. Staphylococcal antimicrobial peptides: relevant properties and potential biotechnological applications. Curr Pharm Biotechnol. 2009;10:38–61. doi: 10.2174/138920109787048580. [DOI] [PubMed] [Google Scholar]
- 61.Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. 2011;131:1974–80. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–9. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 63.Sugimoto S, Iwamoto T, Takada K, Okuda K, Tajima A, Iwase T, et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bac-teriol. 2013;195:1645–55. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cogen AL, Yamasaki K, Muto J, Sanchez KM, Alexander LC, Tanios J, et al. Staphylococcus epidermidis antimicrobial delta-toxin (phenol-soluble modulin gamma) cooperates with host antimicrobial peptides to kill group A Streptococcus. PLoS ONE. 2010;5:e8557. doi: 10.1371/journal.pone.0008557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shu MW, Yu Y, Kuo J, Coda S, Jiang A, Gallo Y, et al. Fermentation of Propioni-bacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Der-matol. 2010;130:2211–21. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wanke I, Steffen H, Christ C, Krismer B, Gotz F, Peschel A, et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol. 2011;131:382–90. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, MacLeod DT, Di Nardo A. Commensal bacteria lipoteichoic acid increases skin mast cell antimicrobial activity against vaccinia viruses. J Immunol. 2012;189:1551–8. doi: 10.4049/jimmunol.1200471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–82. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuki T, Yoshida H, Akazawa Y, Komiya A, Sugiyama Y, Inoue S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J Immunol. 2011;187:3230–7. doi: 10.4049/jimmunol.1100058. [DOI] [PubMed] [Google Scholar]
- 72.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abeck D, Mempel M. Staphylococcs aureus colonization in atopic dermatitis and its therapeutic implications. Br J Dermatol. 1998;139:13–6. doi: 10.1046/j.1365-2133.1998.1390s3013.x. [DOI] [PubMed] [Google Scholar]
- 74.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hata TR, Gallo RL. Antimicrobial peptides, skin infections, and atopic dermatitis. Semin Cutan Med Surg. 2008;27:144–50. doi: 10.1016/j.sder.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 77.Rieg S, Steffen H, Seeber S, Humeny A, Kalbacher H, Dietz K, et al. Deficiency of dermicidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J Immunol. 2005;174:8003–10. doi: 10.4049/jimmunol.174.12.8003. [DOI] [PubMed] [Google Scholar]
- 78.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–60. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 79.Schon MP, Boehncke WH. Psoriasis. N Engl J Med. 2005;352:1899–912. doi: 10.1056/NEJMra041320. [DOI] [PubMed] [Google Scholar]
- 80.Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fahlen A, Engstrand L, Baker BS, Powles A, Fry L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304:15–22. doi: 10.1007/s00403-011-1189-x. [DOI] [PubMed] [Google Scholar]
- 82.Fry L, Baker BS, Powles A, Fahlen A, Engstrand L. Is chronic plaque psoriasis triggered by microbiota in the skin? Br J Dermatol. 2013 doi: 10.1111/bjd.12322. [DOI] [PubMed] [Google Scholar]
- 83.Uckay I, Pittet D, Vaudaux P, Sax H, Lew D, Waldvogel F. Foreign body infections due to Staphylococcus epidermidis. Ann Med. 2009;41:109–19. doi: 10.1080/07853890802337045. [DOI] [PubMed] [Google Scholar]
- 84.Otto M. Staphylococcus epidermidis – the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–30. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Program NCS, Liechty KW, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proceed Natl Acad Sci U S A. 2010;107:14799–804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Adv Exp Med Biol. 2012;946:55–68. doi: 10.1007/978-1-4614-0106-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]