SUMMARY
Iron excess is closely associated with tumorigenesis in multiple types of human cancers, with underlying mechanisms yet unclear. Recently, iron deprivation has emerged as a major strategy for chemotherapy, but it exerts tumor suppression only on select human malignancies. Here, we report that the tumor suppressor protein p53 is downregulated during iron excess. Strikingly, the iron polyporphyrin heme binds to p53 protein, interferes with p53-DNA interactions, and triggers both nuclear export and cytosolic degradation of p53. Moreover, in a tumorigenicity assay, iron deprivation suppressed wild-type p53-dependent tumor growth, suggesting that upregulation of wild-type p53 signaling underlies the selective efficacy of iron deprivation. Our findings thus identify a direct link between iron/heme homeostasis and the regulation of p53 signaling, which not only provides mechanistic insights into iron-excess-associated tumorigenesis but may also help predict and improve outcomes in iron-deprivation-based chemotherapy.
INTRODUCTION
Iron is essential for cell survival, proliferation, and metabolism, a fact highlighted by the association of dysregulated iron metabolism with a myriad of human disorders including cancer and diabetes (Andrews, 2008; Fleming and Ponka, 2012; Rouault, 2005; Simcox and McClain, 2013). In particular, decades of epidemiological and experimental studies have established that iron excess, due to either genetic factors or excessive dietary intake, is implicated in multiple types of human cancers (Torti and Torti, 2013; Toyokuni, 2009). Hereditary hemochromatosis (HH) is a genetic disorder of iron overload, with clinical complications including liver cirrhosis and a 20- to 200-fold increased risk for hepatocellular carcinoma (Elmberg et al., 2003; Niederau et al., 1985) or other cancer types (Osborne et al., 2010; Pietrangelo, 2010; Radulescu et al., 2012). Meanwhile, tumors reprogram iron metabolism to achieve a growth advantage or metastasis, resulting in the emergence of iron deprivation, via iron chelation or application of transferrin receptor-neutralizing antibodies, as a major chemotherapeutic strategy. However, preclinic and clinical studies have demonstrated that iron deprivation only suppresses select human malignancies while having no effect on other cancer types, with underlying mechanisms of the selectivity yet elusive (Buss et al., 2004; Yamasaki et al., 2011). Therefore, a thorough interrogation of the mechanisms of how iron excess contributes to tumorigenesis and the molecular basis of the selective efficacy of iron deprivation would not only further our understanding of cancer biology but also improve the design of targeted chemotherapy for better clinical outcomes.
Heme is an iron polyporphyrin that constitutes the prosthetic group for proteins functioning in a myriad of fundamental biological processes, including respiration, energetic homeostasis, signal transduction, xenobiotic detoxification, iron metabolism, mRNA processing, and control of circadian rhythm (Boon et al., 2005; Dioum et al., 2002; Faller et al., 2007; Gilles-Gonzalez and Gonzalez, 2005; Hu et al., 2008; Ishikawa et al., 2005; Rajagopal et al., 2008; Yang et al., 2010). The switch between ferrous (Fe2+) and ferric (Fe3+) states of iron in the metallopolyporphyrin, termed as heme and hemin, respectively, underlies its unique roles in transducing redox and gas signaling in vivo. Heme was identified as a ligand for transcriptional factors such as NPAS2 (Dioum et al., 2002), E75 (Reinking et al., 2005), and Rev-erb α (Yin et al., 2007), where heme was shown to play a regulatory role by modulating the respective protein functionalities. We have also found that arginyl-tRNA protein transferase (Ate1), a key component of the N-end rule pathway in the ubiquitin (Ub)-proteasome system, binds to heme and allows the N-end rule pathway to act as a sensor of heme and redox state (Hu et al., 2005, 2008; Kwon et al., 2002; Varshavsky, 2012). Because heme is identified as a prosthetic group in an expanding body of proteins in multiple pathophysiological processes, it is conceivable that we might be still at an early stage in understanding the regulatory roles of heme.
Tumor suppressor p53 suppresses tumorigenesis and regulates DNA-damage repair, cell-cycle arrest, and tumor responses to chemotherapy (Baker et al., 1989; Espinosa et al., 2003; Kastan et al., 1991; Liu et al., 2011; Vogelstein et al., 2000; Vousden and Prives, 2009). Recent work also identified p53 as a cellular hub in regulating and responding to cell metabolism (Jiang et al., 2013; Maddocks and Vousden, 2011). p53 protein is also a major regulator of cellular responses to redox signaling. Thus far, a few small molecules, such as NAD+ and ADP, have been identified as physiological ligands for p53 protein, modulating the transcription of a set of p53 target genes in response to changes in cell redox state and energy metabolism (McLure et al., 2004).
It is yet unknown whether p53 might bind to any other cellular small molecules, either metabolites or signaling messengers, and directly sense cellular redox signaling. Given the prominent roles of tumor suppressor p53 in regulating tumorigenesis and cellular responses to genotoxic stresses, we set out to examine whether and how iron excess might affect p53 signaling and thus contribute to tumorigenesis associated with iron overload. We also used cell and animal models to investigate the molecular determinants underlying the selective efficacy of iron deprivation-based chemotherapy.
RESULTS
Tumor Suppressor p53 Protein Is Downregulated during Iron Excess
Patients with HH have defects in the control of iron entry into circulation, which allows for the toxic accumulation of iron in parenchymal cells of vital organs (Pietrangelo, 2010; Weiss, 2010). The known genetic factors for human HH are mutations in the genes encoding the human hemochromatosis protein (Hfe), transferrin receptor 2 (Tfr2), hemojuvelin (Hjv), ferroportin (Fpn), and more rarely hepcidin (Hamp). It was estimated that 10% of the white population worldwide carries disease-associated mutations in Hfe, with disease penetrance of 2%–38% among male and 1%–10% among female carriers (Fleming and Ponka, 2012). A 20- to 200-fold higher risk for hepatocellular carcinoma or many other types of cancer was reported for patients with hemochromatosis with iron overload, highlighting a strong association between increased iron metabolism and human malignancy (Torti and Torti, 2013). However, the underlying mechanism remains unclear.
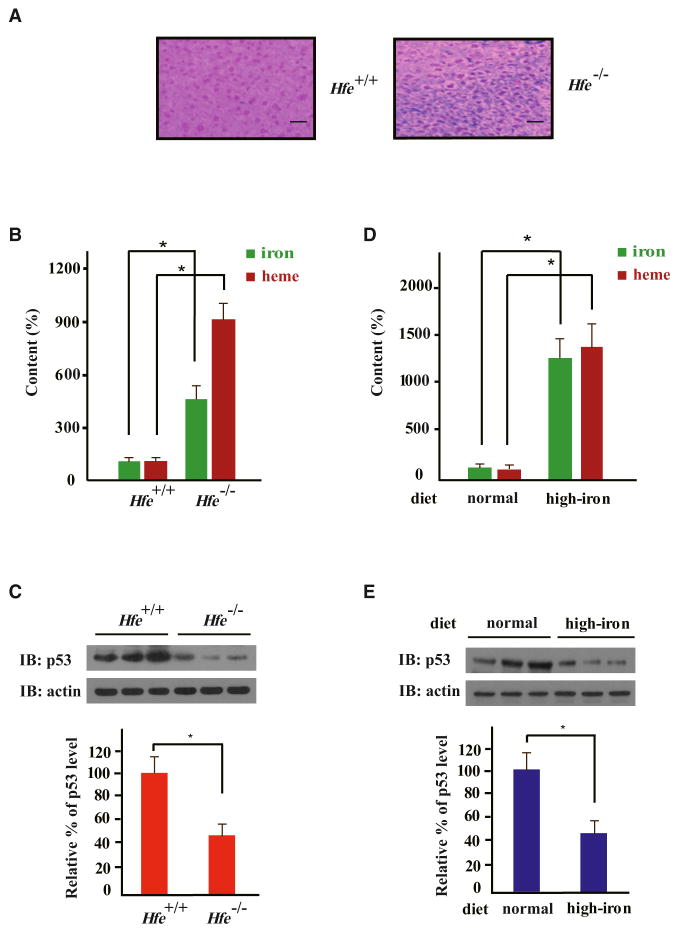
Mice with homozygous deletion of the human hemochromatosis (Hfe) gene (Hfe−/−) faithfully recapitulate most human HH symptoms, including significant liver iron overload as assessed by Perls’ Prussian Blue staining (Zhou et al., 1998) (Figure 1A). Using a 3,3′,5,5′-tetramethylbenzidine (TMB) assay to quantitatively assess liver heme content, we found that Hfe−/− mouse liver lysate had 9-fold more heme than lysate from wild-type livers (3.71 ± 0.4 nmol/mg protein versus 0.41 ± 0.12 nmol/mg protein) (Figure 1B). We next assessed liver p53 protein content and found that total Hfe−/− liver lysates had significantly lower endogenous p53 protein levels than wild-type livers (Figure 1C). The level of p53 protein in primary hepatocytes from Hfe−/− mice was also markedly lower than that in wild-type hepatocytes (Figure S1A). Moreover, compared to wild-type mice on a normal diet, wild-type mice fed with a high iron diet had considerably lower p53 protein levels (Figures 1D and 1E). Thus, iron overload in mice, due to either a genetic perturbation of iron metabolism or a high iron diet, correlates with a significant reduction in p53 protein levels.
Figure 1. Tumor Suppressor p53 Protein Is Downregulated in Iron Excess.
(A) Perls’ Prussian Blue staining of Hfe+/+ and Hfe−/− mouse liver sections (representative sections shown). Scale bars represent 100 μm.
(B–E) Homeostatic levels of iron, heme, and endogenous p53 protein in the livers of Hfe−/− and Hfe+/+ mice (B and C) or Hfe+/+ mice fed on a normal or high-iron diet were measured (D and E). Iron or heme content was determined using an unsaturated iron-binding capacity or TMB assay, respectively (B and D), and protein levels were measured by immunoblotting (IB) with the indicated antibodies (C and E). In (B) and (D), iron or heme levels in the livers of wild-type mice fed a normal diet were set to 100%. Protein levels were normalized by setting the static endogenous p53 levels in the livers of Hfe+/+ mice (C) and normal-diet mice (E) as 100%. Comparative quantification of band intensities was performed with ImageJ. Bar graphs are shown as mean ± SEM (n = 3). *p < 0.05.
Hemin Directly Modulates p53 Stability without Involving the Transcription Factor Hypoxia-Inducible Factor 1 α
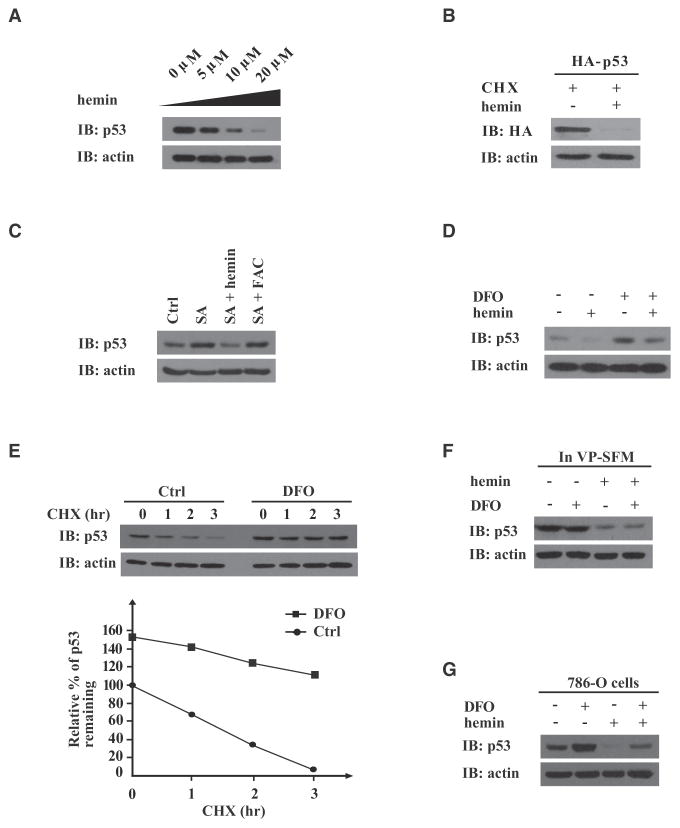
We next demonstrated that hemin treatment could downregulate endogenous p53 protein in both mouse primary hepatocytes and human hepatocarcinoma HepG2 cells in a hemin dose-dependent manner (Figures S1B and 2A). Treatment with ferric ammonium citrate (FAC; 100 μg/ml, 6 hr) apparently also reduced p53 levels (Figure S1C). The p53 reduction seemed to be independent of transcription because hemin also decreased exogenously expressed p53 protein in the presence of the protein translation inhibitor cycloheximide (CHX) (Figure 2B). Furthermore, inhibition of heme biogenesis in HepG2 cells with succinylacetone, a 5-aminolevulinate synthase (ALAS) inhibitor, led to increased p53 (Figure 2C). This effect could be reversed by hemin, but not FAC, suggesting that hemin may directly downregulate endogenous p53 protein.
Figure 2. Hemin Destabilizes p53 Protein Independently of HIF-1α.
Immunoblots (IB) were probed with the indicated antibodies.
(A) HepG2 cells were treated with hemin at indicated concentrations for 8 hr.
(B) HepG2 cells transiently expressing HA-tagged human p53 were pretreated with CHX (25 μg/ml) for 6 hr before hemin addition (15 μM, 8 hr).
(C) HepG2 cells were cultured in the absence (Ctrl) or presence of succinylacetone (1 mM, 24 hr), with or without hemin (10 μM, 6 hr) or FAC (100 μg/ml, 6 hr) treatment.
(D) HCT116 p53+/+ cells were treated with or without DFO (100 μM, 24 hr) and with or without hemin (10 μM, 8 hr).
(E) HepG2 cells were pretreated with or without DFO (100 μM, 24 hr), then subjected to CHX (25 μg/ml) treatment for the indicated times. Relative amounts of remaining endogenous p53 protein (%) were plotted against the indicated time course (lower panel).
(F) HepG2 cells, adapted to serum-free growth conditions by growing in VP-SFM (Invitrogen) for 8 hr, were treated with DFO (100 μM, 24 hr), hemin (10 μM, 24 hr), or both.
(G) HIF-1α and vHL null 786-O cells were treated with DFO (100 μM, 24 hr), hemin (10 μM, 8 hr), or both.
Treatment of HepG2 cells with the iron chelator deferoxamine (DFO) significantly increased the steady-state level of endogenous p53 protein in a manner independent of protein synthesis (Figures 2D and 2E). The HepG2 cells were cultured in normal media that contained serum iron, and hemin could reverse the stabilizing effect of DFO treatment on endogenous p53 protein (Figure 2D). When the cells were adapted to serum- and iron-free media (VP-SFM), DFO itself could no longer stabilize endogenous p53 protein, but hemin treatment still efficiently destabilized p53 protein in a manner that could not be reversed by iron chelation (Figure 2F). This strongly suggests that hemin directly modulates the stability of endogenous p53 protein, without involving iron ions that may be released from heme-oxygenase-catabolized heme.
A previous study has shown that DFO may promote p53 transcription through stabilizing hypoxia-inducible factor 1 α (HIF-1α) (An et al., 1998). To test if we are observing such a phenomena, we probed for p53 protein levels in human renal cancer 786-O cells that are null in both vHL (von Hippel-Lindau tumor suppressor) and HIF-1α (Kaelin, 2004) and found that hemin destabilized endogenous p53 protein and iron chelation stabilized p53 protein in a hemin treatment-reversible manner (Figure 2G). These data collectively suggested that the effect of hemin or iron chelation on the homeostasis of p53 protein involves posttranslational mechanisms that do not significantly involve the transcription factor HIF-1α.
Tumor Suppressor p53 Protein Directly Binds to Heme In Vitro
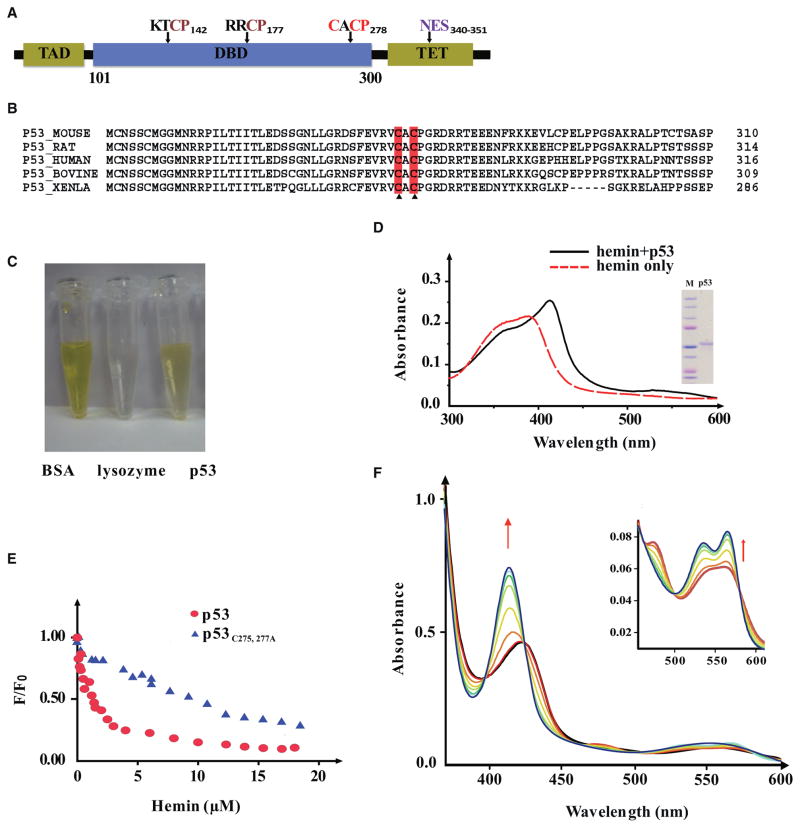
An examination of the p53 amino acid sequences across several species revealed that all of them bear three putative heme regulatory motifs (HRMs), consisting of Cys-Pro (CP) sequences that occur in a subset of heme-binding proteins (Zhang and Guarente, 1995) (Figures 3A and 3B). A TMB assay verified that the His6-tagged p53 protein, freshly purified from bacteria, contained heme (Figure 3C), whereas a similarly tagged human Ub did not (data not shown). Matrix-assisted laser desorption-ionization (MALDI) mass spectra analysis further confirmed that free heme (m/z 616 ± 2 Da) was associated with freshly purified p53 protein (Figure S2A). In addition, hemin immobilized on agarose beads, but not agarose beads alone, recovered endogenous mouse and human p53 proteins from multiple cell types (Figure S2C). However, p53 protein fractions lost heme during dialysis (Figure S2E), suggesting that heme might associate with p53 non-covalently. p53 proteins, as well as ATE1, bound to heme with a dissociation constant (KD) in the low micromolar range and lost their associated heme at a rate faster than BSA, whose affinity to heme is at nanomolar range (Figure S2E). Under the same conditions, lysozyme, which does not bind to heme, readily lost heme even more quickly (Figure S2E). The rates of heme loss from the heme-binding proteins were thus conversely related to how tightly the proteins might bind to heme.
Figure 3. Tumor Suppressor p53 Protein Directly Interacts with Heme.
(A) Schematic domain structure of human p53 protein, with putative HRMs marked by their amino acid sequences. TAD, transcription activation domain; TET, tetramerization domain.
(B) Sequence alignment of the CACP motif in p53 proteins across species.
(C) Gel filtration of p53 protein. BSA was used as a positive control of heme binding and lysozyme as the negative control.
(D) UV-Vis spectrum of heme-p53 complexes (after gel filtration). Inset shows the purity of the p53 protein.
(E) A tryptophan fluorescence quenching assay of hemin with wild-type and mutant p53 determined that heme associates with wild-type human p53 protein at a KD of ~1.20 μM and with p53C275,277A at a KD of ~15.70 μM.
(F) UV-Vis spectra analyses were performed with p53-heme complexes before (red line) and after infusion of CO. Arrow indicates the spectral shift upon infusion of increasing CO until saturation (cyan line). Inset panel zooms in on the 450–625 nm range.
We next incubated heme with purified human p53 protein (see Figure 3D for purity of the protein) to reconstitute the heme-p53 complex in vitro, using gel filtration in size-exclusion chromatography (SEC) to remove unbound heme. UV-visible (UV-Vis) spectra analysis of the recovered heme-p53 complexes manifested a Soret peak at 413 nm, diagnostic of heme-protein binding (Ponka, 1999) (Figure 3D). The TMB assay again confirmed the presence of heme in the post-SEC heme-p53 complexes. A tryptophan fluorescence quenching assay further indicated that the p53 bound to heme at a 1:1 molecular ratio, with a KD of ~1.20 μM (Figure 3E), suggesting a fairly strong interaction between heme and p53 protein. Consistent results were obtained by surface plasmon resonance analysis (Figure S2D).
A previous study has demonstrated that hemoglobin, NPAS2, E75, or other heme-binding proteins gain gas-sensing properties only upon complexing with heme (Reinking et al., 2005). To test whether heme-p53 complexes also respond to gas, the solution containing the preformed hemin-p53 complex was degassed and Argon purged to maximally remove dissolved oxygen (O2). Subsequent addition of dithionite solution and carbon monoxide (CO) infusion led to substantial blue shifts in the UV-Vis spectra: with a λmax of Soret peak I shifting from 423 to 414 nm and a λmax of Soret peak II from 475 to 535 nm (Figure 3F). Interestingly, nitric oxide (NO) infusion induced distinct changes in the UV spectra of p53-heme, whereas O2 infusion caused little or no change. These data suggest that the heme-binding property of p53 protein might confer gas responsiveness to p53, with specificity comparable to that observed with many other heme-binding proteins (Dioum et al., 2002; Reinking et al., 2005).
The C-Terminal HRM in p53 Protein Is Required for Heme Binding
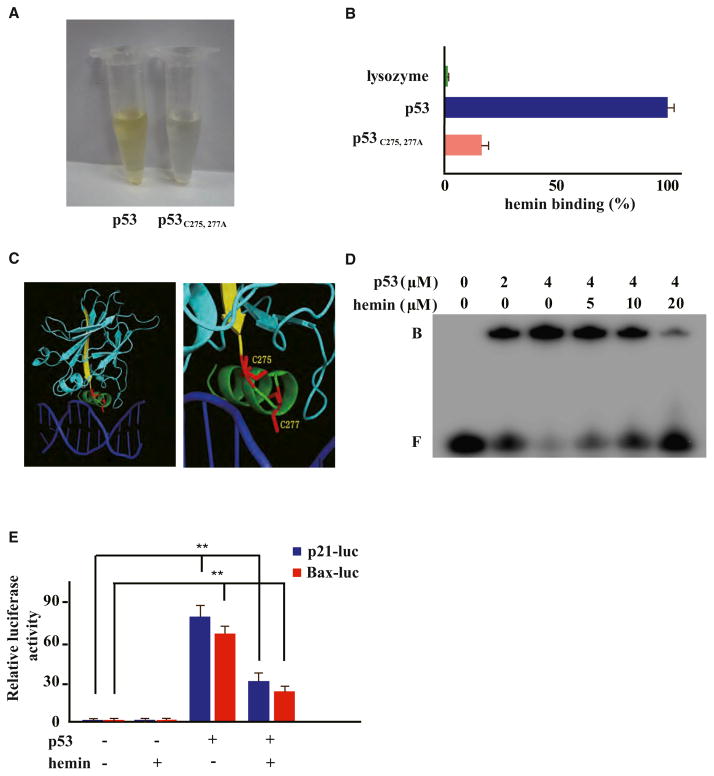
To further dissect the interaction between p53 and heme, we examined the three HRM-like CP motifs that are conserved in p53 (Figures 3A and 3B). Among them, the C-terminal CP motif (Cys277Pro278 in human p53) resides within a redox-sensitive CXC motif at the C terminus of the highly conserved DNA-binding domain (DBD) of p53 protein (Cho et al., 1994; Friedman et al., 1993; Hainaut and Milner, 1993) (Figure 3B). We performed alanine (Ala) substitutions at Cys141-Pro142, Cys176-Pro177, and/or Cys277-Pro278 to generate single or joint mutants of the CP motifs in p53. Both the TMB assay and UV spectra analyses of postgel filtration heme-protein complexes found no significant differences between the wild-type and mutant p53s, regardless of single or joint mutation in any of the three CP motifs (data not shown). Interestingly, Ala substitution of the Cys residues in the C275AC277P stretch manifested in substantially lower heme affinity than wild-type p53 (Figures 4A and 4B). A fluorescence quenching assay further indicated that the corresponding KD value for p53C275, 277A-heme binding rose to ~15.7 μM, over 10-fold higher than that of wild-type p53 (KD ~1.2 μM) (Figure 3E). Meanwhile, mass spectra analysis showed that little or no heme could be detected in association with p53C275, 277A, which was expressed and freshly recovered from E. coli (Figure S2B). This again suggested a weaker binding of the p53C275, 277A mutant to heme that is naturally available in E. coli culture. Altogether, we have identified Cys275 and Cys277 in human p53 protein as two key residues required for p53-heme interaction.
Figure 4. Heme Interacts with Tumor Suppressor p53 through Its C275AC277P Motif and Interferes with p53-DNA Interactions.
(A and B) Recombinant wild-type or p53C275,277A mutant protein (10 μM) was incubated with hemin (50 μM) on ice, followed by gel filtration and TMB assay (Pierce).
(C) A close-up view of the C275 and C277 residues of human p53 in the p53 core domain-DNA (Cho et al., 1994) (derived from PDB 1TSR).
(D) EMSA of the indicated hemin amounts, p53 protein, and double-stranded DNA probes containing p53RE consensus sequences. Band designations are free DNA (F) and bound DNA (B; p53-DNA complex).
(E) HCT116 p53−/− cells transiently expressing HA-tagged wild-type p53 were cotransfected with indicated luciferase reporter plasmids, then treated with or without hemin (10 μM, 6 hr). Error bars indicate ± SEM (n = 3). **p < 0.01.
Heme Interferes with p53-DNA Interaction In Vitro and In Vivo
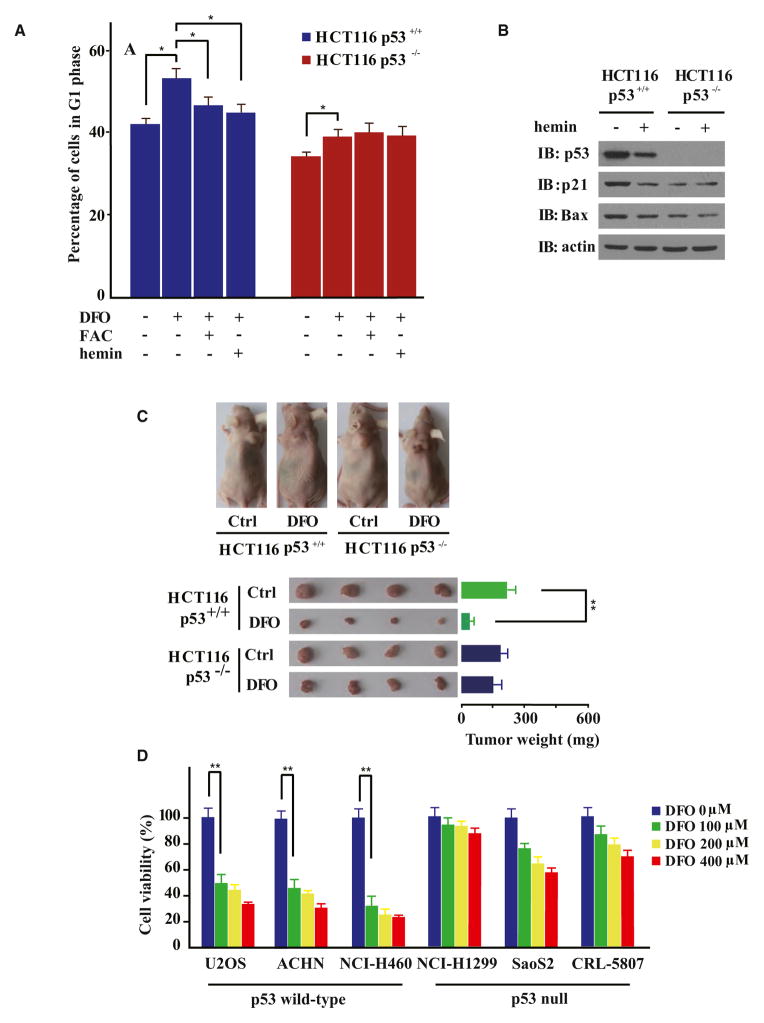
Interestingly, sporadic mutations at Cys275 and Cys277 were found in human patients with cancer (Frebourg et al., 1995) and caused a loss of expression of a p53 transcriptional reporter (Figure S3). We next sought to investigate whether heme-binding would directly affect p53-DNA interactions by assaying the impact of hemin on an electrophoretic mobility shift assay (EMSA) of recombinant p53 protein and double-stranded DNA probes containing the consensus p53-responsive element (p53RE) (Jayaraman and Prives, 1995). Hemin interfered with p53-p53RE interactions in a dose-dependent manner, with 20 μM hemin almost completely abolishing the gel shift (Figure 4D). Hemin, when applied at 10 μM for 6 hr, furthermore appeared to significantly interfere with p53-mediated transcription of luciferase reporters under the control of p21 or Bax promoter-derived p53RE sequences (Figure 4E). Immunoblotting indicated that hemin treatment also led to a reduced expression of endogenous p21 and Bax proteins in a p53-dependent manner (Figure 6B), showing that heme-p53 interactions may potentially play a biological role.
Figure 6. Iron Deprivation Suppresses Multiple Human Tumors in a p53-Dependent Manner.
(A) Flow cytometry analyses of HCT116 p53+/+ or HCT116 p53−/− cells treated with or without DFO (100 μM, 24 hr), FAC (100 μg/ml, 8 hr), or hemin (10 μM, 8 hr) as indicated. Data are represented as mean ± SEM (n = 3). *p < 0.05.
(B) Hemin treatment (10 μM, 6 hr) destabilized p53 and downregulated endogenous p21 and Bax expression in a p53-dependent manner.
(C) Nude mice were subcutaneously seeded with HCT116 p53+/+ and HCT116 p53−/− cells and treated with DFO (500 mg/kg/d over 6 consecutive days). Tumor masses were dissected from treated mice and weighed. Data are represented as mean ± SEM (n = 4). **p < 0.01.
(D) p53 null (NCI-H1299, SaoS2, and CRL-5807) and wild-type p53 (U2OS, ACHN, and NCI-H460) human cancer cell lines were seeded in a 96-well plate at a density of 2 × 103 cells per well and subjected to DFO treatment at indicated concentrations for 72 hr. Cells were stained with Hoechst 33258 for 1 hr, and Hoechst-positive cells were counted in fluorescence images. The data represent at least three independent experiments. Error bars indicate ± SEM. **p < 0.01.
Heme Destabilizes p53 Protein Mainly through the Ub-Proteasome System
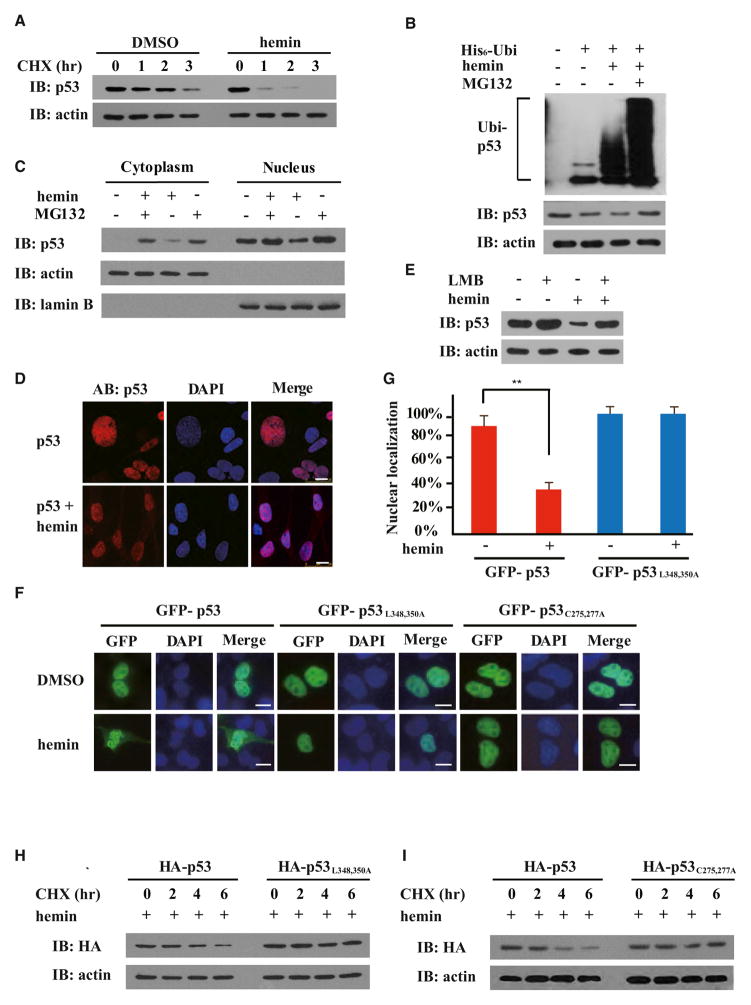
Because we found that the homeostatic level of endogenous p53 protein decreased in a hemin dose-dependent manner (Figures 2A and S1B), we further investigated the potential functional consequences of the heme-p53 interaction in a CHX-chase experiment to examine the stability of endogenous p53 in cells with or without hemin treatment. To this end, the hepatoma cell line HepG2 was first adapted to growth in VP-SFM (8 hr) and subsequently treated with hemin at concentrations comparable to that in previous work (Hu et al., 2008; Wu et al., 2009). In a 3 hr time course experiment, the p53 protein half-life in hemin-treated cells was about 2-fold shorter than that in untreated controls, indicating that hemin treatment accelerated the degradation of p53 in cells (Figures 5A and S4). In addition, hemin-triggered destabilization of p53 protein was concurrent with increased p53 ubiquitylation and could be efficiently blocked by the proteasome inhibitor MG132, but not by the autophagy inhibitor bafilomycin A (BAF), suggesting that hemin-induced p53 degradation occurs mainly through the Ub-proteasome system (Figures 5B and S4).
Figure 5. Heme Destabilizes p53 Protein through Triggering Its Nuclear Export.
Immunoblots (IB) were probed with the indicated antibodies.
(A) HepG2 cells were treated with or without hemin (10 μM, 10 hr) in the presence of CHX (25 μg/ml) for the indicated times.
(B) HepG2 cells transiently expressing His6-tagged Ub (His6-Ubi) were treated with hemin (10 μM), MG132 (10 μM), or both for 6 hr. Endogenous p53 protein was immunoprecipitated with anti-p53 in modified RIPA buffer, followed by immunoblotting with anti-His6.
(C) Cytoplasmic and nuclear fractions were prepared from HepG2 cells treated with or without hemin (10 μM, 6 hr), using actin and lamin B as respective references. Where indicated, 10 μM MG132 was added.
(D) Immunofluorescence microscopy of p53 in HepG2 cells with or without hemin (10 μM, 5 hr).
(E) Hep2G cells were treated with or without LMB (5 ng/ml) and hemin (10 μM).
(F and G) Both GFP-p53L348, 350A (nuclear export sequence mutant) and GFP-p53C275,277A (the p53 mutant impaired in heme binding) were resistant to hemin-induced nuclear export (10 μM, 6 hr). The percentages of cells with predominantly nuclear localization of p53 over the total number of GFP-positive cells in a field of view were quantified in (G). The scale bars represent 10 μm. Data are shown with mean ± SEM (n = 5). **p < 0.01.
(H) HepG2 cells expressing HA-tagged wild-type p53 or HA-tagged p53L348,350A mutant were treated with hemin (10 μM, 6 hr) in the presence of CHX (25 μg/ml) for the indicated times.
(I) HepG2 cells expressing HA-tagged wild-type p53 or HA-tagged p53 C275, 277A mutant were treated with hemin (10 μM, 6 hr) in the presence of CHX (25 μg/ml) for the indicated times.
Heme Triggers Nuclear Export of p53 Protein Involving Its C-Terminal Nuclear Export Sequence
Subcellular localization of p53 protein is critical for regulating its stability because degradation of p53 protein largely takes place in the cytosol (Brooks and Gu, 2011; Liu et al., 2011; Vousden and Prives, 2009). Fractionation experiments indicated that hemin treatment led to p53 protein redistribution in the cell: the level of p53 protein dropped in the nuclear fraction with a concomitant increase in the cytosolic fraction (Figure 5C). Treatment with the proteasome inhibitor MG132 seemed to result in p53 protein accumulation in both cytosolic and nuclear fractions, with or without hemin treatment. Immunofluorescence microscopy directly showed that hemin treatment significantly promoted nuclear export of endogenous or C-terminally GFP-tagged p53 protein in HepG2 cells (Figures 5D and 5G). When hemin was applied to cells pretreated with leptomycin B (LMB), a nuclear export inhibitor that alkylates and inhibits chromosomal region maintenance (CRM1)/exportin 1 (XPO1), hemin-triggered nuclear export and degradation of p53 were both blocked (Figures 5E and S5A), suggesting that hemin-triggered nuclear export of p53 is an upstream event required for heme-mediated p53 destabilization.
Because mammalian p53 proteins contain both N- and C-terminal nuclear export signals (NESs) (Stommel et al., 1999), we introduced amino acid mutations in each NES to determine which NES may be involved in hemin-triggered nuclear export. Although mutations in the N-terminal p53 NES did not affect hemin-induced p53 export in HepG2 cells (data not shown), p53L348,350A, which contains Leu348Ala and Leu350Ala substitutions that inactivate the C-terminal NES in p53, was found to exclusively localize to the nucleus, with or without hemin treatment (Figure 5F). Consistently, p53L348,350A was also resistant to hemin-accelerated degradation (Figure 5H), suggesting that hemin-induced degradation of nuclear p53 protein, if it occurs, contributes little to the hemin-triggered decrease in total cellular p53 protein. Therefore, the C-terminal p53 NES may play a critical role in heme-induced nuclear export of p53. Potentially, heme binding to p53 may unmask the C-terminal p53 NES and promote its interaction with CRM1/XPO1, followed by nuclear export, ubiquitylation, and protea-some-dependent p53 degradation in the cytosol. In further support of this model, glutathione S-transferase (GST)-tagged CRM1 pulled down preconstituted p53-heme complexes more efficiently than p53 protein alone (Figure S5B). In addition, p53C275, 277A, which harbors mutations that disrupt heme binding, was found to be resistant to hemin-induced nuclear export and degradation (Figures 5F and 5I), suggesting that the p53-heme interaction is required for heme-triggered p53 nuclear export and cytosolic degradation.
Iron Deprivation Suppresses Growth and Tumorigenicity of Human Colon Carcinoma Cells in a p53-Depedent Manner
The modulation of iron/heme metabolism has emerged as a major strategy for cancer chemotherapy (Ashcroft et al., 2000; Buss et al., 2004; Liang and Richardson, 2003; Turner et al., 2005). In iron deprivation-based chemotherapy, iron chelators such as DFO suppress the proliferation of multiple types of tumor cells, arrest the cells in the G1/S phase of the cell cycle, and induce apoptosis. Increased levels of p53 protein, and also p63 and p73, were commonly observed upon iron chelation (Ba et al., 2011), but the underlying mechanisms remain unknown.
A past study has indicated that DFO induces G1/S cell-cycle arrest in both HCT116 p53+/+ and HCT116 p53−/− colorectal carcinoma cell lines (Bunz et al., 1998). We applied hemin or iron to DFO-treated cells and only observed rescue from DFO-induced G1/S arrest in HCT116 p53+/+ cells (Figure 6A). Consistent with the hemin-accelerated degradation of p53, addition of hemin was also found to downregulate steady-state levels of p21 and Bax in HCT116 p53+/+ cells, but not in the isogenic p53−/− cells (Figure 6B). Altogether, with our findings that DFO treatment led to p53 protein stabilization in a hemin-reversible manner (Figures 2D and 2G), p53 may mediate at least a portion of the cellular responses to iron or heme metabolism perturbations, likely through modulating the transcription factor activity of p53 and lowering the availability of total p53 protein.
To more directly assess the roles that p53 might play in iron deprivation-based chemotherapy, a tumorigenicity assay was performed with nude mice that bore either HCT116 p53+/+ or p53−/− cells. Both p53+/+ and p53−/− HCT116 cells formed tumors in nude mice with comparable efficiency (Figures 6C and S6). DFO administration (500 mg/kg/d) was found to suppress the growth of tumors formed by p53+/+ HCT116 cells by over 80%. Strikingly, the growth of p53−/− HCT116-derived tumors only decreased by 12% in response to DFO, strongly suggesting that stabilization of p53 might underlie the selective tumor-suppressing effects of iron deprivation in these cells.
Finally, we tested for the tumor-suppressing effects of DFO on six immortal human cancer cell lines that are either p53 null (NCI-H1299, SaoS2, and CRL-5807) or bearing sequence-confirmed wild-type p53 (U2OS, ACHN, and NCI-H460). DFO treatment strongly suppressed proliferation of cancer cell lines expressing wild-type p53 in a DFO dose-dependent manner (over 50%–70% inhibition at 100 μM DFO; approximately 70%–80% inhibition at 400 μM DFO). In contrast, similarly treated p53 null cancer cell lines exhibited no more than 30% inhibition of cell proliferation (Figure 6D). Thus, the presence of endogenous wild-type p53 appears to critically underlie the tumor-suppressing effects of iron deprivation on the cell types examined in this study.
DISCUSSION
Iron is essential for the normal functions of cells, but excess iron has been a strong risk factor for a variety of human malignancies, with mechanisms yet unclear (Torti and Torti, 2013). Given the critical roles that tumor suppressor p53 protein plays in regulating tumorigenesis, cell death, drug responses, as well as cancer cell metabolism (Baker et al., 1989; Maddocks and Vousden, 2011; Tavana and Gu, 2013; Vogelstein et al., 2000), we set out to examine homeostatic levels of p53 protein upon iron excess or deprivation. We found that p53 protein levels in the livers and primary hepatocytes of Hfe−/− mice were significantly lower than that in wild-type mice (Figures 1A–1C and S1A). A similar phenomenon was observed in wild-type mice fed a high iron diet (Figures 1D and 1E). These data strongly suggest that iron excess, a condition of heme overload, leads to downregulation of endogenous p53. The finding that heme directly binds to p53 protein with a micromolar KD further consolidates the direct link between iron metabolism and p53 signaling (Figure 3). The gas-sensing property of the in vitro p53-heme complex (Figures 3F and S2F) has opened the possibility that p53, like other transcription factors, E75 and NPAS2, may at least partially mediate cellular responses to gas signaling if p53 protein indeed binds to heme under normal physiological conditions. Whether this is indeed the case warrants further study.
Our CHX-chase experiments have demonstrated that iron/heme excess downregulates homeostatic levels of p53 protein mainly through accelerating p53 protein degradation. Hemin likely unmasks the C-terminal NES to allow p53 interaction with CRM1/XPO1, nuclear export, and subsequent cytosolic degradation through the Ub-proteasome system (Figures 4 and 5). Mapping out C275AC277P as the heme-interacting region in p53 has provided further molecular detail of the p53-heme interaction (Figure 4). Because C275AC277P resides near the C terminus of the DBD of p53, we also tested and discovered that heme binding interferes with p53-DNA interactions, thus affecting the transcription factor activity of p53 (Figure 4). In particular, the transcription of p21 and Bax, two p53 target genes, was found to be attenuated in cells upon hemin treatment (Figure 6B). Taken together, iron and heme excess may negatively regulate p53 signaling in the afflicted cells through both destabilizing p53 protein and interfering with its binding to the target DNA. Our findings may thus provide mechanistic insight into iron excess-associated tumorigenesis.
Finally, our experiments with HCT116 p53+/+ and p53−/− cells strongly suggested that iron deprivation induces cell-cycle arrest and suppresses tumor formation and cell proliferation in a p53-dependent manner (Figures 6A, 6C, 6D, and S6). As such, the presence of wild-type p53 signaling in tumors might critically affect the clinical outcome of iron deprivation-based therapy. Future clinical studies of iron deprivation-based chemotherapy may choose to monitor the p53 status of patient tumors to determine whether only tumors with wild-type p53 signaling can benefit.
Previous studies have reported on p53-independent tumor-suppressing activities of putative iron chelators (Abeysinghe et al., 2001; Whitnall et al., 2006). Tachpyridine, first synthesized as a potent iron chelator, was reported to induce apoptosis in cultured cancer cells in a manner nonresponsive to p53 overexpression. Tachpyridine induced p53 protein accumulation but did not induce downstream p21 expression. Interestingly, a later study revealed that tachpyridine also chelates zinc ion as efficiently as iron (Zhao et al., 2004) and, thus, potentially deprives p53 of zinc ion, which is necessary for p53’s DNA-binding activities. Thus, tachpyridine may prevent p21 upregulation despite inducing p53 upregulation, explaining the lack of evidence for p53 pathway involvement in tachpyridine’s tumor-suppressing activities. Further study is warranted to investigate the mechanisms underlying reports of p53-independent antitumor effects by other structurally unrelated iron chelators (Whitnall et al., 2006). These previous findings, together with our findings, demonstrate that putative iron chelators, with potentially complex actions perturbing several different pathways, may correlate with iron chelation and p53 upregulation to varying extents. Potentially, cancers, with varying genetic backgrounds within patient populations and across cancer types, may also respond differently to iron chelation depending on the functionality of their individual p53 signaling networks.
The other p53 family proteins, p63 and p73, are upregulated in cell models of iron deprivation-based chemotherapy (Ba et al., 2011) and contain the same CXCP motif we identified to be essential for heme-p53 binding (Figure S7A). Indeed, hemin-agarose affinity chromatography efficiently recovered p63 and p73 proteins overexpressed in HepG2 cells (Figure S7B). Remarkably, hemin also destabilized the p63 and p73 proteins (Figure S7C), reminiscent of our previous observations with p53 protein. Thus, the p53 protein family contains the heme-binding CXCP motif, binds to heme, and undergoes accelerated degradation upon hemin treatment. Conceivably, heme-induced nuclear export and destabilization of p53 family proteins, along with ensuing functional changes, may occur at the organism level. Iron overload and accumulation of heme in afflicted cells or tissues may eventually lead to the production of reactive oxygen species (ROS) and damage intracellular structures including proteins and DNA (Torti and Torti, 2013), where normal function of p53, “the guardian of the genome,” could be essential to mitigating these insults and help cell survival. However, heme-induced nuclear export and destabilization of p53 and other proteins might exacerbate the insults, thus contributing to the pathogenesis of iron excess-associated disorders such as hemochromatosis, e.g., iron excess-promoted tumorigenesis.
In sum, our findings have revealed a direct link between iron/heme homeostasis and the control of the stability, localization, and function of p53 (and potentially the p53 protein family). Therefore, we have not only shed insight on the mechanisms underlying tumorigenesis associated with iron/heme excess but also provided a molecular basis for iron deprivation-based chemotherapy. It would be interesting to explore whether heme-accelerated degradation of p53 might play a role in other pathogenic features of hemochromatosis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
To express human p53 or its mutants bearing Cys to Ala and Pro to Ala substitutions in one or multiple CP motifs, cDNA encoding wild-type or mutant human p53 was cloned into the pHUE vector to obtain proteins with histidine × 6 (His6)-Ub and FLAG tags at their N and C terminus, respectively. Proteins expressed in E. coli were purified, and the N-terminal His6-Ub tag was removed through Usp2CC cleavage as previously described (Hu et al., 2005). GST-tagged CRM1 was constructed and purified following procedures described before (Wu et al., 2013).
Gel Filtration, Spectroscopy Analysis, and TMB Assay of Hemin-Protein Complexes
Purified FLAG-tagged human p53 (10 μM) or its mutants (10 μM), chicken egg white lysozyme (10 μM) or BSA, were incubated with hemin (50 μM) in binding buffer (20 mM HEPES [pH 7.4], 10% glycerol, and150 mM NaCl) for 10 min on ice, then passed through pre-equilibrated NAP-5 columns (GE Healthcare) to remove the unbound hemin as previously described (Hu et al., 2008). UV-Vis spectra analyses of heme only or the heme-protein complexes were carried out on Varian or Hitachi U-3010 spectrophotometers. Hemin content was determined (in triplicate) using the TMB-based assay (Pierce), according to the manufacturer’s instructions.
Animal Manipulations
Hfe knockout mice (Hfe−/−) of C57/Bl/6-129/Ola genetic background were a kind gift from Dr. Nancy C. Andrews (Departments of Medicine and Pediatrics, Harvard Medical School, Boston). All animals were handled according to protocols reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
Measurements of Iron and Heme Contents in Mouse Liver Lysates or Primary Hepatocytes
Experimental iron overload was achieved by feeding 4-week-old B6 wild-type male mice a regular diet supplemented with 8.3 g/kg carbonyl iron (Sigma-Aldrich) for 3 weeks. Liver samples were weighed, and iron content was measured using the unsaturated iron-binding capacity assay as previously described by Ba et al. (2011). Liver samples containing equal amounts of total protein (30 μg) were subjected to the TMB assay to determine hemin content. Isolation of primary hepatocytes from Hfe+/+ and Hfe−/− mice was carried out as described before (Wang et al., 2009). Hemin contents were measured (in triplicate) using the TMB-based assay for hepatocytes from Hfe +/+ versus Hfe−/− mouse littermates.
Preparation of Hemin Solution and Hemin Treatment of Mammalian Cells
Stock hemin (Fe3+ heme) solution was freshly prepared in 0.1 N NaOH for each experiment. The hemin concentration was determined using the extinction coefficient ε385 = 5.84 × 104 M(−1)cm(−1). Mammalian cells were adapted to serum-free growth conditions by growing them in VP-SFM (Invitrogen) as described before (Hu et al., 2008). For cell-based experiments, hemin (freshly prepared in DMSO) at indicated concentrations was added to cells (80%–90% confluence) in VP-SFM (serum-free) medium, with little cytotoxicity observed for the first 12 hr.
Gas-Sensing Assay
The p53 proteins in stock solution were exchanged into the binding buffer (20 mM HEPES, 10% glycerol, and 150 mM NaCl [pH 7.4]) and preserved at reduced status by adding Immobilized TCEP (Tris-2′-carboxyethyl-phosphine) Disulfide Reducing Gel (Thermo Scientific). Immediately before use, the TCEP gels were pelleted to give clear p53 protein solutions. p53 proteins (10 μM) were then incubated with 50 μM hemin in binding buffer on ice for 15 min and desalted using NAP-5 columns to remove free hemin. The solutions (0.5 ml) containing recovered p53-hemin complexes were kept in sealed, airtight cuvettes (VWR) on ice, followed by vacuuming and purging with argon for over 30 min to remove residual oxygen dissolved in the solution. Dithionate (at a final concentration of 1.0 mM in binding buffer) was injected to reduce iron in hemin from the Fe3+ to Fe2+ status. Balloons inflated with CO (Sigma-Aldrich; ≥99.0%), NO (Sigma-Aldrich; 98.5% passed through 1 N KOH), or O2 (Sigma-Aldrich; ≥99.6%) were attached to the cuvettes with long needles inserted into the solutions. UV-Vis spectra were recorded at 1.5 min intervals until no shifts were observed, indicating the maximal formation of p53-heme-gas complexes.
Luciferase Reporter Assay
HCT116 p53−/− cells expressing indicated genes were seeded in 24-well plates and transfected with the luciferase reporter plasmids. Luciferase reporter assays were carried out using a dual luciferase assay kit (Promega). Results were obtained from at least three independent experiments (each in triplicate).
EMSA
EMSA was performed as described before by Jayaraman and Prives (1995) with slight modifications. The double-stranded DNA probes contained the consensus p53RE sequence (5′-AGG CAT GTC TAG GCA TGT CT-3′ and 5′-AGA CAT GCC TAG ACA TGC CT-3′). The probes were labeled with 32P at the 3′ end with T4 ligase (New England Biolabs). Recombinant human p53 protein (4 μM) was then incubated with hemin at increasing concentrations on ice for 30 min to allow for protein-hemin complex formation. After 30 min incubation on ice, the reaction mixtures were subjected to 6% PAGE analysis, resolving the DNA-protein complexes from the free probes. Gels were then dried and exposed to a phosphorimager (Kodak) before scanning on a FLA 9000 Fuji scanner.
Tryptophan Fluorescence Quenching Assay
Fluorescence measurements were performed at room temperature (~23°C) using a Cary Eclipse spectrofluorometer (Varian) as described before (Hu et al., 2008) with slight modifications.
Tumorigenicity Assay
Female nude mice (Bi-kai Biotech) aged 5 weeks old were injected with 1 × 106 HCT116 p53+/+ or HCT116 p53−/− cells suspended in Matrigel (BD Biosciences) as described before by Fasano et al. (1984). Two days after tumor cell seeding, mice bearing evident tumors were randomly divided into control and DFO treatment groups (four mice per group). DFO was dissolved in water and injected into tumors at a dose of 500 mg/kg/d over 6 consecutive days. Animals were euthanized with carbon dioxide, tumor masses were isolated, and tumor weight was measured.
Ubiquitylation Assays
HepG2 cells transfected with His6-tagged Ub plasmids were treated with hemin (10 μM), MG132 (10 μM), or both for 6 hr. Cells were lysed, and a protein ubiquitylation assay was carried out as described previously with slight modifications (Lee et al., 2008).
Flow Cytometry and Fluorescence Microscopy
All procedures were carried out as described before by Pack et al. (2008), with more details in Supplemental Information.
Multiple Sequence Alignments and Protein Data Bank Coordinates
Protein sequence alignment was performed with Clustal X v.2.0 according to the provider’s instruction (Larkin et al., 2007). Coordinates of the p53-DNA complex were extracted from the Protein Data Bank (PDB 1TSR) (Cho et al., 1994) and visualized on PyMOL (DeLano Scientific).
Statistical Analysis
All values are expressed as mean ± SEM. The two-tailed unpaired Student’s t test was used to assess the significance of differences between two data sets, with statistical significance reached when p < 0.05.
Full methods are available in the Supplemental Information.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Alexander Varshavsky (California Institute of Technology) for his generous support, encouragement in initiating the project, and advice on manuscript preparation. The authors are particularly thankful to Dr. Bert Vogelstein (Johns Hopkins University) for his critical reading of the manuscript, generous support, and sharing key reagents. The plasmid encoding GST-CRM1/XPO1 was a gift from Dr. Chuanmao Zhang (Peking University). This work was supported by funding from the National Science Foundation of China to R.H. (31270828 and 31070678) and grants from the Ministry of Science and Technology, China (2010CB912101, 2012CB910800, and 2013CB910900 to R.H. and 2011CB915501 to K.R.). R.H. was also supported by a Sanofi-aventis SIBS Young Investigator award and funding from the Cancer Center of Xuhui Central Hospital (CCR2012003), Shanghai Institute of Neurosciences (SKLN-201206), and Shanghai Municipal Department of Science and Technology (PJ[2010] 00123). We are grateful to Drs. Dangsheng Li and Lin Li (SIBCB), Zhenggang Liu (National Cancer Institute), and Douglas Rees (California Institute of Technology) for helpful discussions. We also thank Dr. Yongguang Gao for his help in image processing.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.02.042.
References
- Abeysinghe RD, Greene BT, Haynes R, Willingham MC, Turner J, Planalp RP, Brechbiel MW, Torti FM, Torti SV. p53-independent apoptosis mediated by tachpyridine, an anti-cancer iron chelator. Carcinogenesis. 2001;22:1607–1614. doi: 10.1093/carcin/22.10.1607. [DOI] [PubMed] [Google Scholar]
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba Q, Hao M, Huang H, Hou J, Ge S, Zhang Z, Yin J, Chu R, Jiang H, Wang F, et al. Iron deprivation suppresses hepato-cellular carcinoma growth in experimental studies. Clin Cancer Res. 2011;17:7625–7633. doi: 10.1158/1078-0432.CCR-10-3099. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nat Chem Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 regulation by ubiquitin. FEBS Lett. 2011;585:2803–2809. doi: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Buss JL, Greene BT, Turner J, Torti FM, Torti SV. Iron chelators in cancer chemotherapy. Curr Top Med Chem. 2004;4:1623–1635. doi: 10.2174/1568026043387269. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Elmberg M, Hultcrantz R, Ekbom A, Brandt L, Olsson S, Olsson R, Lindgren S, Lööf L, Stål P, Wallerstedt S, et al. Cancer risk in patients with hereditary hemochromatosis and in their first-degree relatives. Gastroenterology. 2003;125:1733–1741. doi: 10.1053/j.gastro.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Fasano O, Birnbaum D, Edlund L, Fogh J, Wigler M. New human transforming genes detected by a tumorigenicity assay. Mol Cell Biol. 1984;4:1695–1705. doi: 10.1128/mcb.4.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med. 2012;366:348–359. doi: 10.1056/NEJMra1004967. [DOI] [PubMed] [Google Scholar]
- Frebourg T, Barbier N, Yan YX, Garber JE, Dreyfus M, Fraumeni J, Jr, Li FP, Friend SH. Germ-line p53 mutations in 15 families with Li-Fraumeni syndrome. Am J Hum Genet. 1995;56:608–615. [PMC free article] [PubMed] [Google Scholar]
- Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- Hu RG, Sheng J, Qi X, Xu Z, Takahashi TT, Varshavsky A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- Hu RG, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kato M, Hori H, Ishimori K, Kirisako T, Tokunaga F, Iwai K. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol Cell. 2005;19:171–181. doi: 10.1016/j.molcel.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Jayaraman J, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau tumor suppressor gene and kidney cancer. Clin Cancer Res. 2004;10:6290S–6295S. doi: 10.1158/1078-0432.CCR-sup-040025. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee Y, Lee MJ, Park E, Kang SH, Chung CH, Lee KH, Kim K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SX, Richardson DR. The effect of potent iron chelators on the regulation of p53: examination of the expression, localization and DNA-binding activity of p53 and the transactivation of WAF1. Carcinogenesis. 2003;24:1601–1614. doi: 10.1093/carcin/bgg116. [DOI] [PubMed] [Google Scholar]
- Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med (Berl) 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure KG, Takagi M, Kastan MB. NAD+ modulates p53 DNA binding specificity and function. Mol Cell Biol. 2004;24:9958–9967. doi: 10.1128/MCB.24.22.9958-9967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederau C, Fischer R, Sonnenberg A, Stremmel W, Trampisch HJ, Strohmeyer G. Survival and causes of death in cirrhotic and in noncirrhotic patients with primary hemochromatosis. N Engl J Med. 1985;313:1256–1262. doi: 10.1056/NEJM198511143132004. [DOI] [PubMed] [Google Scholar]
- Osborne NJ, Gurrin LC, Allen KJ, Constantine CC, Delatycki MB, McLaren CE, Gertig DM, Anderson GJ, Southey MC, Olynyk JK, et al. HFE C282Y homozygotes are at increased risk of breast and colorectal cancer. Hepatology. 2010;51:1311–1318. doi: 10.1002/hep.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack M, Trumpfheller C, Thomas D, Park CG, Granelli-Piperno A, Münz C, Steinman RM. DEC-205/CD205+ dendritic cells are abundant in the white pulp of the human spleen, including the border region between the red and white pulp. Immunology. 2008;123:438–446. doi: 10.1111/j.1365-2567.2007.02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrangelo A. Hereditary hemochromatosis: pathogenesis, diagnosis, and treatment. Gastroenterology. 2010;139:393–408. doi: 10.1053/j.gastro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- Radulescu S, Brookes MJ, Salgueiro P, Ridgway RA, McGhee E, Anderson K, Ford SJ, Stones DH, Iqbal TH, Tselepis C, Sansom OJ. Luminal iron levels govern intestinal tumorigenesis after Apc loss in vivo. Cell Rep. 2012;2:270–282. doi: 10.1016/j.celrep.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, Hall C, Uhm S, Mathew MK, Fleming MD, Paw BH, et al. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature. 2008;453:1127–1131. doi: 10.1038/nature06934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Rouault TA. Linking physiological functions of iron. Nat Chem Biol. 2005;1:193–194. doi: 10.1038/nchembio0905-193. [DOI] [PubMed] [Google Scholar]
- Simcox JA, McClain DA. Iron and diabetes risk. Cell Metab. 2013;17:329–341. doi: 10.1016/j.cmet.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana O, Gu W. The Hunger Games: p53 regulates metabolism upon serine starvation. Cell Metab. 2013;17:159–161. doi: 10.1016/j.cmet.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 2009;100:9–16. doi: 10.1111/j.1349-7006.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Koumenis C, Kute TE, Planalp RP, Brechbiel MW, Beardsley D, Cody B, Brown KD, Torti FM, Torti SV. Tachpyridine, a metal chelator, induces G2 cell-cycle arrest, activates checkpoint kinases, and sensitizes cells to ionizing radiation. Blood. 2005;106:3191–3199. doi: 10.1182/blood-2005-03-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang Q, Jiang L, Wang J, Li S, Yu Y, You J, Zeng R, Gao X, Rui L, Li W, Liu Y. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. 2009;49:1166–1175. doi: 10.1002/hep.22774. [DOI] [PubMed] [Google Scholar]
- Weiss G. Genetic mechanisms and modifying factors in hereditary hemochromatosis. Nat Rev Gastroenterol Hepatol. 2010;7:50–58. doi: 10.1038/nrgastro.2009.201. [DOI] [PubMed] [Google Scholar]
- Whitnall M, Howard J, Ponka P, Richardson DR. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc Natl Acad Sci USA. 2006;103:14901–14906. doi: 10.1073/pnas.0604979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Jiang Q, Clarke PR, Zhang C. Phosphorylation of Crm1 by CDK1-cyclin-B promotes Ran-dependent mitotic spindle assembly. J Cell Sci. 2013;126:3417–3428. doi: 10.1242/jcs.126854. [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Terai S, Sakaida I. Deferoxamine for advanced hepatocellular carcinoma. N Engl J Med. 2011;365:576–578. doi: 10.1056/NEJMc1105726. [DOI] [PubMed] [Google Scholar]
- Yang F, Xia XA, Lei HY, Wang ED. Hemin binds to human cytoplasmic arginyl-tRNA synthetase and inhibits its catalytic activity. J Biol Chem. 2010;285:39437–39446. doi: 10.1074/jbc.M110.159913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-er-balpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Planalp RP, Ma R, Greene BT, Jones BT, Brechbiel MW, Torti FM, Torti SV. Role of zinc and iron chelation in apoptosis mediated by tachpyridine, an anti-cancer iron chelator. Biochem Pharmacol. 2004;67:1677–1688. doi: 10.1016/j.bcp.2003.12.036. [DOI] [PubMed] [Google Scholar]
- Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE, et al. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.