SUMMARY
Beach sand is a habitat that supports many microbes, including viruses, bacteria, fungi and protozoa (micropsammon). The apparently inhospitable conditions of beach sand environments belie the thriving communities found there. Physical factors, such as water availability and protection from insolation; biological factors, such as competition, predation, and biofilm formation; and nutrient availability all contribute to the characteristics of the micropsammon. Sand microbial communities include autochthonous species/phylotypes indigenous to the environment. Allochthonous microbes, including fecal indicator bacteria (FIB) and waterborne pathogens, are deposited via waves, runoff, air, or animals. The fate of these microbes ranges from death, to transient persistence and/or replication, to establishment of thriving populations (naturalization) and integration in the autochthonous community. Transport of the micropsammon within the habitat occurs both horizontally across the beach, and vertically from the sand surface and ground water table, as well as at various scales including interstitial flow within sand pores, sediment transport for particle-associated microbes, and the large-scale processes of wave action and terrestrial runoff. The concept of beach sand as a microbial habitat and reservoir of FIB and pathogens has begun to influence our thinking about human health effects associated with sand exposure and recreational water use. A variety of pathogens have been reported from beach sands, and recent epidemiology studies have found some evidence of health risks associated with sand exposure. Persistent or replicating populations of FIB and enteric pathogens have consequences for watershed/beach management strategies and regulatory standards for safe beaches. This review summarizes our understanding of the community structure, ecology, fate, transport, and public health implications of microbes in beach sand. It concludes with recommendations for future work in this vastly under-studied area.
Keywords: beach sand, fecal indicator bacteria, psammon, pathogens, fate, water quality
INTRODUCTION
The organisms inhabiting supratidal and intertidal (also called supralittoral) sands, and those located just above the margin of a water body have historically been termed the psammon (Neel 1948). The psammon can be divided by relative size. The macropsammon is perhaps the most familiar to the beachgoer in the form of mollusks, annelids, and crustaceans. Less familiar are the meiopsammon which are near-microscopic animals that are often collectors, grazers and predators (e.g. copepods, nematodes, and flatworms). Even less understood are the sand dwelling microbes or the micropsammon - the topic of this review. Here, we restrict our discussion to the micropsammon that inhabit the area at the margin or just above the margin of a water body including the intertidal areas of marine environments, the supratidal/supralitoral areas of marine or freshwater beaches, respectively, and the swash zone.
Despite the familiarity of sand as a defining characteristic for many beaches around the world, surprisingly little is known about the micropsammon. Very recently, the micropsammon has received some attention in terms of composition, community structure, ecology and human health implications; however, these areas are often treated separately. An integrative approach that considers both the physical and biological components of these unique ecosystems, which in turn provides the basis for inferences about individual pathogens and health effects for humans, is required to understand the implications of the micropsammon to human health. In this review, we initiate the process of integrating knowledge from these realms.
Some discussion of the terms used in this article will be useful to the reader. We limit our discussion to exposed or unsubmerged sand sediment including the swash zone (area of wave run-up and return), and the intertidal zone (between the high tide and low tide marks) (Figure 1). “Fate” was used as early as 1915 to describes bacterial survival in the face of environmental stressors (Weinzirl and Newton 1915). We use fate as a general term to include the many happenstances that may befall a microbial population in the environment, including population replication, prolonged persistence, transport, and death. The autochthonous microbial community consists of the microbes that are native to the sand habitat, while allochthonous microbes are those that are contributed from external sources (e.g. animals defecating on the beach; people swimming in the water; atmospheric deposition). In the review that follows, public health implications of beach sand microbes are couched in the context of the environment and microbial community around them. Particular emphasis is placed on evaluating the possibility of monitoring beach sand to assess possible health risks and as a means to better predict the microbiological safety of recreational waters.
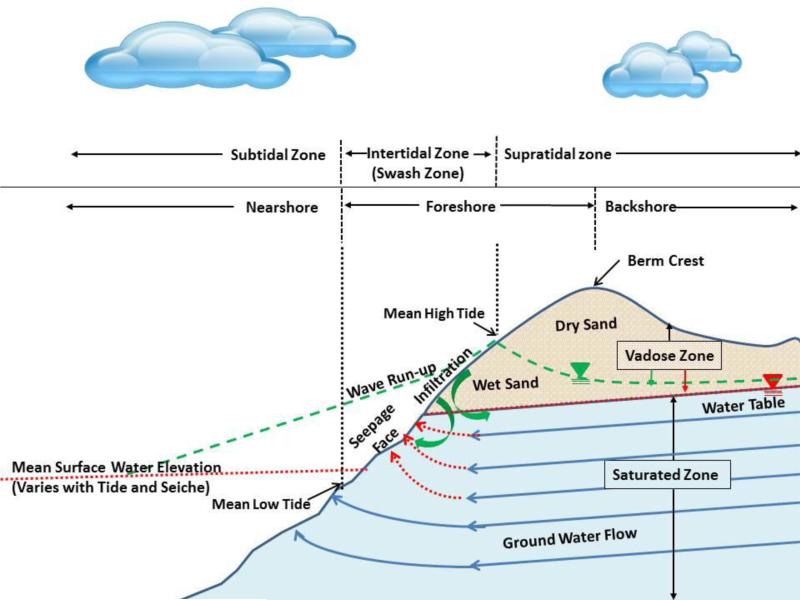
Figure 1.
Beach morphology emphasizing the wave impacted shoreline including the fresh water definition of the foreshore and marine water definition of the intertidal zone. This figure illustrates the seepage face for times when the mean surface water elevation is below the groundwater table (shown by red dotted lines) and also illustrates infiltration that occurs when the surface water level rises above the groundwater table (shown by green dashed lines) as typically occurs during wave run-up. The inverted triangles mark the lines that define the water table for each of these conditions.
SAND MICROBIAL COMMUNITIES
Microbial Community Characteristics
Microbial communities in the sand micropsammon have received relatively little attention compared to those in soil, water, and bottom/submerged sediment. Clearly, bacteria and fungi can proliferate in sand, e.g. direct microscopic counts found greater than 107 total bacteria/g sand (Khiyama and Makemson 1973), and the concentration of culturable fungi isolated from sand ranged from 1.5 to 7.6 × 106 CFU/g (Larrondo and Calvo 1989) at 42 Mediterranean beaches. Studies focused on community analysis found Proteobacteria and Bacteroidetes dominated biofilm-associated communities in supratidal sands from South Florida beaches (Piggot et al. 2012), and community structure varied by location (supratidal, intertidal, or subtidal). Metagenomic studies on microbial communities in the environment have focused on habitats such as the water column or sediments e.g. (Lozupone and Knight 2007), although the 2010 Deepwater Horizon oil spill in the Gulf of Mexico resulted in a study that generated some data on bacterial communities in beach sand (Kostka et al. 2011). The concentration of bacterial 16S rRNA genes in non-oil impacted sand was ~107 copies/g. Members of the Gram-negative Gammaproteobacteria were observed most frequently (33% of samples), but sequences from the phylum Bacteroidetes (14%) and order Chromatiales (10%) were also identified in sand. Analysis of sand microbial communities in Hawaii found greater bacterial diversity in backshore sand compared to foreshore sand, nearshore sand, and water (Cui et al. 2013). Pseudomonas spp. and Bacteroidetes were among the dominant taxa identified.
The authors (Sadowsky and C. Staley) have recently completed some metagenomic analyses on the sand microbiome. 16S rDNA analysis was performed on sand taken from three sites: an estuarine beach in Tampa, FL; a freshwater lake in Saint Paul, MN; and a marine site in Tampa, FL. The most abundant phyla among all three sites were Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria. The most abundant families at all sites included Rhodobacteraceae, Flavobacteriaceae, Flammeovirgaceae, and Campylobacteraceae. Alpha diversity was high among all sites; however, sand from the marine site had considerably greater richness and higher non-parametric diversity indices than the other sites. The microbial community in each sample was distinct via principal coordinate analysis, and analysis of molecular variance (AMOVA) revealed significant differences in microbial community structure among all sites (P < 0.001).
Sources of Allochthonous Microbes to Sand Ecosystems
Many of the microbes found in sand are autochthonous and are adapted to life in sand microbial communities. Allochthonous microbes, introduced from outside the control volume boundary, may include FIB (E. coli, fecal coliforms and enterococci) and pathogens derived from sewage or direct fecal deposition by animals. The source of allochthonous bacteria to sand ecosystems is important from both ecological and public health perspectives, as the pathogens associated with fecal material differ depending upon the host source. The taxa and concentration of microbes in sand are undoubtedly influenced by a myriad of factors, moisture, nutrient availability and composition, physical habitat and nature of the microbial community.
The fate of allochthonous microbes in sand can follow several pathways, which are outlined in Figure 2. Many will die within hours of introduction to sand habitats, however some persist with no or minimal replication for days to months due to permissive conditions and/or their physiological capabilities. A subset of these microbes may establish replicating populations, at which point they are considered “naturalized.” If the naturalized microbes establish long-term, replicating populations, they may be considered part of the autochthonous microbial community. Examples of this process include E. coli populations that reproduce in extra-intestinal habitats such as soil (Byappanahalli and Fujioka 2004; Byappanahalli and Fujioka 1998) and periphyton (Ksoll et al. 2007), stranded algae (Badgley et al. 2011; Byappanahalli et al. 2003b; Olapade et al. 2006; Vanden Heuvel et al. 2010; Whitman et al. 2003), pitcher plants (Whitman et al. 2005) and plankton-amended sand (Byappanahalli et al. 2006b), enterococci populations associated with seaweed from marsh (Grant et al. 2001), and a ubiquitous, persistent Enterococcus casseliflavus strain isolated from water, sediment, and submerged aquatic vegetation in a Florida lake (Badgley et al. 2010).
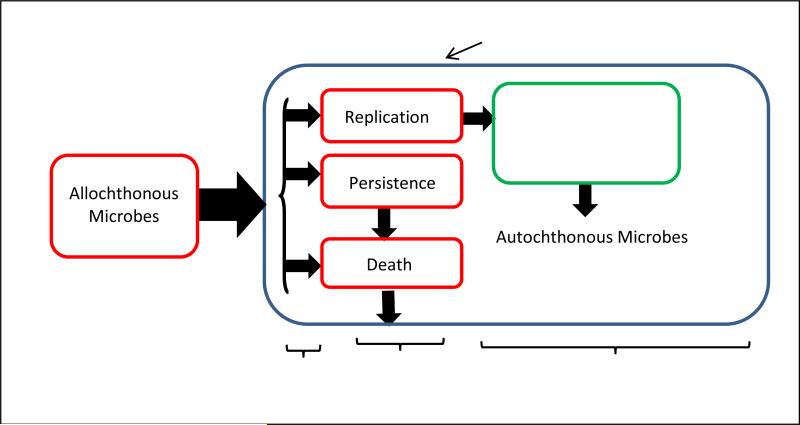
Figure 2.
Fate of allochthonous microbes following introduction into sand habitats. Microbes may die rapidly, persist for days or months with no or minimal growth, or they may form replicating populations, in which case they are “naturalized.” If these naturalized populations are permanently established, they become part of the autochthonous microbial community.
Fecal-derived microbes can reach beach sand via many sources, including direct fecal deposition on sand (e.g. shore birds, dogs) (Kinzelman et al. 2008; Noble et al. 2006), point source (wastewater) pollution to water (Vijayavel et al. 2010) that is subsequently transmitted to sand, and from non-point source pollution that is discharged directly to sand (e.g., stormwater and contaminated groundwater) (Salmore et al. 2006; Sauer et al. 2011; Zhu et al. 2011), or is discharged to water and then transmitted to sand (Piggot et al. 2012) (Table 1). Landscape factors within the watershed can influence fecal indicator bacteria concentrations in source waters and at beaches, e.g. forested headwaters can be a source of fecal indicator bacteria to bathing waters downstream in subtropical and temperate environments (Byappanahalli et al. 2003a; Dunkell et al. 2011; Flood et al. 2011; Frenzel and Couvillion 2002; Fujioka et al. 1988; Mallin et al. 2000; Whitman et al. 2006). Several studies have shown that the degree of urbanization within a watershed is the strongest predictor of fecal indicator abundance, although not necessarily indicative of human fecal pollution (Flood et al. 2011), because impervious surfaces can concentrate runoff laden with fecal indicators from numerous sources.
Table 1.
Examples of sources of FIB and pathogens to swimming water and sand. Note dw = dry weight and ww = wet weight.
| Source | Microbe | Observed Concentrations | Citation |
|---|---|---|---|
| Studies That Specifically Evaluated Sand | |||
| Freshwater Cladophora | Escherichia coli | 1.0 x 108 CFU/g dw of Cladophora | (Whitman et al. 2003) |
| Marine kelp |
Escherichia coli Enterococci |
~1 x 100 to 1 x 104 CFU/g dw of kelp ~3.2 x 100 to 5.6 x 103 CFU/g dw of kelp |
(Imamura et al. 2011) |
| Plankton | Escherichia coli | ~1.0 x 103 to 1.0 x 105 CFU/100g of sand mixed with plankton | (Byappanahalli et al. 2006b) |
| Detritus |
Escherichia coli Enterococci |
3.2 x 102 to 1.8 x 103 MPN/100 ml of detritus mixed with water 3.0 x 101 to 1.0 x 102 CFU/100 ml of detritus mixed with water |
(Haack et al. 2003) |
| Gulls |
Escherichia coli Enterococci |
1.0 x 105 to 1.0 x 109 CFU/g ww of feces 1.0 x 104 to 1.0 x 108 CFU/g ww of feces |
(Fogarty et al. 2003) |
| Geese |
Escherichia coli Enterococci |
4.2 x 103 MPN/g ww of feces 5.0 x 102 CFU/g ww of feces |
(Haack et al. 2003) |
| Pigeons | Fecal coliforms Escherichia coli Enterococci |
1.6x 108 CFU/g of feces 1.7x 108 CFU/g of feces 4 x 105 CFU/g of feces |
(Oshiro and Fujioka 1995) |
| Dogs | Enterococci | 3.9 x 107 CFU/g dw of feces | (Wright et al. 2009) |
| Beach sand | Fecal coliforms Total streptococci Fecal streptococci Clostridium perfringens Pseudomonas aeruginosa Escherichia coli Enterococci Aeromonas Yeasts Fungi Dermatophytes |
3x 102 to 2.4 x 104 CFU/g of sand 4x 100 to 1.1 x 107 CFU/g of sand 4 x 100 to 1.1 x 106 CFU/g of sand 1.4 x 101 to 1.1 x 107 CFU/g of sand 8 x 100 to 2.4 x 107 CFU/g of sand 1.1 x 104 CFU/ 100ml of elutriated sand ~1 × 102 to 1 × 103 CFU/100g dw of sand <4 × 100 to 1.6 × 105 CFU/100g of sand 9 × 100 to 7.2 × 103 CFU/ 100g dw of sand 1.1 × 103 to 9.3 × 105 cell equivalents/100 ml of sand pore water 8.7 × 100 CFU/g of sand 4.5 × 100 CFU/g of sand 1.7 × 100 CFU/g of sand |
(Mendes et al. 1993) (Whitman and Nevers 2003) (Alm et al. 2003) (Oshiro and Fujioka 1995) (Yamahara et al. 2007) (Khan et al. 2009) (Sabino et al. 2011b) |
| Riparian sands | Escherichia coli | 1.0 × 103 MPN/100 ml of elutriated sand | (Byappanahalli et al. 2003a) |
| Littoral water | Escherichia coli | 126 CFU/100 ml of lake water | (Ge et al. 2012a) |
| Studies that Describe Additional FIB Sources that Can Impact Sand | |||
| Lyngbya |
Escherichia coli Enterococci Clostridium perfringens |
3.2 × 103 MPN/g dw of Lyngbya 1.6 × 103 MPN/g dw of Lyngbya 1.6 × 103 MPN/g dw of Lyngbya |
(Vijayavel et al. 2013) |
| Hydrilla | Enterococci | 8.6 × 102 CFU/ 100 g ww of submerged aquatic vegetation | (Badgley et al. 2010) |
| Ducks | Enterococci | 1.5 × 104 to 7.9 × 106 CFU/g ww of feces | (Anderson et al. 1997) |
| Riparian soils |
Escherichia coli Enterococci Clostridium perfringens Total Vibrio Fecal coliforms Fecal streptococci |
1.7 × 103 to 2.4 × 105 CFU/g dw of soil 1 × 102 CFU/g dw of soil 6 × 102 CFU/g dw of soil 2.5 × 105 CFU/100 g ww of soil 1.4 × 104 CFU/100g ww of soil 9.5 × 104 CFU/100g ww of soil |
(Hardina and Fujioka 1991) (Desmarais et al. 2002) (Cui et al. 2013) (Elmanama et al. 2005) |
| Aerosols | Heterotrophic bacteria Hemolitic bacteria Staphylococci Escherichia coli Aeromonas hydrophila Pseudomonas Mesophilic bacteria Psychrophilic bacteria Microfungi |
3.8 × 105CFU/m3 of air 4.8 × 105 CFU/m3 of air 8.5 × 103 CFU/m3 of air 1.4 × 103 CFU/m3 of air 5.6 × 104 CFU/m3 of air 1.3 × 105 CFU/m3 or air 3.0 × 103 CFU/m3 of air 3.2 × 103 CFU/m3 of air 2.5 × 103 CFU/m3 of air |
(Filipkowska et al. 2000) (Grisoli et al. 2009) |
| Storm water |
Escherichia coli
Enterococci |
1.0 × 105 MPN/CFU/100 ml of storm water 1.0 × 103 to 1.0 × 105 MPN/100 ml of storm water |
(Marsalek and Rochfort 2004) (Tiefenthaler et al. 2011) |
| Bathers shedding |
Enterococci
Staphylococcus aureus |
6.0 × 105 CFU/ person per 15 minute swim 6.3 × 105 CFU/ person per 15 minute swim |
(Elmir et al. 2007) (Plano et al. 2011) |
| Discharge from boats | Fecal coliforms Escherichia coli Enterococci |
1.0 × 108 CFU/100 ml of graywater 1.3 × 107 CFU/100 ml of graywater 4.9 × 106 CFU/100 ml of graywater |
(US EPA 2008) |
Wildlife can significantly contribute to the fecal bacteria population within water and soils of a watershed (Alderisio and DeLuca 1999; Hussong et al. 1979; Lévesque et al. 1993), and even in an urbanized watershed the wildlife has been documented as a dominant source of bacteria during rain events (Whitlock et al. 2002). In some cases, the input from specific wildlife in the watershed has been implicated in the contamination of beaches (Oshiro and Fujioka 1995), and molecular methods have enabled the identification of specific wildlife sources that have the greatest impact at beaches e.g. (Hansen et al. 2011). Fecal indicator bacteria from different animal sources may differentially persist in waters and sediments (Anderson et al. 2005), adding another layer of complexity to pollution events at beaches when there are diverse sources within the watershed.
Sources of FIB in sand have been inferred in the absence of direct evidence for a particular contaminant source (Table 1). By measuring the concentration of enterococci in dog, shore bird, shrimp and human waste and incorporating the number of individuals observed per unit time at the beach, dogs were estimated to be the greatest contributors to enterococci levels at one study beach (Wright et al. 2009). Whitman and Nevers (2003) found the number of gulls on a beach on one day was correlated with E. coli concentrations in foreshore sand and beach water on the following day. In Florida, bird counts and enterococci levels were correlated in subtidal sands, but not in supratidal or intertidal regions (Piggot et al. 2012). Microbial source tracking (MST) studies have provided more direct evidence of the source of FIB in beach sand. Edge and Hill (2007) and Edge et al. (2010) applied multiple lines of evidence, including observations of fecal droppings, and E. coli DNA fingerprinting and antimicrobial resistance analyses, to identify birds (e.g. Canada geese and gulls) as the predominant source of E. coli in sand at Lake Ontario beaches. Humans and waterfowl were found to be the main contributors to E. coli concentrations in sand in other studies (Fogarty et al. 2003; Ishii et al. 2007). Bonilla et al. (2007) showed that one gull dropping caused elevated enterococci levels in sand over an area of 3 m2.
Bird feces may also be important sources of pathogens to beach sand. Preliminary surveillance for pathogens in beach sand at the Lake Ontario beach predominantly impacted by bird fecal droppings (Edge and Hill 2007) commonly detected Campylobacter (Khan et al. 2013)(. Salmonella genomic analysis showed close association between isolates from gulls, sand and adjacent swimming water (Whitman et al. 2001). In some cases, humans themselves have been implicated as sources of microbes for sand (Elmir et al. 2009; Graczyk et al. 2007). Staphylococcus aureus and yeasts associated with human hosts in sand were significantly correlated with human activity at a Mediterranean beach (Papadakis et al. 1997).
3. FATE, ECOLOGY AND POPULATION BIOLOGY/GENETICS
Fate (replication, persistence, and death) of the micropsammon is influenced by factors that are extrinsic (e.g. physical-chemical stressors, nutrient and water availability, competition, predation) and intrinsic (e.g. microbial species or strain) to the many microbes that inhabit beach sand, either transiently or consistently. Although study of the entire micropsammon would be most useful, much of the work on microbial fate in the context of sandy beaches has focused on FIB. Conventional wisdom was that upon release to the environment, indicator bacteria would die off at some undetermined rate; yet Ostrolenk et al. (1947) noted that E. coli might be an inferior indicator of sanitary conditions due to the possibility of multiplication outside the host gastrointestinal tract. As early as 1967, researchers obtained evidence of fecal coliform replication in soil following rainfall (Van Donsel et al. 1967). More recently, evidence has steadily accumulated that certain E. coli and Enterococcus phylotypes can replicate in the environment (reviewed in (Byappanahalli et al. 2012a; Ishii and Sadowsky 2008).
Examination of the occurrence and persistence of FIB and pathogens in beach sands is an extension of the early work that demonstrated that lake and river bottom sediments were a reservoir of FIB (Burton et al. 1987; Davies et al. 1995; Francy and Darner 1998; LaLiberte and Grimes 1982; Obiri-Danso and Jones 1999). Some of the earliest reports on the persistence of FIB in shoreline sands of freshwater beaches came from studies on the Laurentian Great Lakes (Alm et al. 2003; Francy et al. 2003; Haack et al. 2003; Whitman et al. 2001; Whitman and Nevers 2003). These studies documented FIB in sand at densities that were orders of magnitude higher than in water at the same beaches. Persistent FIB have been reported in submerged, foreshore, and backshore sand (Byappanahalli et al. 2006b; Whitman and Nevers 2003; Zehms et al. 2008), including those in cold northern environments (Ishii et al. 2007).
The evolution of thought about the replication potential of FIB that occupy “secondary” habitats (e.g. sand, water, soil) is worthy of consideration here, as it impacts the conceptualization of their role in the sand microbial community. The tropical soils of Hawaii and Guam were an early focus of research on the replication of FIB in secondary habitats (Byappanahalli and Fujioka 1998; Byappanahalli et al. 2012b; Fujioka et al. 1999; Fujioka 2001; Hardina and Fujioka 1991). E. coli was shown to replicate in soil collected from south Florida river banks (Solo-Gabriele et al. 2000). A 2003 workshop consensus concluded that FIB can multiply and persist in soil, sediment, and water in some tropical/subtropical environments (Hawaii, Guam, Puerto Rico, south Florida) (Fujioka and Byappanahalli 2003). Numerous studies have since demonstrated this phenomenon, even in temperate soils that experience wide seasonal variability in temperature (Brennan et al. 2010; Byappanahalli et al. 2006a; Ishii et al. 2006). E. coli and enterococci have since been shown to grow in such diverse habitats as marine and freshwater macrophytic algae (Whitman et al. 2003), periphyton (Ksoll et al. 2007), plankton-amended sand (Byappanahalli et al. 2006b), bromeliads (Bermudez and Hazen 1988; Rivera et al. 1988), pitcher plants (Whitman et al. 2005), pulp mill waste (Gauthier and Archibald 2001), Australian reservoir (Ashbolt et al. 1997), soils (Byappanahalli et al. 2003a; Ishii et al. 2006), and silt (Solo-Gabriele et al. 2000). These works and others challenged the paradigm that FIB in secondary habitats such as sand are always primarily of fecal origin.
Alm et al. (2006) showed that in autoclaved mesocosm sand studies, E. coli grew at 19° C from 2 CFU/g to over 2 × 105 CFU/g sand in 48 hr and persisted at that level for 35 days. In situ diffusion studies showed persistence of culturable E. coli at 5 logs MPN/100 g in Lake Huron beach sands for 45 days. Lee et al. (2006) showed remarkable replication in both overlying water and autoclaved sand in microcosm experiments suggesting that enclosed beaches favored increased FIB replication. Wetting and drying of sand was found very important to replication of FIB in marine beaches with a doubling time of 1.1 to 3.1 per day (Yamahara et al. 2009). Evidence for autochthonous FIB replication is more difficult due to multiple in situ sources and variation in nature. Nonetheless circumstantial evidence supports multiplication in sand. Whitman et al. (2003) monitored FIB in upland beach sand before and after replenishment and found that E. coli returned to its former concentration (104 MPN/100 g) within 2 weeks. Despite recurring foreshore removal by storms, Whitman and Nevers (2003) were able to demonstrate population homeostasis of E. coli in foreshore sands (4-5 log MPN/g), compared to much wider variation in submerged sands and at various water depths.
Genotyping of E. coli populations in human feces and septic systems revealed distinct populations in the two environments (Gordon et al. 2002), leading the authors to conclude that certain E. coli types are better adapted to survival in secondary habitats than others. Later work demonstrated that encapsulated E. coli were capable of replicating bloom proportions in two Australian lakes, leading the authors to propose that these strains are capable of a “free-living” lifestyle (Power et al. 2005). Work conducted in temperate soils and other secondary habitats shows certain E. coli genotypes, termed “naturalized,” to be capable of replication in extra-intestinal habitats (Ishii et al. 2007; Ishii and Sadowsky 2008).
Abiotic Factors that Influence Fate
Many environmental factors influence the fate of microbes in sand, including abiotic factors such as moisture, temperature, sunlight, and nutrients, and biotic factors such as competition, and predation. Some of these factors have been explored in beach sand, while for others the effect must be inferred from other environments.
Moisture and Rainfall. Water activity (aw), or the availability of free water molecules, is a critical life requirement for microbes (Atlas and Bartha 1997). Most bacteria prefer aw of 0.97 or above; however, bacteria such as Staphylococcus spp. can grow at aw of 0.85, and halophiles such as the archaeon Halobacterium tolerate aw 0.75. Some fungi are even more xerotolerant, growing at 0.60 (Atlas and Bartha 1997). Production of organic solutes such as trehalose may mediate resistance to desiccation in E. coli strains adapted to survive in soils and sand (Zhang and Yan 2012). Mika et al. (2009) found that desiccation was a potent inactivating factor for E. coli, but not enterococci, in sewage-contaminated sand. However, more water is not always better; e.g. Solo-Gabriele et al. (2000) found that soil hydrated to 14% moisture with brackish water harbored higher E. coli concentrations than soil with 34% moisture. Differential tolerance to desiccation was observed for FIB in soils under laboratory conditions (25°C), where E. coli levels decreased markedly in response to decreasing moisture, while enterococci levels remained relatively consistent (Byappanahalli and Fujioka 2004).
Moisture content of sand varies widely depending upon factors such as location on the beach, grain size, and depth to the water table. In beach sand, water is located in the interstitial spaces between sand grains (pore water). Foreshore sand, nearest the water, generally has reported moisture content between 12 and 25% (Alm et al. 2003; Beversdorf et al. 2007; Ishii et al. 2007; Sampson et al. 2006; Whitman and Nevers 2003). Average moisture content in sand at a Florida marine beach was 8.4% for dry, backshore sand, 20.4% for wet sand, and 24.7% for water-inundated sand (Shah et al. 2011). Microbial levels in unsaturated sands may experience more variability due to moisture fluctuation compared to microbes below the water table that inhabit a consistently moist environment.
FIB have been recovered from all areas of beaches, ranging from relatively dry backshore sand to the moist sand in the swash/intertidal zone (Wright et al. 2011), and at depths ranging from the surface to the water table. In the study described above (Shah et al. 2011), an inverse correlation was found between FIB (e.g. enterococci, fecal coliforms, E. coli) and moisture content, indicating that ~8% is enough moisture to promote survival of bacteria, yeasts, and nematodes. In general, wet foreshore sand at freshwater beaches contains a greater density of FIB than sand submerged under lake water or dry backshore sand (Beversdorf et al. 2007; Whitman and Nevers 2003; Zehms et al. 2008). However, three studies at a marine beach in Florida found higher concentrations of enterococci or E. coli in supratidal sand, above the high tide mark, compared to sands with higher moisture content in the intertidal zone (Abdelzaher et al. 2010; Enns et al. 2012; Phillips et al. 2011a). One of their explanatory hypotheses for this result was that protozoan predators may not survive well in dryer sands, leading to greater survival of enterococci.
Another study showed that when seawater was added to sand collected from the supratidal zone, enterococci replication occurred as measured by either culturable or quantitative PCR (qPCR) methods (Yamahara et al. 2009). Rainfall can also produce a large increase in culturable E. coli in sand (Beversdorf et al. 2007; Kleinheinz et al. 2009; Sinigalliano et al. 2007); however, neither antecedent rainfall nor moisture was correlated with enterococci concentrations in sand in a study of several Florida beaches (Piggot et al. 2012). Several hypotheses, which are not mutually exclusive, can be advanced to explain the positive response of sand-dwelling FIB to rainfall: (1) rainfall may transport microbes from the watershed to the sand; (2) microbes may be resuscitated from a viable but non-culturable state when moisture increases; or (3) the microbes may multiply in response to increased moisture. At a specific beach, the zone with the highest densities of FIB may be the one where the moisture content of the sand is within the optimal range to support either persistence or replication. Alternatively, the moisture content of the sand may influence protozoa that graze on bacteria, leading to greater FIB levels in zones where the moisture content is not suitable for protozoa. Clearly, the complex relationship between moisture and microbial levels in sand is not well understood.
Sunlight Irradiation. The damaging wavelengths of sunlight, particularly those in the ultraviolet (UV) range below 300 nm, contribute to microbial inactivation in aquatic environments (Davies-Colley et al. 1994; Romero et al. 2011). Although short-wavelength UVC light is the most microbicidal, this wavelength is effectively absorbed by ozone and other constituents of the atmosphere (http://www.who.int/uv/uv_and_health/en/). In contrast, UVB light (280-320 nm) directly damages the genome (Schuch and Menck 2010; Sutherland 1981). UVA radiation and full-spectrum sunlight are also damaging, particularly when coupled with exogenous activators such as humic acids (Romero et al. 2011). Whitman and co-workers (2004) determined that E. coli levels in Lake Michigan were higher in the morning and on cloudy days compared to the afternoon or on sunny days, and that insolation rather than UV radiation alone was correlated with E. coli inactivation. Similarly, E. coli levels in marine water were also greater at 8 am than noon, presumably due to greater insolation (Hamilton et al. 2010).
The sand environment probably provides E. coli and other bacteria with protection from the inactivating effects of irradiation. Mika et al. (2009) found that exposure to sunlight was not a significant factor in the decline of E. coli concentrations in sand over an eight day period. Another study found that exposure to UV radiation did not affect the densities of E. coli in sand compared to controls maintained in the dark (Beversdorf et al. 2007). Although Imamura et al. (2011) found that E. coli and enterococci levels remained higher in dark microcosms compared to those exposed to sunlight, the microcosms were incubated on a rooftop and sand temperature may well have been a factor in the differential rate of decline of the FIB.
Temperature. Temperature affects E. coli persistence and E. coli replication in sand differently. E. coli may persist longer in beach sand in cooler temperatures, as studies of soil or sand inoculated with E. coli and incubated at temperatures between 4°C and 37°C showed that the decay rate of E. coli was lower at the cooler temperatures (Ishii et al. 2006; Sampson et al. 2006). A study of sewage-contaminated sand (Mika et al. 2009) found that E. coli and enterococci survived very poorly at sand temperatures above 50° C. Higher temperatures may promote an increase in E. coli densities during summer months (Edge and Hill 2007; Francy et al. 2003; Ishii et al. 2007; Twinning et al. 1993; Whitman and Nevers 2003; Zehms et al. 2008), suggesting the possibility of replication at warmer temperatures. Laboratory microcosm and field incubation studies show that E. coli is capable of growing in sand at ambient temperatures (Alm et al. 2006; Byappanahalli et al. 2006b). E. coli densities increased transiently over a wide range of temperatures from 4°C to 44.5°C in a study in which sand was exposed to controlled temperatures in the laboratory or ambient temperatures outdoors. Although the significance of the increase was not determined, ambient temperatures that ranged from 23 - 32° C achieved the greatest level of replication (Beversdorf et al. 2007).
Evidence suggests that E. coli “overwinters” in sand at some freshwater beaches, even in temperate climates where freezing weather regularly occurs. E. coli densities in sand from Lake Erie beaches in February were as high as those in summer (Francy et al. 2003). E. coli were cultured from Lake Huron sand in December when the lake was frozen and snow covered the beach (Kon et al. 2007) and were also recovered from frozen sand in Lake Superior (Ishii et al. 2007). E. coli at levels as low as 2 cells/g sand was recovered from frozen sand on a Lake Superior beach in Duluth-Superior Harbor (Johnson and Sadowsky, unpublished). Monthly samples taken over an 18-month period along southern Lake Michigan showed diminished concentrations for E. coli in winter but continued persistence in both fore- and backshore (near the groundwater table) sands (Byappanahalli et al. 2006b). E. coli was, however, undetectable in sand at northern Lake Michigan beaches sampled in January (Zehms et al. 2008), suggesting that the presence of E. coli in sand during winter months at some beaches may be attributed to continuous sources rather than to overwintering.
Nutrient Availability
Nutrient availability influences the survival of E. coli in freshwater beach sand. A study conducted in shoreline sand from Lake Huron measured total organic carbon, total phosphorus, and nitrogen species along with E. coli concentrations in inoculated microcosms and diffusion chambers, and found that nutrients were adequate to support replication of E. coli on the scale of five orders of magnitude (Alm et al. 2006). Additional nutrients can encourage further replication of E. coli. When sand was amended with plankton in laboratory experiments, E. coli initially increased about 2 log and then gradually decreased, but remained 1 log higher than initial concentrations (Byappanahalli et al. 2006b). Generally, more complex carbon sources prolonged the replication of E. coli in microcosms relative to rapidly metabolizable substrates such as lactose (Ishii et al. 2010). Another study reported that survival of E. coli and enterococci in microcosms was greater when wrack (macroalgae that has washed onto the shore) was applied to the surface of the sand (Imamura et al. 2011).
Biotic Factors that Influence Fate
Predation by microfauna such as protozoa and nematodes on bacteria is an important top-down control on populations in many environments (reviewed in (Jousset 2012)). Bacterial competition for nutrients and other resources also shapes microbial community structure and influences the fate of both autochthonous and allochthonous community members (Korajkic et al. 2013; Stocker 2012; Wanjugi and Harwood 2013). Alm et al. (2006) found that E. coli in sterile sand grew to high densities in diffusion chambers, while levels in ambient sand adjacent to diffusion chambers were very low, suggesting that the autochthonous microbiota contributed to the removal of E. coli from the community. In another study, E. coli survival was significantly increased by removing competing bacteria from sand, but not by inhibition of protozoan predation with cycloheximide (Feng et al. 2010).
Biofilms
Biofilms consist of bacteria, and sometimes other microbes such as algae and protozoa, attached to particles by an extracellular matrix whose main component is generally polysaccharides. This matrix is frequently termed extracellular polymeric substances (EPS) and can range from a loose slime to a complex structure with water channels enabling oxygenation deep within the biofilm. The adhesive structures of biofilm EPS can contribute to intertidal sediment stability (Yallop et al. 2000). Biofilms, which may be quite complex and include many microbial phyla, contribute to microbial survival in many environments, ranging from the human body to hydrothermal vents (reviewed in (Hall-Stoodley et al. 2004). Microbial communities can expand as the biofilm matures and cells can slough off together if resources become limited, but the structure of the mature biofilm generally limits the exchange of cells between sand and the porewater. Biofilm can provide several advantages to enteric bacteria introduced to the aquatic environment, including protection from physical or chemical stressors, protection from predation, and the acquisition of advantageous genes through horizontal gene transfer within the biofilm.
For allochthonous bacteria introduced to sands via water, two habitat spaces are broadly available: the porewater and the surface of the sand grains. Despite potentially vigorous interaction between water, porewater, and sand (e.g., with wave run-up at a beach, infiltrating sand, and then draining out), these three environmental compartments host distinct bacterial communities. Pyrosequencing studies of the bacterial diversity in the tidal flats of the North Sea show that only 2-3% of the unique bacterial constituents are present in all three habitats (Gobet et al. 2012). Furthermore, total abundance of sand-associated bacteria is much greater than pore water bacteria, which has been estimated as having <0.2% of the total cell abundance found in sands (Gobet et al. 2012; Rusch et al. 2003). This partitioning between microbial communities on sand and in pore water can primarily be explained by the formation of biofilm on sand grains, as well as attachment to fine particulate matter.
Sands covered in biofilm could contribute to the retention of waterborne pathogens at beaches. In laboratory studies, E. coli were flushed through sands before and after the formation of biofilm. Sands retained approximately 9% of E. coli cells in pore water without biofilm, but in sands with a developed biofilm 47% of E. coli cells were retained under similar flow conditions (Wang et al. 2011). Beyond that, a significant proportion of FIB and pathogens may enter the beach environment already attached to particles and possibly protected within particle-associated biofilm (Fries et al. 2006; Suter et al. 2011). In the New River Estuary, 38% of FIB in the water column were particle-bound (Fries et al. 2006). In the Lower Hudson River Estuary, a larger fraction of enterococci (52.9%) in the water column were associated with particles than the fraction of the total bacterial population (23.8%) associated with suspended particles (Suter et al. 2011). Only 10% of the enterococci in beach sand could be recovered from pore water, suggesting that the remainder were attached to sand grains (Phillips et al. 2011b). A study conducted at eight saltwater beaches in Florida found consistent biofilm presence on the quartz/calcium carbonate sand grains common at these beaches. Enterococci density in supratidal sand was related to extracellular polysaccharide (EPS) levels in a non-linear manner, peaking at ~7 µg EPS/g sand; however, a similar relationship was not found in sands from the intertidal or subtidal zones (Piggot et al. 2012). The knowledge that bacteria in aquatic environments generally “prefer” attachment to particles to a planktonic state is decades old (reviewed in (Costerton et al. 1987); therefore the question of the extent to which waterborne pathogens exist in biofilms in sand is a critical issue for the public health of beach users.
Population Biology and Genetics
While studies that have extensively explored the population biology and genetics of bacteria in sand are scarce, some evidence exists for self-sustaining naturalized populations of FIB. The dominant source(s) of E. coli in sand may influence the potential for persistence or replication. The observed increase of E. coli densities in sand during the summer at freshwater beaches could be due, in part, to shifts in contributions from various sources (e.g., at a Lake Superior beach). E. coli in samples collected in spring originated from treated wastewater effluent, but as the seasons proceeded to summer and fall, the percentage of E. coli coming from Canada geese and ring-billed gulls increased (Ishii et al. 2007). Whitman and Nevers (2003) found that E. coli population levels in foreshore sands of Lake Michigan beaches remained roughly steady over six months, and newly introduced sands were quickly recolonized, suggesting either continual input from birds and wastewater, or that populations were in equilibrium with the carrying capacity of the habitat.
Genotyping is a useful tool for exploring the relatedness of bacterial strains in the sand environment (Ishii and Sadowsky 2008). While Byappanahalli et al. (2006b) did not see evidence of the selection of a specific genotype of E. coli in sand, other studies have reported the repeated recovery of certain genotypes, suggesting replication and/or differential survival. When analyzed by repetitive extragenic palindromic PCR (REP-PCR), 34 of 160 (21%) sand isolates from Lake Michigan could be placed into six clonal groups (Beversdorf et al. 2007). REP-PCR analysis of E. coli recovered from Lake Huron foreshore interstitial water also revealed dominant strains of E. coli (Kon et al. 2007), and “naturalized” E. coli strains were found in Lake Superior sand by using a modified rep-PCR DNA fingerprinting technique (Ishii et al. 2007). Multiple isolates recovered from the same sampling location were identical or very similar, and different sites on a beach had distinct dominant strains. Edge and Hill (2007) applied REP-PCR to indicate that E. coli populations in Lake Ontario beach sand were a unique subset of the predominantly bird-derived E. coli that were likely more adapted to persisting in beach sand. They also found that the E. coli populations in the adjacent beach water were predominantly derived from beach sand rather than directly from bird fecal droppings. E. coli recovered from intertidal sand and the water column of six Lake Huron and St. Clair River beaches also revealed extensive genetic diversity by multilocus enzyme electrophoresis and multilocus sequence typing (MLST), yet several genotypes were recovered from separate sites at different times (Walk et al. 2007). Multilocus sequence typing suggested that natural selection favored the retention of certain genotypes of E. coli within the beach sand environment. One of the most common sequence types (ET-1) was isolated seven times at five of the six beaches, at all depths of sand sampled, and at separate times over 35 months, suggesting repeated isolation of a widespread genotype that is in high frequency at the beach (Walk et al. 2007).
Methicillin-resistant Staphylococcus aureus (MRSA) and Staphylococcus spp. isolated from beach water and intertidal sands in Washington State were typed by several phenotypic and genotypic methods, including antimicrobial susceptibility and MLST (Soge et al. 2009). Four of the five MRSA strains isolated were similar to hospital isolates, rather than to strains associated with community-acquired isolates (Soge et al. 2009).
Viable but Nonculturable Bacteria
With the exception of studies where qPCR is specifically mentioned, all of the findings discussed in this section were derived from experiments in which bacteria were cultured on selective-differential media. While culturing bacteria has many advantages, including the knowledge that the cells counted are living and the sensitivity to detect one target cell, many bacteria enter a state termed viable but nonculturable (VBNC) when they are physiologically stressed (reviewed in (Grimes et al. 1986; Oliver 2010). In this state, FIB and enteric pathogens remain metabolically active and have the potential to infect a host and/or to become culturable when they encounter more favorable conditions (resuscitation) (Alam et al. 2007; Heim et al. 2002; Pommepuy et al. 1996). Furthermore, VBNC-inducing stresses vary from one species to the next and include salinity, nutrient level, and temperature, to name a few. Quantitative PCR, which detects viable and nonviable cells, as well as free environmental DNA, generally measures higher levels of target bacteria than the corresponding culture-dependent method (Ahmed et al. 2012; Chase and Harwood 2011; Khan et al. 2009; Lavender and Kinzelman 2009). Because regulatory, monitoring, and many clinical applications of microbiology detect FIB and pathogens by culture methods, the VBNC phenomenon represents a potential confounding factor in any microbiology experiment, and should be further explored in the beach sand environment.
4. TRANSPORT OF MICROBES TO, THROUGH, AND FROM SAND
In addition to allochthonous sources (Section 2) fate-related processes (Section 3), which influence persistence and replication of microbes, the concentrations of specific microbes within the micropsammon are influenced by transport processes that move microbes from one reservoir to another.
The reservoirs where the micropsammon reside vary considerably in scale (Ginn et al. 2002). At the small scale, the reservoirs include the sand matrix and interstitial water, which contain microbial communities that are adhered to the sand matrix. Above the water table, interstitial water may or may not entirely fill the pore space among sand grains which contain the biofilms. At a larger scale, the reservoirs include the nearshore waters, the wave impacted shoreline (i.e. the foreshore), the beach sand area not impacted by wave action (i.e. the backshore), and the air space immediately above the beach sand. For marine waters, the zones are defined in terms of tidal ranges and include the subtidal, the intertidal, and the supratidal zones. In the vertical direction, reservoirs include sand comprising the vadose zone (partially saturated) above the water table, at or below the water table, or permanently inundated (i.e. located below the nearshore or subtidal water as shown in Figure 1). Small-scale transport processes can be integrated to describe the transport of microbes in the larger scale reservoirs within the beach environment.
Given these definitions of different microbial reservoirs, transport of microbes within the sand environment can then be defined to occur:
Through interstitial flow within the sand interstitial spaces
Through sediment transport for microbes attached to sand
Through the exchange of microbes to and from the sand matrix
Through the replication of the microbial population and the overall growth of biofilms.
The microbial transport via all of these processes is influenced by the rate of fluid flow (e.g. water flow via surface runoff, groundwater flow, surface to subsurface infiltration/exfiltration, waves, and wind) throughout the beach environment. Sediment transport at the larger scale manifests itself as drift and/or burial of the micropsammon.
Interstitial Flow
Interstitial flow of water through the sand pore spaces can occur under saturated conditions through groundwater flow or under unsaturated conditions within the vadose zone (the partially saturated sand zone located above the water table). The interstitial transport of microbes has been extensively evaluated through column experimentation (Logan et al. 2001; Rijnaarts et al. 1996) in the context of groundwater sources of drinking water (Díaz et al. 2010; Robertson and Edberg 1997) and in the context of bioremediation of dissolved chemical compounds (Ginn et al. 2002; Murphy and Ginn 2000). Rare, however, are studies that focus on interstitial flow of microbes through beach sands. In controlled laboratory studies that utilized washed quartz sand, Chen and Walker (2012) found that different fecal indicator bacteria have different behaviors during interstitial flow. They found that E. faecalis would preferentially attach at the air/water interface whereas E. coli showed similar affinity to the air/water interface and to the sand surface. In natural sand column experiments, Phillips et al. (2011b) observed that interstitial flow accounted for about 10% of the bacterial indicator (enterococci) transported through beach sands. Yamahara et al. (2007) also found that interstitial flow carries bacteria but in their case they observed nearly 100% of the bacteria transported through interstitial pore flow. The discrepancy in the observations may be due to differences in sand column preparation and/or sand characteristics. Intact columns retrieved from the field may behave differently than reconstituted columns prepared in the laboratory. We suspect that quorum sensing among bacteria may be playing a role in their release from the sand matrix.
Within the larger-scale beach environment, groundwater may flow from the aquifer to the open water body or vice versa depending upon the relative elevations between the exposed water surface and water table. The rate at which the water moves through the groundwater system is dependent upon hydraulic conductivity of the sand, (in general between 10−2 to 10−1 cm/s) and water table gradient; the steeper the gradient the more rapid the flow. In the Great Lakes, groundwater below beaches continuously flows towards and discharges into the lake. Estimates of groundwater discharge fluxes at beaches of the Great Lakes range from approximately 15 to 900 m3 per m of beach per year (Crowe and Meek 2009; Crowe and Milne 2013). In marine systems, Boehm et al. (2004) found that microbes could be potentially transported to the surf zone through tidally driven exchange of groundwater, and de Sieyes et al. (2011) determined that groundwater could transport nutrients to the surf zone. The maximum exchange of groundwater occurred during spring tides when water level gradients were the steepest, however the maximum transport of nutrients occurred during neap tides (de Sieyes et al. 2008) when the water level gradients are most shallow. These nutrients, transported by groundwater, were hypothesized to promote the persistence and population replication of bacteria within the surf zone.
Transport processes in the vertical direction, in the context of water movement, has also been well documented. Infiltration of water from the surface can occur through precipitation, snow melt, accumulation of runoff (Price et al. 2013) or wave run-up (Xin et al. 2010). This water, in turn, can transport nutrients and microbes. Vertical transport of microbes specifically through porous media has been evaluated extensively through soil column experiments. Ripp et al. (2001) have shown that vertical fluctuations in water table elevation can cause the transport of microbes vertically within sand and soil columns. Even without the vertical fluctuations, the groundwater can transport microbes upwards above the groundwater table by capillarity (Dunn et al. 2005), (upward movement, or wicking, of water from the water table under a negative pressure).
Transport of the Sand Matrix
A wealth of well-established sediment transport theory dating back to the late 1800's (Ettema and Mutel 2004) can be used as the basis for understanding and simulating sediment transport in the water environment. Sediment transport includes deposition to the sand environment and the removal of sand particles through resuspension (Nielsen 1992). Resuspension can result in a significant importation of microbes into the water column if their concentrations are high in the sediment.
Recent developments of sediment transport theory have focused on simulating sediment transport in the nearshore zone under the combined influence of current, waves, and in marine-tidal systems (Feng et al. 2013; Ge et al. 2012a). The processes can be dynamic and heterogeneous, given complex concentration distribution patterns in the water column and hydrodynamic conditions in the nearshore (Ge et al. 2010; Ge et al. 2012a; Inman et al. 1971). For example, FIB loading carried by nearshore currents can change with the variability of current velocity and direction within hours, and parts of an embayed beach (approximately 1 km cross-shore and 2 km alongshore) can have different characteristics in retaining FIB from external sources depending upon the embayment infrastructure and the bathymetry (Ge et al. 2012b).
Exchange of Microbes from the Sand Matrix
An understanding of physico-chemical processes of microbial deposition and release from the porous matrix can be obtained from the water filtration literature and colloid filtration theory (Foppen et al. 2007), which defines many mechanisms of filtration including straining (Díaz et al. 2010) and electrostatic interactions (Johnson et al. 2007). Field-scale studies have identified the classic mechanisms of dispersion, preferential flow, and mass transfer to immobile domains as additional important processes (Woessner et al. 2005).
More recent fundamental developments focus on describing surface bio-chemical characteristics and other biotic factors that influence transport. Surface biochemical properties include lipopolysaccharides, proteins and other surface structures that promote the adherence of bacteria to surfaces (Foppen et al. 2010). Murphy and Ginn (2000) link attachment/detachment rates of bacteria to surfaces to changes in metabolic activity. They found that changes in metabolic activity control the partitioning of the microorganism between the aqueous and solid phase. They argue that when describing the transport of bacteria through porous media, both physical processes and biotic processes should be considered, as the interplay will dictate transport. In addition to the physical exchange of bacterial cells between the sand matrix and interstitial pore water, Lovins et al. (1993) found that introduced bacteria (in this case genetically engineered Pseudomonas aeruginosa) were capable of exchanging genes with native bacterial populations as they are transported through soil columns. Such exchange adds another layer of complexity to the overall transport process that influences microbial community composition.
Within the larger scale beach surface environment, the influence of waves can be considerable. Physical processes induced by wave action include shearing effects between the water and solid matrix phase and abrasion between sand particles. Russell et al. (2012) specifically evaluated transport of enterococci from naturally contaminated beach sands to the groundwater table via infiltrating seawater. They found that infiltrating seawater could influence detachment of enterococci from beach sand, transporting them to the groundwater. These detached bacteria could then be discharged to coastal waters via submarine groundwater discharge.
A by-product of wave effects is the transport of microbes to and from the sand. As a possible consequence of wave-induced transport, several studies have found that water quality is related to adjacent sand quality (Beversdorf et al. 2007; Kinzelman et al. 2004; Phillips et al. 2011a; Skalbeck et al. 2010). Alm et al. (2003) found that E. coli densities in the wave-washed swash zone of the beach correlated with densities in adjacent surface water, particularly for the top several centimeters of sand. While E. coli move back and forth between water and sand, the net movement of E. coli is from the foreshore zone of the beach lakeward into the water (Whitman and Nevers 2003). Whitman and Nevers (2003) also found correlations (r = 0.625 with P < 0.001) between foreshore sand and surface water FIB concentration at 45 and 90 cm water depths throughout the day, an indication that this exchange is persistent rather than transient in the nearshore environment. Edge and Hill (2007) used MST techniques to determine that E. coli in beach water at a Lake Ontario beach were predominantly derived from beach sand up to 150 meters offshore. When evaluating genetic characteristics, the FIB found in marine beach waters were more similar to bacteria in sand than to other potential sources (Bonilla et al. 2006), such as wastewater; the combined effects of the detachment of the microbes from the sand and erosion of sediment from the beach surface contribute to nearshore water quality. In addition, exfiltration through the beach face during wave run-up and downwash cycles could also import sand-borne microbes into the swash zone (Li et al. 2002). In a recent study, a mass-balance model predicted that sand was the dominant source of enterococci to nearshore marine waters at a California beach (Russell et al. 2013).
Growth-Induced Transport
The physical growth of biofilms in the subsurface has been evaluated for the purpose of developing biobarriers which are biofilm layers used for the removal or retardation of contaminants within groundwater (Cunningham et al. 1991; Ross et al. 2001). The process involves the irreversible adsorption of the bacteria to a surface from which the bacteria then multiplies and secretes EPS (Perkins et al. 2000). Through this process the microbes change the hydrology of the system by decreasing the hydraulic conductivity (Ross et al. 2001), which in turn impacts the rate at which microbes adsorb and uptake nutrients. Piggot et al. (2012) found that indicator bacteria are found at optimum levels of EPS. They suggest that biofilms are necessary at low levels to promote the survival of enterococci. Too much biofilm, however, inhibits enterococci. Bonilla et al. (2007) observed the spread of FIB in undisturbed beach sand during periods of no rainfall. This spread was attributed to the possible growth of biofilms which, over time, can potentially increase the distribution of microbes throughout the beach environment. Thus bacteria can move within beach sand and other porous environments without a carrier fluid or carrier sand matrix. Their ability to form biofilms allows the micropsammon to spread through environmental systems at a rate governed by their rate of multiplication and EPS production.
5. PUBLIC HEALTH IMPLICATIONS OF SAND MICROBES
Humans receive extensive exposure to sand-associated microbes during recreational activities. These microorganisms may be autochthonous or allochthonous (Section 2). While most of them are harmless, some are pathogenic, and the potential for pathogen occurrence is particularly great when sand is contaminated by human or animal waste. Pathogens that have been reported from sand habitats are discussed below.
Pathogen Occurrence in Sand
While there have been few studies of microorganisms in beach sand compared to beach water, there have been even fewer studies of human pathogenic microorganisms in beach sand. Studies indicate that a variety of potential pathogens have been reported from beach sand (Table 2). While many of the reported pathogens are of fecal origin, importantly, some are not. Some of these pathogens cause disease among individuals with normal immune systems whereas others are considered opportunistic pathogens only capable of causing disease in individuals with weakened immune systems. It is also important to note that almost all of these studies have been based on detecting taxonomic groups (e.g. genera or species) known to contain pathogenic strains of bacteria, protozoa, fungi or viruses in beach sand. While some taxonomic groups may be comprised of mostly pathogenic strains, others may be comprised of many strains that are not associated with causing human disease. Characterization of the virulence characteristics of putative pathogens detected in beach sand, or determining whether they are genetically similar to clinical strains known to cause human disease, has rarely been done. In the future, additional research will be required to more fully evaluate whether pathogens reported from beach sand are strains likely to cause disease in healthy individuals.
Table 2.
Studies identifying pathogenic taxa in beach sands around the world.
| Pathogen taxa | Beach type | Sand type | Location | Detection Percentage | Reference |
|---|---|---|---|---|---|
| Aeromonas spp. | Freshwater lake | Foreshore sand | Hamilton harbour Lake Ontario, Ont. Canada | 100% | (Khan et al. 2009) |
| Campylobacter spp. | Marine coast | Dry sand | California, USA | 13% | (Yamahara et al. 2012) |
| Freshwater lake | Foreshore interstitial sand pore water | Hamilton harbour Lake Ontario, Ont., Canada | (Khan et al. 2013) | ||
| Freshwater lake | Foreshore sand | Lake Simcoe, Ont. Canada | 27% | (Khan and Edge 2013) | |
| Marine coast | Foreshore and dry sand | Northwest and southwest England, U.K. | 45% | (Bolton et al. 1999) | |
| Marine coast | Wet sand | Tel Aviv, Israel | 45% | (Ghinsberg et al. 1994) | |
| Marine bay | sediment | Northwest England, U.K. | (Obiri-Danso and Jones 2000) | ||
| Salmonella spp. | Freshwater lake | Foreshore sand, and sediment | Chicago, Lake Michigan, IL, USA | (Byappanahalli et al. 2009; Whitman et al. 2001) | |
| Marine coast | Foreshore and dry sand | England, U.K. | 6% | (Bolton et al. 1999) | |
| Marine coast | Wet and dry sand | Fortaleza, Brazil | 3% | (Vieira et al. 2001) | |
| Marine coast | Dry sand | California | 15% | (Yamahara et al. 2012) | |
| Marine coast | Swash zone | Gaza Strip | 7% | (Elmanama et al. 2005) | |
| Marine coast | Wet sand | Kuwait | (Shatti and Abdullah 1999) | ||
| Staphylococcus aureus | Marine coast | Wet sand | Greece | 52% | (Papadakis et al. 1997) |
| Marine coast | Foreshore wet sand and dry sand | Seattle, WA, USA | 6% | (Levin-Edens et al. 2011) | |
| Freshwater lake | Dry sand | Seattle, WA, USA | 33% | (Levin-Edens et al. 2011) | |
| Marine coast | Foreshore wet sand | Seattle, WA, USA | (Soge et al. 2009) | ||
| Marine coast | Foreshore sand | Southern California, USA | 53% | (Goodwin et al. 2012) | |
| Marine coast | Dry sand | California | 14% | (Yamahara et al. 2012) | |
| Marine coast | Dry sand | South Florida, USA | (Esiobu et al. 2013) | ||
| Marine coast | Wet and dry sand | South Florida, USA | (Esiobu et al. 2004) | ||
| Marine coast | Wet and dry sand | South Florida, USA | 50% | (Shah et al. 2011) | |
| Lake | Egypt | (Dowidart and Abdel-Monem 1990) | |||
| Marine coast | Chile | (Prado et al. 1994) | |||
| Marine coast | Italy | (Bonadonna et al. 1993) | |||
| Marine coast | Wet sand | Tel Aviv, Israel | (Ghinsberg et al. 1994) | ||
| Escherichia coli pathotypes | Freshwater lake | Swash zone sand | Lake Huron and Lake St. Clair, Michigan, USA | 2% | (Bauer and Alm 2012) |
| Freshwater lake | Wet interstitial sand | Lake Huron, Ont. Canada | 0% | (Kon et al. 2007) | |
| Freshwater lake | Wet and dry sand and sediment | Lake Superior, Minnesta, USA | (Ishii et al. 2007) | ||
| (Shigella) | Marine coast | Bay of Gdansk, Poland | (Dabrowski 1982) | ||
| Pseudomonas aeruginosa | Marine coast | Wet and dry sand | South Florida, USA | (Esiobu et al. 2004) | |
| Marine coast | Wet and dry sand | Portugal | (Mendes et al. 1993) | ||
| Marine coast | Azore Islands | (Mendes et al. 1997) | |||
| Marine coast | Swash zone | Gaza Strip | (Elmanama et al. 2005) | ||
| Marine coast | Dry sand | Sao Paulo, Brazil | (Sanchez et al. 1986) | ||
| Marine coast | Dry sand | South Carolina, USA | (Stevens et al. 2012) | ||
| Freshwater lake | Sediment | Ontario, Canada | (Seyfried et al. 1985) | ||
| Freshwater lake | Sediment | Ontario, Canada | (Palmer 1988) | ||
| Marine coast | Wet sand | Tel Aviv, Israel | (Ghinsberg et al. 1994) | ||
| Vibrio spp. | Marine coast | Wet and dry sand | Ustka, Baltic Sea, Poland | (Mudryk et al. 2013) | |
| Marine coast | Swash zone | Gaza Strip | 22% | (Elmanama et al. 2005) | |
| Marine coast | Wet sand | Tel Aviv, Israel | 13% | (Ghinsberg et al. 1999) | |
| V. vulnificus | Marine coast | Wet and dry sand | South Florida, USA | 37.5% | (Abdelzaher et al. 2010) |
| V. vulnificus | Marine coast | Wet and dry sand | South Florida, USA | 100% | (Shah et al. 2011) |
| V. parahaemolyticus | Marine coast | Wet and dry sand | Fortaleza, Brazil | 20% | (Vieira et al. 2001) |
| V. alginolyticus and V. parahaemolyticus | Marine coast | Submerged sand | Adriatic Sea, Italy | 59% | (Pianetti et al. 2004) |
| V. parahaemolyticus and V. harvey | Marine coast | Africa | 12% | (Aldova 1989) | |
| Cryptosporidium spp. | Marine coast | Wet and dry sand | South Florida, USA | 25% | (Abdelzaher et al. 2010) |
| Marine coast | Wet sand | South Florida, USA | 5.6% | (Shah et al. 2011) | |
| Cryptosporidium spp. and Giardia spp. | Freshwater lake | Foreshore interstitial pore water | Hamilton, harbour Lake Ontario, Ont. Canada | Edge and Neumann (unpublished data) | |
| Giardia spp. | Marine coast | Dry sand | Sao Paulo, Brazil | 1% | (Sato et al. 2005) |
| Enterovirus | Marine coast | Wet and dry sand | South Florida, USA | 1% | (Shah et al. 2011) |
| Marine coast | Black Sea, Romania | 83% | (Nestor et al. 1984) | ||
| Marine coast | Submerged sand | Adriatic Sea, Italy | 23% | (Pianetti et al. 2004) | |
| Fungi | Marine coast | Dry sand | Portugal | 60% | (Sabino et al. 2011a) |
| Marine coast | Wet sand | Greece | (Papadakis et al. 1997) | ||
| Marine coast | Wet and dry sand | South Florida, USA | (Shah et al. 2011) | ||
| Marine coast | Dry sand | South Carolina, USA | (Stevens et al. 2012) | ||
| Marine coast | Swash zone | Gaza Strip | (Elmanama et al. 2005) | ||
| Marine coast | Dry sand | South Carolina, USA | (Stevens et al. 2012) | ||
| Marine coast | Wet and dry and | South Florida, USA | (Vogel et al. 2007) | ||
| Marine coast | Hawaii, USA | (Kishimoto and Baker 1969) | |||
| Marine coast | California, USA | (Dabrowa et al. 1964) | |||
| Marine coast | Casablanca, Morocco | 47.5% | (Abdallaoui et al. 2007) | ||
| Marine coast | Central coast, Portugal | 42% | (Sousa 1990) | ||
| Marine coast | (Izquierdo et al. 1986) | ||||
| Marine coast | Southern France | (Bernard et al. 1988) | |||
| Marine coast | Wet sand | Tel Aviv, Israel | (Ghinsberg et al. 1994) | ||
| Marine coast | Guadeloupe | (Boiron et al. 1983) | |||
| Marine coast | Spain | (Roses Codinachs et al. 1988) | |||
| Marine coast | Mediterranean Sea, Spain | (Larrondo and Calvo 1989) | |||
| Marine coast | Malaga, Spain | (Marino et al. 1995) | |||
| Marine coast | Azore Islands | (Mendes et al. 1997) | |||
| Marine coast | Wet and dry sand | Fortaleza, Brazil | 13% | (Vieira et al. 2001) | |
| Marine coast | Dry sand | Sao Paulo, Brazil | 19% | (Sanchez et al. 1986) | |
| Marine coast | Wet and dry sand | Sao Paulo, Brazil | 18% | (Sato et al. 2005) |
Bacterial Pathogens
A variety of pathogens have been reported in beach sand, including bacterial pathogens with antimicrobial resistance such as methicillin-resistant Staphylococcus aureus (MRSA) (Goodwin and Pobuda 2009; Goodwin et al. 2012; Levin-Edens et al. 2012; Shah et al. 2011; Soge et al. 2009; Yamahara et al. 2012). While hospital settings and the retail food supply are increasingly recognized as important sources of antimicrobial resistant pathogens, the extent of sand-borne exposure is not known. In addition, the public health implications of antimicrobial resistance in FIB (Bennani et al. 2012; de Oliveira and Pinhata 2008; Edge and Hill 2009; Roberts et al. 2009) and naturally occurring heterotrophic bacteria (de Oliveira et al. 2010; Mudryk et al. 2010) found in beach sands is still poorly understood.
Aeromonas spp
Khan et al. (2009) found that both culture and qPCR-based detection methods enumerated higher numbers of Aeromonas bacteria in interstitial pore water of foreshore sand than in adjacent surface water at two freshwater beaches on Lake Ontario. Foreshore sand was found to serve as a reservoir for higher numbers of aeromonads, similar to this phenomenon for FIB like E. coli. Khan et al. (2009) did not specifically confirm the pathogenicity of any Aeromonas isolates recovered from beach sand, however outbreaks of Aeromonas hydrophila have been attributed to recreational exposures to mud fields (Vally et al. 2004).
Campylobacter spp
Campylobacter has been commonly reported from a variety of beach sands. Campylobacter was detected in 82/182 (45%) sand samples collected at each of the four UK marine beaches investigated by Bolton et al. (1999). The frequency of detection was higher (50%, n = 92) at the two beaches that were not compliant with the EC Bathing Water Directive standard, compared to the two compliant beaches (40%, n=90). Campylobacter was detected more commonly in wet sand 1-2 m from the water's edge, than in dry sand from just below the high water mark. The highest detection frequency (77%) for Campylobacter occurred in the wet sand at one of the non-compliant beaches (n=26). However, Campylobacter was also found to be common (50%) in dry sand at one compliant beach where mean water content of the sand was only 4-11%. Bolton et al. (1999) detected C. jejuni, C. coli, C. lari, and urease positive thermophilic campylobacters at each beach. C. jejuni was most common at the two non-compliant beaches, while C. lari was most common at the two compliant beaches. Many of the Campylobacter isolates were subtypes frequently isolated from patients with Campylobacter diarrhea in England.
Obiri-Danso and Jones (2000) also detected Campylobacter in sediments at three marine beaches in Morecambe Bay in northwestern England. Campylobacter geometric mean numbers in these sediments were about 3 MPN/cm3, which were one to two orders of magnitude lower than the numbers of FIB in the same sediments. There was no relationship between occurrence of Campylobacter and FIB presence or density. Campylobacter were isolated more frequently from sediments in colder months and were generally absent in the spring and summer. No C. jejuni or C. coli were detected. Most isolates were urease positive thermophilic campylobacters and C. lari suggesting an avian rather than sewage source.
Ghinsberg et al. (1994) detected Campylobacter, including confirmed isolates of C. jejuni, in 52/115 (45%) of sand samples collected from bathing beaches in Israel. Campylobacter densities ranged between 13 and 20 CFU/g sand and were higher than in adjacent surface water. Yamahara et al. (2012) investigated the occurrence of bacterial pathogens in dry sand at 53 California marine beaches. Campylobacter spp. was detected in sand at 13% of these beaches, and while it was found to be more commonly associated with higher sand moisture, it had no significant relationship to any indicator organism. Campylobacter species have been commonly detected in foreshore beach sand at some freshwater beaches in the Great Lakes. For example, C. jejuni and C. lari have been commonly detected in beach sand at Bayfront Park and Pier 4 Beaches in Hamilton Harbour (Lake Ontario) that are impacted by bird fecal droppings (Khan et al. 2013); Edge, unpublished data). Like marine studies, the frequency of detection and numbers of Campylobacter were higher in beach sand than adjacent surface water at these two beaches. Campylobacter species were also detected in foreshore beach sand at several Lake Simcoe Beaches in southern Ontario (Khan and Edge 2013). Campylobacter was detected more commonly in beach sand interstitial samples (27%) than adjacent ankle (9%) or chest (5%) depth surface water samples at these beaches. Among 67 beach sand interstitial samples from Lake Simcoe beaches, Khan and Edge (2013) found C. jejuni (18%) most common, followed by C. lari (10 %); C. coli were not detected. Campylobacter concentrations in Lake Simcoe beach sands were low, occurring at minimum detection levels of 3-30 cells/L of interstitial pore water.
Escherichia coli (E. coli) pathotypes
While there have been an increasing number of studies investigating the occurrence of E. coli in beach sand, these studies have rarely looked at E. coli as a pathogen. While E. coli is often seen as a commensal microorganism, a variety of E. coli pathotypes can be recognized based largely on their associated clinical effects in humans. Kaper et al. (2004) categorized E. coli pathogens into eleven different pathotypes, ranging from EHEC enterohemorrhagic strains (e.g. E. coli O157:H7) to ExPEC strains causing extraintestinal diseases such as urinary tract infections.
A few studies have reported on the occurrence of E. coli pathotypes at recreational beaches, however, these studies have been largely limited to beach water rather than beach sand. While there have been a growing number of studies reporting on the large numbers of E. coli that can be recovered from beach sand, there has been little investigation into what proportion of these could cause human infections. Bauer and Alm (2012) reported the detection of an E. coli O157:H7 isolate from beach sand at a Lake Huron beach in Michigan, USA. Dabrowski (1982) isolated closely related Shigella bacteria from marine beach sand in Poland. However, Goodwin et al. (2009) did not detect E. coli O157:H7 in Florida beach sand. Harrison and Kinra (2004) did not detect E. coli O157 in beach sand as part of an outbreak investigation in the U.K. E. coli O157:H7 was found to survive in simulated U.K. marine beach sand for at least 5 days under both dry conditions and regular wetting-drying tidal cycles (Williams et al. 2007).
Bauer and Alm (2012) found that genes coding for pathogen attachment proteins intimin (eae) and bundle-forming pilus (bfp) were commonly detected in E. coli isolates from beaches along Lake Huron and Lake St. Clair. The eae gene was detected in 94/121 (78%) of E. coli enrichments from beach sand samples across seven beaches. However, the toxin gene stx1 was not detected in any sand sample, and the stx2 gene was only detected in 2/121 (1.7%) of sand samples. Bauer and Alm (2012) suggested that the higher frequencies of attachment genes rather than toxin genes in E. coli from beach sand could be associated with enabling greater E. coli attachment and persistence in the beach swash zone. They also raised concern that beach sand could be serving as a reservoir for pathogenicity genes that could contribute to the emergence of novel pathogens.
Conversely, Ishii et al. (2007) detected hemolysin production and the attachment protein intimin (eae) gene that is associated with E. coli pathogenicity in only one of 3557 isolates from beach sand and surface water samples at a Lake Superior beach in Minnesota. Shiga toxin genes (stx1 and stx2) were not detected. Kon et al. (2007) also did not detect any pathotypes from DNA microarray studies of E. coli isolates from Lake Huron beach sand. All 50 E. coli isolates that were examined by Kon et al. (2007) possessed incomplete pathotype gene sets, and only three isolates possessed a single tetracycline resistance gene. However, a caveat for DNA-based analyses of E. coli isolates is that the culture isolation step is often performed at 44.5°C which reduces the likelihood of detecting some E. coli pathotype strains such as O157:H7.
Pseudomonas aeruginosa
Pseudomonas aeruginosa has been reported from beach sediments at Great Lakes beaches in Ontario, Canada (Palmer 1988; Seyfried et al. 1985), as well as in beach sand at a subtropical marine beach in Florida, U.S. (Esiobu et al. 2004) and from dry sand at South Carolina marine beaches (Stevens et al. 2012). Ghinsberg et al. (1994) found P. aeruginosa at higher levels in beach sand than in beach water along the Israeli coast. More than 103 P. aeruginosa CFU/g sand were measured at some beaches. Mendes et al. (1993) commonly detected P. aeruginosa in beach sands at marine beaches in Portugal, and concentrations were measured as high as 2.4 × 107 cells/g sand. P. aeruginosa was also commonly detected in beach sand at beaches in the Azore Islands, reaching over 103 MPN/g sand (Mendes et al. 1997). Sanchez et al. (1986) detected P. aeruginosa commonly in beach sand at eight marine beaches in Sao Paulo, Brazil, and numbers were much higher in the sand than adjacent beach water. Concentrations exceeded 104/100 g, and numbers better correlated with total coliforms than FIB in sand. Elmanama et al. (2005) detected Pseudomonas aeruginosa in almost all 130 sand samples analyzed from the swash zone at marine beaches along the Israeli coast. They found P. aeruginosa concentrations as high as 900 CFU/100 g sand and considered the widespread occurrence of this microorganism as alarming. Mohammed et al. (2012) suggested P. aeruginosa might be useful to assess sanitary conditions of beach sand in the absence of ideal indicators of non-enteric health risks.
Salmonella spp
A number of studies have detected Salmonella in beach sand. Salmonella was found in sand at three of four marine beaches in England (Bolton et al. 1999), although two of the beaches only had a single Salmonella detection. Salmonella was detected in 10/182 (6%) of all sand samples. There was a higher detection frequency of Salmonella detection (9%, n=92) at two beaches that were not compliant with the EC Bathing Water Directive standard, compared to two compliant beaches (2%, n=90). Salmonella was detected in both wet sand 1-2 m from the water's edge and dry sand just below the high water mark. Bolton et al. (1999) isolated six different Salmonella serotypes from the beach sand, including two isolates of S. enteritidis (phage types 5 and 8), and two isolates of S. typhimurium (phage types 99 and 154).
Yamahara et al. (2012) investigated the occurrence of bacterial pathogens in dry sand at 53 California marine beaches using qPCR techniques. Salmonella was detected in sand at 15% of these beaches, and while it was found more associated with higher sand moisture, its occurrence was only correlated with culturable E. coli. Byappanahalli et al. (2009) detected Salmonella in beach sand and sediment at 63rd St. Beach on Lake Michigan. These beach sands were suggested to be a reservoir for exchange of Salmonella with filamentous Cladophora algae on the beach. Salmonella (serotype agona) was detected in only one dry sand sample (n=30) and one wet sand sample (n=30) out of 60 sand samples collected across three marine beaches in Brazil (Vieira et al. 2001). Elmanama et al. (2005) detected Salmonella in 9/130 (7%) of sand samples from the swash zone at marine beaches along the Israeli coast. They found Salmonella more common in beach sand than the adjacent beach waters. Shatti and Abdullah (1999) detected Salmonella in several wet beach sand samples from a Kuwait beach impacted by wastewater discharges. While Campylobacter was detected by Obiri-Danso and Jones (2000) in UK marine beach sediments, Salmonella was not detected in their study. Salmonella was also not detected in 171 sand samples from marine beaches in Sao Paulo, Brazil (Sanchez et al. 1986) or in 39 submerged sand samples from two marine Italian beaches (Pianetti et al. 2004).
Staphylococcus aureus
Staphylococcus aureus is an opportunistic pathogen, although some strains are capable of causing disease in healthy individuals. Staphylococcus species have been reported from beach sands in Egypt (Dowidart and Abdel-Monem 1990), Chile (Prado et al. 1994), and Italy (Bonadonna et al. 1993). Ghinsberg et al. (1994) found S. aureus at higher levels in beach sand than in beach water along the Israeli coast, with more than 103 S. aureus CFU/g sand measured at some beaches. Similarly, Papadakis et al. (1997) analyzed wet sand samples from two marine beaches in Greece, and S. aureus was detected at both beaches. S. aureus was recovered more often from the beach sand than adjacent beach water. Sand samples contained higher levels of S. aureus in the summer months, and this was attributed to higher numbers of bathers at these beaches as S. aureus counts in sand were correlated with the number of swimmers at the more popular beach. Papadakis et al. (1997) drew attention to the importance of pathogens like S. aureus in beach sand, particularly for children, and that FIB may not be good indicators of health risks from non-fecal pathogens.
S. aureus has been commonly reported from beach sand at subtropical marine beaches in Florida, U.S. (Esiobu et al. 2013; Esiobu et al. 2004; Plano et al. 2011; Shah et al. 2011). Esiobu et al. (2004) detected S. aureus in wet and dry sand from three marine beaches in southern Florida, where S. aureus was more abundant in sand than adjacent water and occurred at densities as high as 57.5 × 103 per g sand. The numbers of S. aureus were higher in wet beach sand during summer months of more intense beach usage by bathers. Esiobu et al. (2013) detected S. aureus in beach sand at Florida marine beaches, with the highest average densities in dry sand at 3.46 × 105 CFU/g. They reported the occurrence of S. aureus to be associated with hotspots of human use and possible bacterial re-replication. A brief epidemiology survey associated with this study found a slight association between beach use and skin infections, although S. aureus in beach sand was not found to constitute a major health risk. Shah et al. (2011) detected S. aureus more abundantly in beach sand than adjacent water, with levels ranging from 0.5 to 66 CFU/g sand at a Florida beach. Shah et al. (2011) indicated that some indicator bacteria might be useful for predicting the occurrence of this pathogen in subtropical beach sand. Mohammed et al. (2012) demonstrated that S. aureus could proliferate in sterile sand microcosms, but not unsterile beach sand, and suggested that S. aureus might be useful in assessing the sanitary conditions of beach sand in the absence of ideal indicators of non-enteric health risks.
S. aureus has also been detected in beach sand at marine beaches in Washington, U.S. (Levin-Edens et al. 2012; Soge et al. 2009) and California (Yamahara et al. 2012). Yamahara et al. (2012) found S. aureus in dry sand at 14% of 53 marine beaches in California, and its occurrence was correlated with a Bacteroidales human-specific DNA marker. An intensive surveillance for S. aureus was conducted at several California marine beaches by Goodwin et al. (2012). S. aureus was detected in 53% of beach sand samples collected across these beaches over three years (n=358). The mean concentration of S. aureus in beach sand was 187 CFU/100 dry g, although concentrations were as high as 830 CFU/100 dry g at one beach. Goodwin et al. (2012) found S. aureus concentrations in beach sand were correlated with seawater S. aureus concentrations, seawater enterococci concentrations, seawater temperature, and wind strength (inversely). It was suggested that beach sands were a source of S. aureus to adjacent seawaters at these California beaches.
Concerns about the spread of antimicrobial resistance have prompted investigations of the occurrence of methicillin-resistant Staphylococcus aureus (MRSA) in beach sand, although transmission of MRSA cases via sand have been lacking to date. MRSA have been detected in beach sand at a subtropical marine beach in Florida, U.S. (Shah et al. 2011) and temperate marine beaches in the northwest of the United States (Soge et al. 2009). Levin-Edens et al. (2012) investigated beach sand at two marine beaches and one freshwater beach in the northwest of the United States. They detected MRSA in 3/11 (27%) sand samples at the freshwater beach on Lake Washington, and 4/85 (5%) sand samples at the two marine beaches. Yamahara et al. (2012) detected MRSA in beach sand at 3% of 53 marine beaches surveyed in California. Goodwin and Pobuda (2009) detected MRSA across several California beaches at between 0% to 12% of beach sand samples. In a larger follow-up study, MRSA was detected in 10/366 (2.7%) of marine beach sand samples from California beaches (Goodwin et al. 2012).
Vibrio spp
Vibrio bacteria have been reported from beach sand at numerous marine beaches around the world. Vibrio-like bacteria were widespread in wet and dry sand at a marine beach on the Baltic Sea (Mudryk et al. 2013), with many isolates showing antibiotic resistance that was considered a possible public health threat. Elmanama et al. (2005) detected Vibrio in 29/130 (22%) of sand samples from the swash zone at marine beaches along the Israeli coast. They found Vibrio more common in beach sand than the adjacent beach waters. Ghinsberg et al. (1999) detected Vibrio in 18/142 (13%) of wet sand samples from marine beaches in Israel in 1993-94. V. alginolyticus was most common (9%) followed by V. parahaemolyticus (2%) and V. vulnificus (1%). Subsequent analyses of more Israeli beach sand samples found V. vulnificus in 18/624 (3%) sand samples. In both sand surveys, V. vulnificus was more common in beach water than beach sand. V. vulnificus isolates were resistant to polymixin B and colistin. Pianetti et al. (2004) detected Vibrio in 23/39 (59%) submerged sand samples from two marine beaches in Italy. These Vibrio positive samples were comprised of strains of V. alginolyticus (87%) and V. parahaemolyticus (52%). Vibrio vulnificus was also detected from beach sand at a subtropical marine beach in Florida, U.S. (Abdelzaher et al. 2010). Shah et al. (2011) found V. vulnificus was ubiquitous in wet sand, dry sand and inundated sand samples from a beach in southern Florida. Vibrio parahaemolyticus was found in wet and dry sand from two of three marine beaches in Brazil (Vieira et al. 2001), although it was only detected in 5/60 (12%) of sand samples analyzed. V. parahaemolyticus and V. harvey were reported in African sands by Aldova (1989).
Protozoan Pathogens
Cryptosporidium spp. was detected in one dry beach sand sample (12 oocysts/100g dry sand) and one wet beach sand sample (6 oocysts/100g wet sand) at a subtropical marine beach in Florida, U.S. (Abdelzaher et al. 2010). A single wet sand sample (out of 36 wet, dry and inundated sand samples) was positive for Cryptosporidium (0.63 oocysts/g sand) at a subtropical marine beach in Florida, USA (Shah et al. 2011). While Abdelzaher et al. (2010) and Shah et al. (2011) detected Cryptosporidium in beach sand samples, Giardia spp. was not detected. Sato et al. (2005) detected several Giardia lamblia cysts in dry sand from a Brazilian beach. Sanchez et al. (1986) did not detect any protozoan cysts in marine sand samples from Brazil.
Cryptosporidium and Giardia have been detected from interstitial pore water in foreshore beach sand at Bayfront Park Beach in Hamilton Harbour (Lake Ontario, Canada) (Edge and Neumann, unpublished data). This beach is impacted by bird fecal droppings (Edge and Hill 2007), and preliminary genotyping results indicated that oocysts were the baileyi genotype typically associated with birds and not likely to be infectious for humans.
Fungal Pathogens
Inhaled fungal spores are a well-known cause of allergies and asthma, including seasonal asthma resulting in episodic events in late summer and autumn. In some places the rate and severity of asthma in the population have been linked to airborne levels of the mold spores Alternaria and Cladosporium, with severe episodes requiring hospitalization. These molds can be linked to serious disease in those who are immuno-depressed or who have hyper-reactive immune systems. Inhaled conidia will in some cases express itself in the violence of acute respiratory infections even to immune-competent hosts, such as in the cases of Histoplasma, Coccidiodes, Paracoccidioides and Cladophialophora.
Studies have detected a range of fungi in beach sands from around the world. Kishimoto and Baker (1969) commonly found dermatophytes in Hawaiian beach sands, and Dabrowa et al. (1964) reported pathogenic fungi species from the California coast. A variety of yeasts were detected in beach sand in Guadeloupe (Boiron et al. 1983). Bernard et al. (1988) isolated potentially pathogenic keratinophylic fungi and Candida albicans from beach sand in the south of France. Sousa (1990) detected dermatophytes in 42% of Portuguese sand samples, with Trichophyton mentagrophytes, T. rubrum, and Microsporum nanum most common.
A number of studies have been conducted at Brazilian beaches. Sanchez et al. (1986) isolated C. albicans from 32 of 171 (19%) sand samples from marine beaches in Sao Paulo, Brazil, and found its occurrence was more correlated with total coliforms in sand than other FIB. Vieira et al. (2001) detected yeasts in the wet and dry sand at each of three marine beaches investigated in Brazil. Yeasts were detected from 26 to 41% of sand samples at these three beaches, with C. albicans detected most frequently. Higher numbers were isolated from dry sands. Sato et al. (2005) detected C. albicans in about 18% of wet and dry sand samples from marine beaches in Sao Paulo, Brazil, at a maximum concentration of 34 000 CFU/g in dry sand .
Around the Mediterranean, Ghinsberg et al. (1994) found fungi and C. albicans in higher numbers in beach sand than beach water along the Israeli coast. Papadakis et al. (1997) detected yeasts (e.g. Candida species) and molds (e.g. Aspergillus species) in wet beach sand at two marine beaches in Greece. Yeasts, likely of human origin, were present in the sand than in the adjacent water during the summer. The number of yeasts of human origin in beach sand was correlated with the numbers of swimmers at the more popular beach. Elmanama et al. (2005) detected yeasts in almost all 130 sand samples analyzed from the swash zone at marine beaches along the Gaza Strip. They found yeast concentrations as high as 2300 CFU/100 g sand. Abdallaoui et al. (2007) identified 70 fungi species in marine beach sand from Morocco including C. albicans, Aspergillus sp., and Penicillium sp. Abdallaoui et al. (2007) suggested that the keratinophilic fungi detected could favor the incidence of dermatomycoses among beachgoers, although no epidemiological study has yet been done in order to confirm this. Larrondo and Calvo (1989) surveyed beach sand at 42 beaches in Spain and most commonly detected Penicillium, Cladosporium, Aspergillus, Acremonium, Altenaria, and Fusarium. Fungal density was found as high as several hundred thousand CFU/g sand. A variety of fungi have also been isolated from Spanish beach sands including particularly, Penicillium, Aspergillus, and Cladosporium (Izquierdo et al. 1986; Roses Codinachs et al. 1988). Mendes et al. (1997) found the predominant fungi in beach sand at beaches on the Azores Island were potentially pathogenic fungi (maximum about 60 CFU/g sand) and the allergenic and/or environmental saprophytic fungi (maximum about 70 CFU/g sand). Keratinolytic fungi (levels < 10 CFU/g sand) and Candida species (maximum about 10 CFU/g sand) were not common.
In the United States, a variety of potentially pathogenic yeasts were isolated from beach sand at a subtropical marine beach in Florida (Shah et al. 2011). These colonies were identified as Candida guilliermondi, C. tropicalis, C. albicans, C. parapsilosis, and C. glabrata. Yeast cell counts were generally more elevated in beach sand than adjacent beach water, and Shah et al. (2011) indicated that some FIB may be useful for predicting the occurrence of pathogenic yeasts in subtropical beach sand. Vogel et al. (2007) found yeast concentrations at Florida beaches were highest in dry beach sand, reaching an average of 37, 720 CFU/100 g dry sand at the busiest bathing beach. DNA sequencing identified 21 yeast species from the beach sand samples, the most common being Candida tropicalis and Rhodotorula mucilaginosa. Mean fungal concentrations in dry beach sand at South Carolina (USA) marine beaches varied between 109 CFU/g dry sand at low human use beaches and 472 CFU/g dry sand at high use commercial beaches (Stevens et al. 2012). The fungi were grown at 37°C and were considered potential pathogens, particularly for immune compromised individuals. Two opportunistic human pathogens, Rhodotorula mucilaginosa and Pichia/Candida guilliermondi, were confirmed by sequencing PCR products.
Much work has been conducted to investigate the occurrence of fungi in beach sand in Portugal. An extensive study of 33 marine beaches in Portugal detected fungi (Aspergillus fumigatus, A. niger, Chrysosporium sp., Fusarium sp., Scytalidium sp., Scedosporium sp., and Scopulariopsis sp.) in 60.4% of 495 dry sand samples (Sabino et al. 2011a). Yeasts were detected in 25.4% of sand samples, of which 67.5% were Candida sp. (mean 5.8 CFU/g). Potentially pathogenic fungi were found in 47.9% of the sand samples with a predominance of the genus Aspergillus (mean 0.87 CFU/g). Dermatophytes were detected in 14.3% of samples with a predominance of the genus Trichophyton (mean 1.5 CFU/g). A positive correlation was found between yeasts and total coliforms in beach sand; however, no other correlations were found with FIB.
Brandão et al. (2002) found increased amounts of some filamentous fungi and yeasts during the bathing season, associated with human activity. Many of the swimmers may be asymptomatic, causing contamination of bathing waters and sands. Tidal cycles and runoff during periods of rain can be natural sources of contamination and means of transport. In one study (Sabino et al. 2011b), yeasts of environmental origin revealed increased virulence when compared with clinical strains. Anderson (1979) found human pathogenic fungi could survive in beach sand microcosms sufficiently to be potential sources of infection at public beaches in Hawaii. This in vitro study showed that Trichosporon cutaneum, Candida albicans, Microsporum gypseum and Trichophyton mentagrophytes could survive at least one month in nonsterile sand. Another similar study found five species of dermatophytes (Epidermophyton floccosum, Microsporum canis, M. gypseum, Trichophyton mentagrophytes and T. rubrum) and Scopulariopsis brevicaulis survived from 25 to 360 days (Carrillo-Muñoz et al. 1990). This study showed that the survival of fungi in the sands can be longer than enteric bacteria due to their ability to form resistant spores.
Viral pathogens
There have been few studies of the occurrence of enteric viruses in beach sand. Nestor et al. (1984) detected low numbers of enterovirus in sand at marine beaches on the Romanian Black Sea. Pianetti et al. (2004) detected enteric viruses in 9/39 (23%) submerged sand samples from two marine beaches in Italy. The enteric virus positive samples were comprised of reovirus (67%) and enterovirus (59%). The enteroviruses were further identified to coxackievirus B4, coxackievirus B3, and poliovirus types 1 and 3. Shah et al. (2011) detected enterovirus in beach sand at a subtropical marine beach in Florida, U.S.; however, enterovirus was only detected in one dry sand sample (1.4/100 g sand; n=12), and one inundated sand sample (0.2/100g sand) at this beach. Goodwin et al. (2009) did not detect adenovirus in several dry Florida beach sand samples.
Health Risks from Beach Sand Microbes
Although disease outbreaks have been associated with accidental ingestion of sand from recreational sandboxes (Doorduyn et al. 2006; Staff et al. 2012), outbreaks attributed specifically to exposure to beach sand have not been reported. A growing number of studies are detecting pathogens in beach sands from around the world, however, and it will be important to understand the comparative prevalence of different pathogens in beach sand and their associated health risks. A challenge in comparing pathogen prevalence in beach sand is that pathogen occurrence is likely associated with the local proximity of contamination sources (e.g. bathers or fecal pollution) as well as different environmental persistence, transport, and ecological characteristics of pathogens. Different detection methods will also bias comparisons of pathogen occurrence in beach sand between different studies.
Some studies have investigated pathogen occurrence in beach sand and concluded there was little associated health risk. For example, Chabasse et al. (1986) conducted a bacteriological, parasitological, and mycological investigation of beach sand on a lake in France and concluded that beach sands did not show any infectious hazards. Conversely, other studies have detected pathogens in beach sand and suggested they pose a health risk (Elmanama et al. 2005; Sanchez et al. 1986; Shah et al. 2011; Stevens et al. 2012; Yamahara et al. 2012). Concerns with exposure to fungi in beach sand are also being raised.
In order to understand the significance of pathogen occurrence in beach sand, it is important to understand potential for exposure and to conduct risk assessments and epidemiological studies. A study by Whitman et al. (2009) investigated the potential for exposure to pathogens in sand by analyzing the transferability of bacterial and viral indicator organisms from beach sand to human hands and their rate of removal through rinsing. E. coli and MS2 coliphage were readily transferred from beach sand to hands but could be removed adequately with hand rinsing.
An additional approach for estimating health risks in recreational beach settings is quantitative microbial risk assessment (QMRA) (Ashbolt et al. 2010). Shibata and Solo-Gabriele (2012) applied QMRA to estimate health risks from exposure to sand at a beach in South Florida. Applying the acceptable level of risk of gastrointestinal illness in U.S. marine recreational waters (19 cases per 1000 swimmers) to beach sand, they calculated there would be acceptable risks associated with < 10 Cryptosporidium oocysts/g sand, < 5 enterovirus MPN/g sand, and < 106 Staphylococcus aureus CFU/g sand. Pathogen concentrations measured in the sand at this beach were orders of magnitude below these calculated reference levels, suggesting health risks from sand exposure were relatively low.
Most epidemiological studies at recreational beaches have focused on measuring the human health risks associated with exposure to beach water rather than beach sand, even though people often have more contact with sand than bathing water. Early studies did not find consistent associations between illness and fecal contamination in beach sand or sand contact activities (Marino et al. 1995; Seyfried et al. 1985). For example, Marino et al. (1995) did not find a significant relationship between the densities of dermatophytic fungi and Candida albicans in beach sand and incidence of dermatitis in beachgoers at two marine beaches in Spain.
Preliminary investigations at Florida beaches provided some indication of potential for health risks associated with contact with beach sands. Bonilla et al. (2007) conducted a pilot epidemiology study associated with their microbiological study of beach sand. Bonilla et al. (2007) reported that beach user time spent in contact with wet sand (midway between water level and high tide line) and time spent in the water at a Florida marine beach were associated with increased risk of gastrointestinal illness. Beach user time spent in contact with dry sand (5m above high tide line) was not associated with increased illness at this beach. Esiobu et al. (2013) detected S. aureus in beach sand at three Florida marine beaches. A brief epidemiology survey conducted in this study found a slight association between beach use and skin infections, although S. aureus in beach sand was not considered to constitute a major health risk
In a study by Heaney (2009), over 26,600 beachgoers were interviewed at seven beaches across the United States; the resulting report provided one of the first comprehensive epidemiological investigations of the risk of illness associated with specific sand contact activities. Digging in the sand was associated with a modest, but significant, increased risk of gastrointestinal illness and diarrhea. Being buried in the sand was more strongly associated with risk of gastrointestinal illness and diarrhea than digging in the sand. Children under 10 years old were most associated with an increased risk of diarrhea from digging in beach sand. There was no increased risk of nonenteric illnesses associated with sand activities, although dermatological alterations were not considered in this study. Risk of enteric illness associated with beach sand contact varied between different beaches. Heaney et al. (2012) investigated two of the seven U.S. beaches in more detail and found that increased concentrations of enterococci (measured by both culture and qPCR methods) in wet sand were associated with increased risk of gastrointestinal and diarrhea illness from digging in sand and being buried in sand. However, the authors noted that because most of those individuals who dug or were buried in the sand also swam, it was difficult to estimate the independent effects of sand and water exposure.
Implications for Beachgoers, Beach Managers, and Beach Policy Makers
Beachgoers
Many beachgoers spend a significant portion of their time on the beach itself rather than in the water, particularly in temperate areas around the world. Recreational activities at the beach can involve a variety of opportunities for exposure to sand from simply sitting/lying and strolling to playing in interstitial pore water, building sand castles, throwing sand, and being buried in beach sand. Heaney et al. (2009) collected data from over 26,600 beachgoer interviews as part of an epidemiology investigation at seven beaches across the United States. They indicated that 10,776 beachgoers (40%) reported digging in sand while at the beach and 2,474 (9%) reported being buried in sand. A higher proportion of individuals reported getting sand in their mouth from being buried in sand compared to those only digging in the sand. It was more common for children less than 10 years old to dig in the sand or be buried in the sand. It is possible that exposure to beach sand may present more significant health risks for some beachgoers. Children can play in the sand more frequently and actively, display more hand-to-mouth activity, and have less developed immune systems for responding to pathogen exposure. Heaney et al. (2009; 2012) found evidence for increased risk of diarrhea and gastrointestinal illness among children exposed to beach sand than adults. In many countries, it is possible that aging populations will result in an increasing number of elderly and immune-compromised individuals exposed to beach sand in the future. These individuals, along with children, may be more at risk of infection from opportunistic pathogens. Shibata and Solo-Gabriele (2012) calculated separate risk estimates for exposure to beach sand for children with an eating disorder called pica characterized by cravings to eat nonfood items.
It is important for beachgoers to consider simple good hygiene practices when having contact with sand at a beach. Whitman et al. (2009) demonstrated that hand rinsing after contact with beach sand can be an effective means of reducing indicator microorganism densities on human hands. They suggested simply rinsing hands before eating or leaving the beach might reduce the incidence of disease. Beachgoers can also reduce health risks to others by ensuring they do not contribute to pathogen loading into beach sand themselves. The shedding of pathogens by beachgoers is considered an under-recognized source of health risks in recreational settings (Ashbolt et al. 2010). Fecal excreta from pets can contaminate beach sand. Beachgoers can also refrain from leaving litter on beach sand or feeding animals near the beach.
Beach Managers
It is recognized that beach sand can serve as an important habitat and reservoir for FIB. The activity of bathers can resuspend submerged sand, and wave action can readily erode and transport foreshore beach sand into adjacent beach waters. These physical processes can lead to transfer of significant loads of FIB into adjacent beach water under certain conditions. Beach managers may need a better understanding of the extent of the reservoir of FIB in beach sands at their beaches in order to understand the occurrence of FIB in beach water samples collected as part of regular beach water quality monitoring programs. It is possible that at some beaches, a considerable load of FIB may be coming from beach sand and may not represent recent sources of fecal pollution. In these cases, the association between levels of FIB in beach water and health risks may not be as strong as when FIB in water are the result of direct fecal contamination events. It may be possible to apply remediation techniques (e.g. grooming, chlorine, iodine or UV treatment) to reduce FIB levels in beach sand and reduce the numbers of beach postings, although these techniques have had variable effects to date and need more study.
In some cases, beach managers may need to understand the implications of FIB and pathogens in beach sand to guide day-to-day decisions to reduce health risks at beaches, both for users and for workers. For example, while beach postings and closures can prevent beachgoers from entering the water, they may also result in increased time spent in contact with beach sand during a beach visit. In other cases, beach managers may need to understand these implications for guiding how to respond to specific pollution contamination events such as sewage spills on beach sand. At present there is little specific guidance for beach managers for controlling access to beach sand or on grooming or remediation approaches for contaminated beach sand.
Regular beach grooming activities can be an important management strategy for removing animal fecal droppings and litter on beach sand. Sand grooming techniques (Kinzelman et al. 2004; Kinzelman et al. 2003) beach slope alterations (Kinzelman and McLellan 2009) and gull control methods (Converse et al. 2012) have helped reduce FIB at Lake Michigan beaches. Such management actions may also reduce pathogen occurrence in beach sand and associated health risks due to beach sand exposure. Bolton et al. (1999) found it surprising that Campylobacter could be detected in as much as 50% of dry sand samples from a beach in England, despite other claims that this pathogen is sensitive to environmental conditions such as low moisture. Mohammad et al. (2012) found that optimal survival of Staphylococcus aureus and Pseudomonas aeruginosa occurred via attachment to intermediate-sized sand particles (850 μm to 2 mm) at a Florida marine beach. They suggested this size range of sand particles could be preferable for formation of micro-niches and should be considered in beach management decisions related to sand replacement, beach nourishment or beach classification schemes.
A large spill of raw sewage occurred onto the beach sand of Manhattan Beach, California, in 2006. Beach mangers decided to confine the spill to the beach rather than let the sewage run off and contaminate nearshore waters. The event identified the lack of guidance for deciding how to control such sewage spills and manage beach sand remediation. It also identified the lack of clean beach sand standards for determining when the public could be allowed access to the beach sand. While it also spurred research to investigate beach disinfection and grooming techniques (Mika et al. 2009), inconsistencies were found in the effectiveness of grooming techniques like sand raking.
Beach Policy Makers
A number of studies have indicated the need to investigate standards for assessing the microbiological quality of sand on bathing beaches (Bolton et al. 1999; Mendes et al. 1993; Sabino et al. 2011a; Shibata and Solo-Gabriele 2012; Whitman et al. 2009). Bolton et al. (1999) indicated that assessment of water quality alone may not be a sufficient basis for determining public health risks from bathing beaches. Some preliminary efforts have been made to propose microbiological standards for sand. Mendes et al (1993) proposed standards for total coliforms (10,000 CFU/g), fecal coliforms (1000 CFU/g), fecal streptococci (100 CFU or MPN/g), and Candida spp. (10 CFU/g). Sabino et al. (2011a) proposed revised standards for potentially pathogenic fungi (17 CFU/g), yeasts (15 CFU/g), and dermatophytes (8 CFU/g) comparing with earlier work reported by the same group (Brandão et al. 2002). Such standards however will probably have to be region specific because positive correlations between level of contamination and region were temperature-dependent in Portugal (Sabino et al. 2011a) – the colder the climate, the longer microorganisms will survive.
However, a number of challenges exist for developing microbiological standards for sand quality. One challenge may be the lack of clear authority in some agencies to develop such standards. For example in the United States, the Clean Water Act covers discharges to surface waters but not necessarily secondary contamination from beach sand. As a result, standards might need to come from individual states or other regulatory agencies. In addition, standard methods and protocols for collecting sand samples and measuring indicator bacteria or pathogens in beach sand have not been developed to date. A further challenge is that much less is known about the role of indicator bacteria in evaluating the quality of beach sand compared to the quality of beach water. Importantly, most indicator bacteria like enterococci and E. coli are associated with fecal pollution and may not be relevant for predicting occurrence of non-fecal pathogens or non-fecal health risks associated with sand.
To date, traditional FIB have proven inconsistent in their ability to predict the occurrence of pathogens and health risks associated with beach sand. Sabino et al. (2011a) investigated occurrence of fungi in beach sand across 33 beaches in Portugal. While they found a positive correlation between yeasts and total coliforms in beach sand, no other correlations were found with FIB. Similarly, there was no discernible relationship between the numbers of Campylobacter and FIB in the sediments of three marine beaches in England (Obiri-Danso and Jones 2000). Yamahara et al. (2012) also found FIB were not consistently associated with pathogens in dry beach sand from 53 California marine beaches. Sands with higher moisture tended to have higher concentrations or more frequent occurrence of pathogens. While there was some evidence of a correlation between Salmonella and E. coli and between Staphylococcus aureus and a human-specific Bacteroidales DNA marker, Campylobacter showed no significant relationship with any FIB in the California sands.
Shah et al. (2011) found that FIB did not correlate consistently with pathogens in subtropical Florida marine beach sand. However, yeasts were significantly correlated with fecal coliforms in beach sand, and red yeasts in particular, were significantly correlated with enterococci. Shah et al. (2011) concluded that indicator microorganisms could predict the presence of some pathogens in subtropical Florida sand and suggested they may be useful for monitoring beach sand quality at non-point source beaches. Goodwin et al. (2012) found that Staphylococcus aureus concentrations in California beach sand were positively correlated with water temperature and S. aureus and enterococci concentrations in adjacent seawater and inversely correlated to wind strength. They indicated this was evidence in support of beach sand being a source of pathogens in adjacent surface water.
Heaney et al. (2012) investigated two U.S. marine beaches and found that increased concentrations of enterococci (measured by both culture and qPCR methods) in wet sand were associated with increased risk of gastrointestinal and diarrhea illness from digging in sand and being buried in sand. However, a culture-based method for enumerating F+ coliphage and qPCR methods for enumerating fecal Bacteroides and Clostridium in sand were inconsistent in identifying an association with increased health risks at these two marine beaches.
To date, the extent of potential health risks from beach sand has been considered inconclusive, and evidence of the need for sand standards has been considered insufficient (Health Canada 2012; World Health Organization 2003). Halliday and Gast (2011) suggested further research into the introduction, distribution, and persistence of FIB and pathogens in beach sand, and the public health implications of these findings, is needed before incorporating beach sands into a beach monitoring framework. At present, guidance is provided for safe hygiene practices and beach management strategies such as grooming and litter removal until health risks associated with sand exposure are better understood (Health Canada 2012; World Health Organization 2003).
SUMMARY AND CONCLUSIONS
Pure sand alone provides neither the nutrients nor metabolic requirements to support replicating microbial populations, as sand grains are generally formed from materials such as silica and calcium carbonate. Sand does provide extensive surface area for adsorption of nutrients, microbial attachment, and a matrix that traps organic matter and water. The ecological niches of microbes are constrained at the level of microenvironments, where pore spaces and sand grain surfaces may provide opportunity for enhanced survival, replication and viable populations, resulting in microbial communities in sand environments. Presumably, advantageous characteristics for sand-dwelling microbes in what many may perceive as a biologically-challenging habitat include rapid colonization through replication and/or accumulation, tolerance to harsh and ever-changing conditions, formation of biofilms, and wide tolerance to variable pore water conditions. Apart from episodic disturbances by wind and water, wetted sands of beaches afford a highly suitable environment for microbes, particularly just above the tide and swash zones.
Summary
Through our review we have demonstrated that beach sands harbor dense and diverse assemblages of microorganisms. Microbiological communities in the sand, i.e. the micropsammon, are being revealed through an accumulating literature focused on measures of specific bacteria coupled with more recent advances in microbial community analysis. The transport, source and fate of organisms highlight the complexity of microbial population ‘budgets,’ both within the beach and adjacent water. Replication, resuscitation, persistence, offshore importation, animal deposition, passive and active movement along the shore, infiltration and exfiltration interacting with differential environmental factors help account for the variation in the characterization of this community in the literature (Figure 3). All of these processes impact the distribution of microbes in the sand environment and can have public health implications through direct exposure of human populations to sand and through indirect exposure to water containing microbes derived from sand.
Figure 3.
Important factors influencing the net gains and losses of foreshore micropsammon particularly the pathogens and FIB. General autochthonous factors are in green while allochthonous trends are in blue. Net gains and losses trends are denoted by + and −.
With respect to habitat, beach sands offer a unique environment for incidental and naturalized microbes. Pore water is an excellent medium for prokaryotes. Sand surfaces themselves not only offer a large surface area for biofilm development but also microbial micro-habitats that provide cover from predators and micro-niches that enhance diversity. Microbial diversity is likely favored by the varied vertical and horizontal zonation. Oxygen varies from near zero below the water table to saturation within the fringe layer. Waves, capillary displacement and groundwater flow supplies the zone with new water and nutrients while removing metabolic wastes. At the larger scale, tides shift shorelines continuously, presenting microbes in the swash zone with unique challenges such as abrasion, exposure to light and continual habitat instability. Thus, a dynamic swash zone has fewer microbes but the band a few meters inland where wrack, debris and berm accumulates often has maximal concentrations that then again diminishes landward as the influence of surface and groundwater diminishes. Backshore sands, while more stable, are also often cooler at depth in respect to more surficial exposed foreshore sands and further removed from surface water organic input, surface- groundwater interchange and recruitment of new microbes. All of these factors influence the distribution of microbes within the beach environment.
Sources of microbes to the micropsammon are rarely singular or simple. For instance, existing background populations—regardless if they are persistent, resident, adapted—can also be supplemented by sewage, human and animal shedding, replication, resuscitation, and latent importation from pre-existing on and offshore reservoirs. While the literature supports long time survival of FIB, more studies are needed to understand potential sinks and other sources (e.g. storm water culverts, algae, deposition zones, riparian runoff). A growing body of evidence is indicating the importance of bird fecal droppings as a potential source of FIB and pathogens in beach sand, particularly around the Great Lakes.
Persistence and Replication
The micropsammonic community must be able to persist and replicate in the harsh ecosystem, characterized by the dynamic setting at the sand-water interface. A convincing body of evidence indicates that many allochthonous microbes form self-sustaining populations in sand. Evidence for this process is found in both the traditional ecological literature as well as through more recent advances. Studies near isothermic and chemically stable artesian springs have shown the gradient of sediment from gravel through sorted sand to fines and detritus and discovered microbial zonation (Byappanahalli et al. 2003a; Whitman et al. 2006). Because these organisms have adapted recently to long ago to the environment, it follows that other opportunistic enteric microbes might exploit or adapt to this habitat. Additional evidence is provided by genomic studies that show multiplication and persistence in the environment (Badgley et al. 2010; Byappanahalli et al. 2006a; Byappanahalli et al. 2012c; Ishii et al. 2006; Whitman et al. 2005) and homeostatic populations whose carrying capacity is limited by carbon, competition, or predation (Byappanahalli and Fujioka 2004; Feng et al. 2010; Hartke et al. 2002; McCambridge and McMeekin 1980; Whitman et al. 2005). Studies have shown that FIB survival and replication is not limited to sand or soils but is also observed in other environments including animal enclosures, bog pitcher plant fluids, bromeliads, pulp mills, detritus, and aquatic plant material. These microbes have been termed naturalized, resident, endogenous, endemic, environmental, ambient, autochthonous, non-enteric, non-fecal, opportunistic, incidental, persistent, psammonic, or phreatic largely depending on the presumed life history, phylogeny, sources, habitat, and emphasis of the author. This diverse terminology needs consolidation, or at least clarification. We propose here the term allochthonous microbes to represent opportunistic introductions generally from other natural or cultural sources. These microbes may then become naturalized if they adapt and establish replicating populations ultimately becoming part of the autochthonous micropsammonic community. We recommend the use of these terms until such time that research better reveals the natural history of these organisms.
Physical and Biological Transport
Levels of microbes within the micropsammon are also governed by physical and biological transport. Transport of microbes into and through the sand is critical to our understanding of the distribution, occurrence, and interchange of this community. Groundwater transport depends on the relative water elevations between the surface water and groundwater table, which are influenced by hydraulic forces acting upon the beach (waves, seiches, and tides). Higher shoreline kinetics may favor sediment transport and a stronger exchange of microbes between the water and sand. Much is known about transport of sediments along coastlines, and while these processes are largely driven by wind and currents within the surf zone, more complex non-turbulent conditions may prevail at the upper fringes of the foreshore where microorganisms persist. Intensive studies in the very nearshore and swash may help explain how microbe laden sands are resuspended, imported and exported from the foreshore and submerged sediment. The resuspension and transport of FIB from beach sand is increasingly recognized as a cause of beach closures. The beach sand/water interface can be a dynamic habitat with fluxes of microbes from beach water into sand, or from sand into adjacent beach water at times. Further studies are needed of the prevalence and conditions leading to this phenomenon, particularly as it can compromise the use of FIB as an indicator of health risks.
In addition to physical processes, biological processes also influence the transport of microbes in sand. Bacteria living on sands secrete EPS which, in turn, may decrease hydraulic conductivity resulting in changes in nutrient fluxes, promoting adsorption and survival of bacteria. Biofilm development may aid in the spread of microbes along hospitable media and may account for the rapid recolonization witnessed in new beach nourishment, late spring/early summer blooms or population density resiliency after storms. Spatial dispersal by replication of microbes can be considered biological transport and it appears to be a more common phenomenon than formerly supposed. The replication of microbes has been encouraged to the point of developing biobarriers which can be utilized for bioremediation.
A wide variety of studies have documented the large numbers of FIB in beach sand. FIB numbers in beach sand can be orders of magnitude higher than in adjacent beach water. At present the public health implications of these high numbers of FIB are not well understood. While there have been some preliminary proposals for sand quality standards (e.g. for fungi), government agencies have yet to develop standards for beach sand quality. A growing number of studies are also reporting the occurrence of bacterial, protozoan, fungal, and viral pathogens in beach sands around the world. Foreshore beach sand has been identified as a reservoir for pathogens as well as FIB. However, tools such as QMRA or epidemiology studies have only recently been applied to consider potential health risks associated with exposure to beach sand. One of the first comprehensive epidemiological investigations of the risk of illness associated with specific sand contact activities found that digging in the sand was associated with a modest, but significant, increased risk of gastrointestinal illness and diarrhea (Heaney et al. 2009). Being buried in the sand was more strongly associated with risk of gastrointestinal illness and diarrhea than digging in the sand. Children under 10 years old were most associated with an increased risk of diarrhea from digging in beach sand. Additional research is urgently needed to better understand potential health risks associated with exposure to beach sand and whether standards are required for sand quality in addition to existing ones widely used for beach water quality.
The dynamics of the micropsammon call into question the implications with respect to public health. The potential presence of pathogens in sand is of interest for beachgoers, public health specialists, regulators, and beach managers. If environmentally adapted populations of FIB prevail, populations of human pathogens may well also exist. The literature shows that the highest density of FIB is in the cooler, moist sands of foreshore: a favorite location for infants and children to play and for seniors to relax. Unfortunately, these are also the age groups most vulnerable to disease. A better understanding of microbial community structure and the fate of pathogens and indicators is needed to evaluate potential impacts of exposure and health risk. Because there is a continual interchange of FIB between beach sand and adjacent surface water, findings should be extended and interpreted within the context of this transition zone. Cleaning nearby offshore fecal pollution sources (e.g. wastewater effluents) may deliver only limited or short-term improvement unless onshore fecal pollution sources (e.g. bird fecal droppings) and the sand micropsammon are also addressed. Knowledge of the sources and coastal dynamics of onshore and nearshore contaminants should be considered before beach design because infrastructure and situation have a large impact in both sand and water quality. Because sand not only has higher densities of microbes and more persistence, long term control may be necessary to achieve prolonged improvement. We have not specifically discussed remediation alternatives, but beach redesign, beach re-contouring, sand grooming, bird deterrence, and increasing water flow within embayment are areas of needed study.
Most impressive upon review of work on beach sand quality are the fundamental questions that still need to be answered. We know relatively little about the biology, ecology, and transport of these microbes and most importantly, we do not know the health implications or how to manage for it, if significant. More work is needed to understand the physical, biotic, and ecological interactions of the micropsammon in the context of controlling populations of microbes of human health significance. Research is needed to better characterize the role of microbial communities in controlling the levels of indicators and pathogens within the micropsammon. More studies are needed to evaluate the interplay of microbes between the sand and the water. One possible scenario includes sand serving as the primary reservoir of microbes but the water serving as the main exposure route. Of interest would be to evaluate the significance of the exchange of microbes between sand and water by conducting holistic epidemiologic studies that evaluate both water and sand exposure routes within the context of the same study. In summary, the micropsammon is a vastly understudied ecosystem that merits additional attention due to its influence on human health through direct exposure to sand or through indirect exposures through water. In order to understand the risks of microbes within the micropsammon, more work is needed in understanding the microbial sources, fate, ecology, and transport processes that control the occurrence of pathogens within the beach environment.
ACKNOWLEDGMENTS
This paper is a direct outcome of “Linking Fecal Indicator Bacteria in Beach Sand and Water” Workshop held during the 2011 Great Lakes Beach Association Conference, Michigan City, IN; September 26-28. We thank Tim Wade of the U.S. EPA for input and helpful comments.
REFERENCES
- Abdallaoui MS, Boutayeb H, Guessous-Idrissi N. Fungal flora from the sand of two beaches of Casablanca (Morroco). Analysis and epidemiological corollary. Journal de Mycologie Médicale. 2007;17:58–62. [Google Scholar]
- Abdelzaher AM, Wright ME, Ortega C, et al. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Applied and Environmental Microbiology. 2010;76:724–732. doi: 10.1128/AEM.02127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Richardson K, Sidhu J, Toze S. Escherichia coli and Enterococcus spp. in rainwater tank samples: comparison of culture-based methods and 23S rRNA gene quantitative PCR assays. Environmental Science and Technology. 2012;46:11370–11376. doi: 10.1021/es302222b. [DOI] [PubMed] [Google Scholar]
- Alam M, Sultana M, Nair GB, et al. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proceedings of the National Academy of Sciences. 2007;104:17801–17806. doi: 10.1073/pnas.0705599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderisio K, DeLuca N. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada geese (Branta canadensis). Applied and Environmental Microbiology. 1999;65:5628–5630. doi: 10.1128/aem.65.12.5628-5630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldova E. Serovars of Vibrio parahaemolyticus. Journal of Hygiene, Epidemiology, Microbiology, and Immunology. 1989;33:219. [PubMed] [Google Scholar]
- Alm EW, Burke J, Hagan E. Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. Journal of Great Lakes Research. 2006;32:401–405. [Google Scholar]
- Alm EW, Burke J, Spain A. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Research. 2003;37:3978–3982. doi: 10.1016/S0043-1354(03)00301-4. [DOI] [PubMed] [Google Scholar]
- Anderson JH. In vitro survival of human pathogenic fungi in Hawaiian beach sand. Medical Mycology. 1979;17:13–22. doi: 10.1080/00362177985380031. [DOI] [PubMed] [Google Scholar]
- Anderson KL, Whitlock JE, Harwood VJ. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Applied and Environmental Microbiology. 2005;71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Turner S, Lewis G. Enterococci in the New Zealand environment: implications for water quality monitoring. Water Science and Technology. 1997;35:325–331. [Google Scholar]
- Ashbolt NJ, Dorsch MR, Cox PT, Banens B. Blooming E. coli, what do they mean? In: Kay D, Fricker C, editors. Coliforms and E. coli, problem or solution? The Royal Society of Chemistry; Cambridge, United Kingdom: 1997. [Google Scholar]
- Ashbolt NJ, Schoen ME, Soller JA, Roser DJ. Predicting pathogen risks to aid beach management: The real value of quantitative microbial risk assessment (QMRA). Water Research. 2010;44:4692–4703. doi: 10.1016/j.watres.2010.06.048. [DOI] [PubMed] [Google Scholar]
- Atlas RM, Bartha R. Microbial Ecology: Fundamentals and Applications. Benjamin-Cummings Pub. Co.; Menlo Park, CA: 1997. pp. 309–313. [Google Scholar]
- Badgley BD, Ferguson J, Vanden Heuvel A, et al. Multi-scale temporal and spatial variation in genotypic composition of Cladophora-borne Escherichia coli populations in Lake Michigan. Water Research. 2011;45:721–731. doi: 10.1016/j.watres.2010.08.041. doi:DOI: 10.1016/j.watres.2010.08.041. [DOI] [PubMed] [Google Scholar]
- Badgley BD, Thomas FIM, Harwood VJ. The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. Environmental Microbiology. 2010;12:1271–1281. doi: 10.1111/j.1462-2920.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- Bauer L, Alm E. Escherichia coli toxin and attachment genes in sand at Great Lakes recreational beaches. Journal of Great Lakes Research. 2012;38:129–133. [Google Scholar]
- Bennani M, Amarouch H, Oubrim N, Cohen N. Identification and antimicrobial resistance of fecal enterococci isolated in coastal Mediterranean environments of Morocco. European Journal of Scientific Research. 2012;70:266–275. [Google Scholar]
- Bermudez M, Hazen TC. Phenotypic and genotypic comparison of Escherichia coli from pristine tropical waters. Applied and Environmental Microbiology. 1988;54:979–983. doi: 10.1128/aem.54.4.979-983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Gueho E, Pesando D. Recherche de dermatophytes et de moisissures pathogènes dans les sables des plages, 1986-87. 1988 [Research on dermatophytes and pathogenic moulds in sea sand, 1986-87] [Google Scholar]
- Beversdorf LJ, Bornstein-Forst SM, McLellan SL. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. Journal of Applied Microbiology. 2007;102:1372–1381. doi: 10.1111/j.1365-2672.2006.03177.x. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Shellenbarger GG, Paytan A. Groundwater discharge: Potential association with fecal indicator bacteria in the surf zone. Environmental Science and Technology. 2004;38:3558–3566. doi: 10.1021/es035385a. [DOI] [PubMed] [Google Scholar]
- Boiron P, Agis F, Nguyen V. [A study of the yeast flora of medical interest in beach of Saint Anne in Guadeloupe]. Bulletin de la Société de Pathologie Exotique et de ses Filiales. 1983;76:351–356. [PubMed] [Google Scholar]
- Bolton F, Surman S, Martin K, Wareing D, Humphrey T. Presence of Campylobacter and Salmonella in sand from bathing beaches. Epidemiology and Infection. 1999;122:7–13. doi: 10.1017/s0950268898001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonadonna L, De Mattia M, Liberti R, Volterra L. Presenza e distribuzione di stafilococchi in ambienti marini [Presence and distribution of staphylococci in the marine environment]. L'Igiene Moderna. 1993;99:706–714. [Google Scholar]
- Bonilla TD, Nowosielski K, Cuvelier M, et al. Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Marine Pollution Bulletin. 2007;54:1472–1482. doi: 10.1016/j.marpolbul.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Bonilla TD, Nowosielski K, Esiobu N, McCorquodale DS, Rogerson A. Species assemblages of Enterococcus indicate potential sources of fecal bacteria at a south Florida recreational beach. Marine Pollution Bulletin. 2006;52:807–810. doi: 10.1016/j.marpolbul.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Brandão J, Wergikosky B, Rosado C, et al. Relatório final do projecto “Qualidade Microbiológica de areias de Praias Litorais”. Associação Bandeira Azul da Europa. 2002 [Google Scholar]
- Brennan FP, O'Flaherty V, Kramers G, Grant J, Richards KG. Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Applied and Environmental Microbiology. 2010;76:1449–1455. doi: 10.1128/AEM.02335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton GA, Jr., Gunnison D, Lanza GR. Survival of pathogenic bacteria in various freshwater sediments. Applied and Environmental Microbiology. 1987;53:633–638. doi: 10.1128/aem.53.4.633-638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli M, Fowler M, Shively D, Whitman R. Ubiquity and persistence of Escherichia coli in a midwestern stream. Applied and Environmental Microbiology. 2003a;69:4549–4555. doi: 10.1128/AEM.69.8.4549-4555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli M, Fujioka R. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Science and Technology. 2004;50:27–32. [PubMed] [Google Scholar]
- Byappanahalli MN, Fujioka RS. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Science and Technology. 1998;38:171–174. [Google Scholar]
- Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment: a review. Microbiology and Molecular Biology Reviews. 2012a;76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Roll BM, Fujioka RS. Evidence for occurrence, persistence, and growth potential of Escherichia coli and enterococci in Hawaii's soil environments. Microbes and Environments. 2012b;27:164–170. doi: 10.1264/jsme2.ME11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Sawdey R, Ishii S, Shively DA, Ferguson J, Whitman RL, Sadowsky MJ. Seasonal stability of Cladophora-associated Salmonella in Lake Michigan watersheds. Water Research. 2009;43:806–814. doi: 10.1016/j.watres.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Shively DA, Nevers MB, Sadowsky MJ, Whitman RL. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiology Ecology. 2003b;46:203–211. doi: 10.1016/S0168-6496(03)00214-9. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Sadowsky MJ, Ishii S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environmental Microbiology. 2006a;8:504–513. doi: 10.1111/j.1462-2920.2005.00916.x. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Ting WTE, Tseng CC, Nevers MB. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. Journal of Water and Health. 2006b;4:313–320. doi: 10.2166/wh.2006.518. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Yan T, Hamilton MJ, Ishii S, Fujioka RS, Whitman RL. The population structure of Escherichia coli isolated from subtropical and temperate soils. Science of the Total Environment. 2012c;417-418:273–279. doi: 10.1016/j.scitotenv.2011.12.041. doi:doi:10.1016/j.scitotenv.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Carrillo-Muñoz A, Torres Rodríguez J, Madrenys-Brunet N, Dronda-Ayza A. Comparative study on the survival of 5 species of dermatophytes and S. brevicaulis in beach sand, under laboratory conditions. Revista Iberoamericana de Micología. 1990;7:36–38. [Google Scholar]
- Chabasse D, Laine P, Simitzis-Le-Flohic A, Martineau B, Hourch ME, Becaud J. Sanitary study of surface water and of the beach of a water sports and leisure complex. Journal of Hygiene. 1986;96:393–401. doi: 10.1017/s0022172400066158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase E, Harwood VJ. Comparison of the effects of environmental parameters on growth rates of Vibrio vulnificus biotypes I, II, and III by culture and quantitative PCR analysis. Applied and Environmental Microbiology. 2011;77:4200–4207. doi: 10.1128/AEM.00135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Walker SL. Fecal indicator bacteria transport and deposition in saturated and unsaturated porous media. Environmental Science and Technology. 2012;46:8782–8790. doi: 10.1021/es301378q. [DOI] [PubMed] [Google Scholar]
- Converse RR, Kinzelman JL, Sams EA, et al. Dramatic improvements in beach water quality following gull removal. Environmental Science and Technology. 2012;46:10206–10213. doi: 10.1021/es302306b. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Cheng K, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annual Reviews in Microbiology. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Crowe AS, Meek GA. Groundwater conditions beneath beaches of Lake Huron, Ontario, Canada. Aquatic Ecosystem Health and Management. 2009;12:444–455. [Google Scholar]
- Crowe AS, Milne J. Relationship between dry and wet beach ecosystems and E. coli levels in groundwater below beaches of the Great Lakes, Canada. In: Ribeiro L, Stigter TY, Chambel A, de Melo MTC, Monteiro JP, Medeiros A, editors. Groundwater and Ecosystems. The Netherlands. 1st edn. Vol. 18. CRC Press; Leiden: 2013. pp. 237–251. [Google Scholar]
- Cui H, Yang K, Pagaling E, Yan T. Spatial and temporal variation in enterococcal abundance and its relationship to the microbial community in Hawaii beach sand and water. Applied and Environmental Microbiology. 2013;79:3601–3609. doi: 10.1128/AEM.00135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham AB, Characklis WG, Abedeen F, Crawford D. Influence of biofilm accumulation on porous media hydrodynamics. Environmental Science and Technology. 1991;25:1305–1311. [Google Scholar]
- Dabrowa N, Landau J, Newcomer V, Plunkett O. A survey of tide-washed coastal areas of southern California for fungi potentially pathogenic to man. Mycopathologia et Mycologia Applicata. 1964;24:137–150. doi: 10.1007/BF02075556. doi:10.1007/bf02075556. [DOI] [PubMed] [Google Scholar]
- Dabrowski J. Isolation of the Shigella genus bacteria from the beach sand and water of the bay of Gdańsk. Bulletin of the Institute of Maritime and Tropical Medicine in Gdynia. 1982;33:49. [PubMed] [Google Scholar]
- Davies-Colley RJ, Bell RG, Donnison AM. Sunlight inactivation of enterococci and fecal coliforms in sewage effluent diluted in seawater. Applied and Environmental Microbiology. 1994;60:2049–2058. doi: 10.1128/aem.60.6.2049-2058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CM, Long JAH, Donald M, Ashbolt NJ. Survival of fecal microorganisms in marine and freshwater sediments. Applied and Environmental Microbiology. 1995;61:1888–1896. doi: 10.1128/aem.61.5.1888-1896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira AJFC, de França PTR, Pinto AB. Antimicrobial resistance of heterotrophic marine bacteria isolated from seawater and sands of recreational beaches with different organic pollution levels in southeastern Brazil: evidences of resistance dissemination. Environmental Monitoring and Assessment. 2010;169:375–384. doi: 10.1007/s10661-009-1180-6. [DOI] [PubMed] [Google Scholar]
- de Oliveira AJFC Pinhata JMW. Antimicrobial resistance and species composition of Enterococcus spp. isolated from waters and sands of marine recreational beaches in Southeastern Brazil. Water Research. 2008;42:2242–2250. doi: 10.1016/j.watres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- de Sieyes NR, Yamahara KM, Layton BA, Joyce EH, Boehm AB. Submarine discharge of nutrient-enriched fresh groundwater at Stinson Beach, California is enhanced during neap tides. Limnology and Oceanography. 2008;53:1434–1445. [Google Scholar]
- de Sieyes NR, Yamahara KM, Paytan A, Boehm AB. Submarine groundwater discharge to a high-energy surf zone at Stinson Beach, California, estimated using radium isotopes. Estuaries and Coasts. 2011;34:256–268. [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Applied and Environmental Microbiology. 2002;68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz J, Rendueles M, Díaz M. Straining phenomena in bacteria transport through natural porous media. Environmental Science and Pollution Research. 2010;17:400–409. doi: 10.1007/s11356-009-0160-2. [DOI] [PubMed] [Google Scholar]
- Doorduyn Y, Van Den Brandhof W, Van Duynhoven Y, Wannet W, Van Pelt W. Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiology and Infection. 2006;134:617–626. doi: 10.1017/S0950268805005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowidart A, Abdel-Monem MH. Effect of chemical pollution on bacterial counts in El-Temsah Lake area, Ismailia, Egypt. J Egyptian Public Health Assoc. 1990;65:305–328. [PubMed] [Google Scholar]
- Dunkell DO, Bruland GL, Evensen CI, Walker MJ. Effects of feral pig (Sus scrofa) exclusion on enterococci in runoff from the forested headwaters of a Hawaiian watershed. Water, Air and Soil Pollution. 2011;221:313–326. [Google Scholar]
- Dunn AM, Silliman SE, Dhamwichukorn S, Kulpa CF. Demonstration of microbial transport into the capillary fringe via advection from below the water table. Journal of Hydrology. 2005;306:50–58. [Google Scholar]
- Edge T, Hill S. P315 Occurrence of ampicillin resistance in E. coli from Lake Ontario beaches and nearby sources of fecal pollution in the Toronto Hamilton area. International Journal of Antimicrobial Agents. 2009;34:S125–S125. [Google Scholar]
- Edge TA, Hill S. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Research. 2007;41:3585–3594. doi: 10.1016/j.watres.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Edge TA, Hill S, Seto P, Marsalek J. Library-dependent and library-independent microbial source tracking to identify spatial variation in faecal contamination sources along a Lake Ontario beach (Ontario, Canada). Water Science and Technology. 2010;62:719–727. doi: 10.2166/wst.2010.335. [DOI] [PubMed] [Google Scholar]
- Elmanama AA, Fahd MI, Afifi S, Abdallah S, Bahr S. Microbiological beach sand quality in Gaza Strip in comparison to seawater quality. Environmental Research. 2005;99:1–10. doi: 10.1016/j.envres.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Elmir SM, Shibata T, Solo-Gabriele HM, et al. Quantitative evaluation of enterococci and Bacteroidales released by adults and toddlers in marine water. Water Research. 2009;43:4610–4616. doi: 10.1016/j.watres.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmir SM, Wright ME, Abdelzaher A, et al. Quantitative evaluation of bacteria released by bathers in a marine water. Water Research. 2007;41:3–10. doi: 10.1016/j.watres.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns AA, Vogel LJ, Abdelzaher AM, et al. Spatial and temporal variation in indicator microbe sampling is influential in beach management decisions. Water Research. 2012;46:2237–2246. doi: 10.1016/j.watres.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esiobu N, Green M, Echeverry A, et al. High numbers of Staphylococcus aureus at three bathing beaches in South Florida. International Journal of Environmental Health Research. 2013;23:46–57. doi: 10.1080/09603123.2012.699027. doi:10.1080/09603123.2012.699027. [DOI] [PubMed] [Google Scholar]
- Esiobu N, Mohammed R, Echeverry A, et al. The application of peptide nucleic acid probes for rapid detection and enumeration of eubacteria, Staphylococcus aureus and Pseudomonas aeruginosa in recreational beaches of S. Florida. Journal of Microbiological Methods. 2004;57:157–162. doi: 10.1016/j.mimet.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Ettema R, Mutel CF. Hans Albert Einstein: Innovation and compromise in formulating sediment transport by rivers. Journal of Hydraulic Engineering. 2004;130:477–487. [Google Scholar]
- Feng F, Goto D, Yan T. Effects of autochthonous microbial community on the die-off of fecal indicators in tropical beach sand. FEMS Microbiology Ecology. 2010;74:214–225. doi: 10.1111/j.1574-6941.2010.00921.x. [DOI] [PubMed] [Google Scholar]
- Feng Z, Reniers A, Haus BK, Solo-Gabriele HM. Modeling sediment-related enterococci loading, transport, and inactivation at an embayed nonpoint source beach. Water Resources Research. 2013;49:693–712. [Google Scholar]
- Filipkowska Z, Janczukowicz W, Krzemieniewski M, Pesta J. Microbiological air pollution in the surroundings of the wastewater treatment plant with activated-sludge tanks aerated by horizontal rotors. Polish Journal of Environmental Studies. 2000;9:273–280. [Google Scholar]
- Flood C, Ufnar J, Wang S, Johnson J, Carr M, Ellender R. Lack of correlation between enterococcal counts and the presence of human specific fecal markers in Mississippi creek and coastal waters. Water Research. 2011;45:872–878. doi: 10.1016/j.watres.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Fogarty LR, Haack SK, Wolcott MJ, Whitman RL. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. Journal of Applied Microbiology. 2003;94:865–878. doi: 10.1046/j.1365-2672.2003.01910.x. [DOI] [PubMed] [Google Scholar]
- Foppen JW, Lutterodt G, Röling WFM, Uhlenbrook S. Towards understanding inter-strain attachment variations of Escherichia coli during transport in saturated quartz sand. Water Research. 2010;44:1202–1212. doi: 10.1016/j.watres.2009.08.034. [DOI] [PubMed] [Google Scholar]
- Foppen JW, van Herwerden M, Schijven J. Transport of Escherichia coli in saturated porous media: Dual mode deposition and intra-population heterogeneity. Water Research. 2007;41:1743–1753. doi: 10.1016/j.watres.2006.12.041. [DOI] [PubMed] [Google Scholar]
- Francy DS, Darner RA. Factors affecting Escherichia coli concentrations at Lake Erie public bathing beaches. US Geological Survey; Columbus, Ohio: 1998. [Google Scholar]
- Francy DS, Gifford AM, Darner RA. Escherichia coli at Ohio bathing beaches--Distribution, sources, wastewater indicators, and predictive modeling. U.S. Geological Survey; Columbus, OH: 2003. [Google Scholar]
- Frenzel SA, Couvillion CS. Fecal-indicator bacteria in streams along a gradient of residential development. Journal of the American Water Resources Association. 2002;38:265–273. [Google Scholar]
- Fries JS, Characklis GW, Noble RT. Attachment of fecal indicator bacteria to particles in the Neuse River Estuary, NC. Journal of Environmental Engineering. 2006;132:1338–1345. [Google Scholar]
- Fujioka R, Sian-Denton C, Borja M, Castro J, Morphew K. Soil: The environmental source of Escherichia coli and enterococci in Guam's streams. Journal of Applied Microbiology Symposium Supplement. 1999;85:83S–89S. doi: 10.1111/j.1365-2672.1998.tb05286.x. [DOI] [PubMed] [Google Scholar]
- Fujioka RS. Monitoring coastal marine waters for spore-forming bacteria of faecal and soil origin to determine point from non-point source pollution. Water Science and Technology. 2001;44:181–188. [PubMed] [Google Scholar]
- Fujioka RS, Byappanahalli MN. Proceedings and report: Tropical water quality indicator workshop. University of Hawaii, Water Resources Research Center; Honolulu, HI.: 2003. pp. 1–95. SR-2004-01. [Google Scholar]
- Fujioka RS, Tenno K, Kansako S. Naturally occurring fecal coliforms and fecal streptococci in Hawaii's freshwater streams. Toxicity Assessment. 1988;3:613–630. [Google Scholar]
- Gauthier F, Archibald F. The ecology of fecal indicator bacteria commonly found in pulp and paper mill water systems. Water Research. 2001;35:2207–2218. doi: 10.1016/s0043-1354(00)00506-6. [DOI] [PubMed] [Google Scholar]
- Ge Z, Nevers MB, Schwab DJ, Whitman RL. Coastal loading and transport of Escherichia coli at an embayed beach in Lake Michigan. Environmental Science and Technology. 2010;44:6731–6737. doi: 10.1021/es100797r. [DOI] [PubMed] [Google Scholar]
- Ge Z, Whitman RL, Nevers MB, Phanikumar MS. Wave-induced mass transport affects daily Escherichia coli fluctuations in nearshore water. Environmental Science and Technology. 2012a;46:2204–2211. doi: 10.1021/es203847n. doi:10.1021/es203847n. [DOI] [PubMed] [Google Scholar]
- Ge Z, Whitman RL, Nevers MB, Phanikumar MS, Byappanahalli MN. Evaluating the role of an embayed beach as a reservoir and a net source of fecal contamination. Limnology and Oceanography. 2012b;57:362–381. [Google Scholar]
- Ghinsberg R, Dror R, Nitzan Y. Isolation of Vibrio vulnificus from sea water and sand along the Dan region coast of the Mediterranean. Microbios. 1999;97:7–17. [PubMed] [Google Scholar]
- Ghinsberg RC, Bar Dov L, Rogol M, Sheinberg Y, Nitzan Y. Monitoring of selected bacteria and fungi in sand and sea water along the Tel Aviv coast. Microbios. 1994;77:29–40. [PubMed] [Google Scholar]
- Ginn TR, Wood BD, Nelson KE, Scheibe TD, Murphy EM, Clement TP. Processes in microbial transport in the natural subsurface. Advances in Water Resources. 2002;25:1017–1042. [Google Scholar]
- Gobet A, Böer SI, Huse SM, et al. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. The ISME Journal. 2012;6:542–553. doi: 10.1038/ismej.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K, Pobuda M. Performance of CHROMagar™ Staph aureus and CHROMagar™ MRSA for detection of Staphylococcus aureus in seawater and beach sand–Comparison of culture, agglutination, and molecular analyses. Water Research. 2009;43:4802–4811. doi: 10.1016/j.watres.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Goodwin KD, Matragrano L, Wanless D, Sinigalliano CD, LaGier MJ. A preliminary investigation of fecal indicator bacteria, human pathogens, and source tracking markers in beach water and sand. Environmental Research Journal. 2009;3:395–417. [PMC free article] [PubMed] [Google Scholar]
- Goodwin KD, McNay M, Cao Y, Ebentier D, Madison M, Griffith JF. A multi-beach study of Staphylococcus aureus, MRSA, and enterococci in seawater and beach sand. Water Research. 2012;46:4195–4207. doi: 10.1016/j.watres.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Bauer S, Johnson JR. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002;148:1513–1522. doi: 10.1099/00221287-148-5-1513. [DOI] [PubMed] [Google Scholar]
- Graczyk TK, Sunderland D, Tamang L, Lucy FE, Breysse PN. Bather density and levels of Cryptosporidium, Giardia, and pathogenic microsporidian spores in recreational bathing water. Parasitology Research. 2007;101:1729–1731. doi: 10.1007/s00436-007-0734-1. [DOI] [PubMed] [Google Scholar]
- Grant SB, Sanders B, Boehm A, et al. Generation of enterococci bacteria in a coastal saltwater marsh and its impact on surf zone water quality. Environmental Science and Technology. 2001;35:2407–2416. doi: 10.1021/es0018163. [DOI] [PubMed] [Google Scholar]
- Grimes D, Atwell R, Brayton P, et al. The fate of enteric pathogenic bacteria in estuarine and marine environments. Microbiological Sciences. 1986;3:324. [PubMed] [Google Scholar]
- Grisoli P, Rodolfi M, Villani S, et al. Assessment of airborne microorganism contamination in an industrial area characterized by an open composting facility and a wastewater treatment plant. Environmental Research. 2009;109:135–142. doi: 10.1016/j.envres.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Haack SK, Fogarty LR, Wright C. Escherichia coli and enterococci at beaches in the Grand Traverse Bay, Lake Michigan: Sources, characteristics, and environmental pathways. Environmental Science and Technology. 2003;37:3275–3282. doi: 10.1021/es021062n. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Reviews Microbiology. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Halliday E, Gast RJ. Bacteria in beach sands: An emerging challenge in protecting coastal water quality and bather health. Environmental Science and Technology. 2011;45:370–379. doi: 10.1021/es102747s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MJ, Hadi AZ, Griffith JF, Ishii S, Sadowsky MJ. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Research. 2010;44:5463–5473. doi: 10.1016/j.watres.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DL, Ishii S, Sadowsky MJ, Hicks RE. Waterfowl abundance does not predict the dominant avian source of beach. Journal of Environmental Quality. 2011;40:1924–1931. doi: 10.2134/jeq2011.0111. [DOI] [PubMed] [Google Scholar]
- Hardina CM, Fujioka RS. Soil: The environmental source of Escherichia coli and enterococci in Hawaii's streams. Environmental Toxicology and Water Quality. 1991;6:185–195. [Google Scholar]
- Harrison S, Kinra S. Outbreak of Escherichia coli O157 associated with a busy bathing beach. Communicable Disease and Public Health. 2004;7:47–50. [PubMed] [Google Scholar]
- Hartke A, Lemarinier S, Pichereau V, Auffray Y. Survival of Enterococcus faecalis in seawater microcosms is limited in the presence of bacterivorous zooflagellates. Current Microbiology. 2002;44:329–335. doi: 10.1007/s00284-001-0018-4. [DOI] [PubMed] [Google Scholar]
- Health Canada . Guidelines for Canadian Recreational Water Quality. Third Edition Ottawa, Ontario: 2012. [Google Scholar]
- Heaney C, Sams E, Wing S, Marshall S, Brenner K, Dufour AP, Wade TJ. Contact with beach sand among beachgoers and risk of illness. American Journal of Epidemiology. 2009;170:164–172. doi: 10.1093/aje/kwp152. [DOI] [PubMed] [Google Scholar]
- Heaney CD, Sams E, Dufour AP, et al. Fecal indicators in sand, sand contact, and risk of enteric illness among beachgoers. Epidemiology. 2012;23:95–106. doi: 10.1097/EDE.0b013e31823b504c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim S, Lleo MM, Bonato B, Guzman CA, Canepari P. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. Journal of Bacteriology. 2002;184:6739–6745. doi: 10.1128/JB.184.23.6739-6745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong D, Damare J, Limpert RJ, Sladen W, Weiner RM, Colwell RR. Microbial impact of Canada geese (Branta canadensis) and whistling swans (Cygnus columbianus columbianus) on aquatic ecosystems. Applied and Environmental Microbiology. 1979;37:14–20. doi: 10.1128/aem.37.1.14-20.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura GJ, Thompson RS, Boehm AB, Jay JA. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiology Ecology. 2011;77:40–49. doi: 10.1111/j.1574-6941.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- Inman D, Tait R, Nordstrom C. Mixing in the surf zone. Journal of Geophysical Research. 1971;76:3493–3514. [Google Scholar]
- Ishii S, Hansen DL, Hicks RE, Sadowsky MJ. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environmental Science and Technology. 2007;41:2203–2209. doi: 10.1021/es0623156. [DOI] [PubMed] [Google Scholar]
- Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Applied and Environmental Microbiology. 2006;72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S, Sadowsky MJ. Escherichia coli in the environment: Implications for water quality and human health. Microbes and Environments. 2008;23:101–108. doi: 10.1264/jsme2.23.101. [DOI] [PubMed] [Google Scholar]
- Ishii S, Yan T, Vu H, Hansen D, Hicks R, Sadowsky M. Factors controlling long-term survival and growth of naturalized Escherichia coli populations in temperate field soils. Microbes and Environments. 2010;25:8–14. doi: 10.1264/jsme2.me09172. [DOI] [PubMed] [Google Scholar]
- Izquierdo J, Piera G, Aledany M, Lucena F. Estudio de la flora fungica de la arena de la playa de Barcelona [A study of the fungal flora of the beaches in Barcelona] 1986 [Google Scholar]
- Johnson WP, Li X, Assemi S. Deposition and re-entrainment dynamics of microbes and non-biological colloids during non-perturbed transport in porous media in the presence of an energy barrier to deposition. Advances in Water Resources. 2007;30:1432–1454. [Google Scholar]
- Jousset A. Ecological and evolutive implications of bacterial defences against predators. Environmental Microbiology. 2012;14:1830–1843. doi: 10.1111/j.1462-2920.2011.02627.x. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Reviews Microbiology. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Khan I, Edge TA. Investigation of the prevalence of thermophilic Campylobacter species at Lake Simcoe recreational beaches. Inland Waters. 2013;3:93–104. [Google Scholar]
- Khan IU, Hill S, Nowak E, Edge TA. Effect of incubation temperature on the detection of thermophilic Campylobacter species from freshwater beaches, nearby wastewater effluents, and bird fecal droppings. Applied and Environmental Microbiology. 2013;79:7639–7645. doi: 10.1128/AEM.02324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IUH, Loughborough A, Edge TA. DNA-based real-time detection and quantification of aeromonads from fresh water beaches on Lake Ontario. Journal of Water and Health. 2009;7:312–323. doi: 10.2166/wh.2009.041. [DOI] [PubMed] [Google Scholar]
- Khiyama HM, Makemson JC. Sand beach bacteria: enumeration and characterization. Applied Microbiology. 1973;26:293–297. doi: 10.1128/am.26.3.293-297.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzelman J, McLellan SL. Success of science-based best management practices in reducing swimming bans—a case study from Racine, Wisconsin, USA. Aquatic Ecosystem Health and Management. 2009;12:187–196. [Google Scholar]
- Kinzelman J, McLellan SL, Amick A, et al. Identification of human enteric pathogens in gull feces at Southwestern Lake Michigan bathing beaches. Canadian Journal of Microbiology. 2008;54:1006–1015. doi: 10.1139/W08-096. [DOI] [PubMed] [Google Scholar]
- Kinzelman J, Pond K, Longmaid K, Bagley R. The effect of two mechanical beach grooming strategies on Escherichia coli density in beach sand at a southwestern Lake Michigan beach. Aquatic Ecosystem Health and Management. 2004;7:425–432. [Google Scholar]
- Kinzelman J, Whitman RL, Byappanahalli MN, Jackson EK, Bagley RC. Evaluation of beach grooming techniques on Escherichia coli density in foreshore sand at North Beach, Racine, WI. Lake and Reservoir Management. 2003;19:349–354. [Google Scholar]
- Kishimoto RA, Baker GE. Pathogenic and potentially pathogenic fungi isolated from beach sands and selected soils of Oahu, Hawaii. Mycologia. 1969:537–548. [PubMed] [Google Scholar]
- Kleinheinz GT, McDermott CM, Hughes S, Brown A. Effects of rainfall on E. coli concentrations at Door County, Wisconsin beaches. International Journal of Microbiology. 20092009 doi: 10.1155/2009/876050. Article ID 876050 doi:10.1155/2009/876050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Weir SC, Howell ET, Lee H, Trevors JT. Genetic relatedness of Escherichia coli isolates in interstitial water from a Lake Huron (Canada) beach. Applied and Environmental Microbiology. 2007;73:1961–1967. doi: 10.1128/AEM.02437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Harwood VJ, Shanks OC, Fout GS, Ashbolt NJ. Differential decay of enterococci and Escherichia coli originating from two fecal pollution sources. Applied and Environmental Microbiology. 2013;79:2488–2492. doi: 10.1128/AEM.03781-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka JE, Prakash O, Overholt WA, et al. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Applied and Environmental Microbiology. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksoll WB, Ishii S, Sadowsky MJ, Hicks RE. Presence and sources of fecal coliform bacteria in epilithic periphyton communities of Lake Superior. Applied and Environmental Microbiology. 2007;73:3771–3778. doi: 10.1128/AEM.02654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLiberte P, Grimes DJ. Survival of Escherichia coli in lake bottom sediment. Applied and Environmental Microbiology. 1982;43:623–628. doi: 10.1128/aem.43.3.623-628.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo JV, Calvo MA. Fungal density in the sands of the Mediterranean coast beaches. Mycopathologia. 1989;108:185–193. doi: 10.1007/BF00436224. [DOI] [PubMed] [Google Scholar]
- Lavender JS, Kinzelman JL. A cross comparison of QPCR to agar-based or defined substrate test methods for the determination of Escherichia coli and enterococci in municipal water quality monitoring programs. Water Research. 2009;43:4967–4979. doi: 10.1016/j.watres.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Lee CM, Lin TY, Lin CC, Kohbodi GA, Bhatt A, Lee R, Jay JA. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Research. 2006;40:2593–2602. doi: 10.1016/j.watres.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Lévesque B, Brousseau P, Simard P, Dewailly E, Meisels M, Ramsay D, Joly J. Impact of the ring-billed gull (Larus delawarensis) on the microbiological quality of recreational water. Applied and Environmental Microbiology. 1993;59:1228–1230. doi: 10.1128/aem.59.4.1228-1230.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Edens E, Bonilla N, Meschke JS, Roberts MC. Survival of environmental and clinical strains of methicillin-resistant Staphylococcus aureus [MRSA] in marine and fresh waters. Water Research. 2011;45:5681–5686. doi: 10.1016/j.watres.2011.08.037. [DOI] [PubMed] [Google Scholar]
- Levin-Edens E, Soge OO, No D, Stiffarm A, Meschke JS, Roberts MC. Methicillin-resistant Staphylococcus aureus from Northwest marine and freshwater recreational beaches. FEMS Microbiology Ecology. 2012;79:412–420. doi: 10.1111/j.1574-6941.2011.01229.x. doi:10.1111/j.1574-6941.2011.01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Barry D, Pattiaratchi C, Masselink G. BeachWin: modelling groundwater effects on swash sediment transport and beach profile changes. Environmental Modelling and Software. 2002;17:313–320. [Google Scholar]
- Logan AJ, Stevik TK, Siegrist RL, Rønn RM. Transport and fate of Cryptosporidium parvum oocysts in intermittent sand filters. Water Research. 2001;35:4359–4369. doi: 10.1016/s0043-1354(01)00181-6. [DOI] [PubMed] [Google Scholar]
- Lovins K, Angle J, Wiebers J, Hill R. Leaching of Pseudomonas aeruginosa, and transconjugants containing pR68. 45 through unsaturated, intact soil columns. FEMS Microbiology Ecology. 1993;13:105–111. [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proceedings of the National Academy of Sciences. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallin MA, Williams KE, Esham EC, Lowe RP. Effect of human development on bacteriological water quality in coastal watersheds. Ecological Applications. 2000;10:1047–1056. [Google Scholar]
- Marino F, Morinigo M, Martinez-Manzanares E, Borrego J. Microbiological-epidemiological study of selected marine beaches in Malaga (Spain). Water Science and Technology. 1995;31:5–9. [Google Scholar]
- Marsalek J, Rochfort Q. Urban wet-weather flows: sources of fecal contamination impacting on recreational waters and threatening drinking-water sources. Journal of Toxicology and Environmental Health, Part A. 2004;67:1765–1777. doi: 10.1080/15287390490492430. [DOI] [PubMed] [Google Scholar]
- McCambridge J, McMeekin TA. Relative effects of bacterial and protozoan predators on survival of Escherichia coli in estuarine water samples. Applied and Environmental Microbiology. 1980;40:907–911. doi: 10.1128/aem.40.5.907-911.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes B, Nascimento MJ, Oliveira JS. Preliminary characterization and proposal of microbiological quality standard of sand beaches. Water Science and Technology. 1993;27:453–456. [Google Scholar]
- Mendes B, Urbano P, Alves C, Lapa N, Morais J, Nascimento J, Oliveira J. Sanitary quality of sands from beaches of Azores islands. Water Science and Technology. 1997;35:147–150. [Google Scholar]
- Mika KB, Imamura G, Chang C, et al. Pilot- and bench-scale testing of fecal indicator bacteria survival in marine beach sand near point sources. Journal of Applied Microbiology. 2009;107:72–84. doi: 10.1111/j.1365-2672.2009.04197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed R, Echeverry A, Stinson C, et al. Survival trends of Staphylococcus aureus, Pseudomonas aeruginosa, and Clostridium perfringens in a sandy South Florida beach. Marine Pollution Bulletin. 2012;64:1201. doi: 10.1016/j.marpolbul.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Mudryk Z, Perliński P, Skórczewski P. Detection of antibiotic resistant bacteria inhabiting the sand of non-recreational marine beach. Marine Pollution Bulletin. 2010;60:207–214. doi: 10.1016/j.marpolbul.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Mudryk ZJ, Kosiorek A, Perliński P. In vitro antibiotic resistance of Vibrio-like organisms isolated from seawater and sand of marine recreation beach in the southern Baltic Sea. Hydrobiologia. 2013;702:141–150. doi:10.1007/s10750-012-1317-4. [Google Scholar]
- Murphy EM, Ginn TR. Modeling microbial processes in porous media. Hydrogeology Journal. 2000;8:142–158. [Google Scholar]
- Neel JK. A limnological investigation of the psammon in Douglas Lake, Michigan, with especial reference to shoal and shoreline dynamics. Transactions of the American Microscopical Society. 1948:1–53. [Google Scholar]
- Nestor I, Costin-Lazår L, Sovrea D, Ionescu N. Detection of enteroviruses in seawater and beach sand. Zentralblatt für Bakteriologie, Mikrobiologie und Hygiene 1 Abt Originale B. Hygiene. 1984;178:527–534. [PubMed] [Google Scholar]
- Nielsen P. Coastal bottom boundary layers and sediment transport. Vol. 4. World Scientific Publishing Company Incorporated; 1992. [Google Scholar]
- Noble RT, Griffith JF, Blackwood AD, et al. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Applied and Environmental Microbiology. 2006;72:1604–1612. doi: 10.1128/AEM.72.2.1604-1612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiri-Danso K, Jones K. Distribution and seasonality of microbial indicators and thermophilic campylobacters in two freshwater bathing sites on the River Lune in northwest England. Journal of Applied Microbiology. 1999;87:822–832. doi: 10.1046/j.1365-2672.1999.00924.x. [DOI] [PubMed] [Google Scholar]
- Obiri-Danso K, Jones K. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognised bathing waters in north west England. Water Research. 2000;34:519–527. [Google Scholar]
- Olapade OA, Depas MM, Jensen ET, McLellan SL. Microbial communities and fecal indicator bacteria associated with Cladophora mats on beach sites along Lake Michigan shores. Applied and Environmental Microbiology. 2006;72:1932–1938. doi: 10.1128/AEM.72.3.1932-1938.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiology Reviews. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- Oshiro R, Fujioka R. Sand, soil, and pigeon droppings: sources of indicator bacteria in the waters of Hanauma Bay, Oahu, Hawaii. Water Science and Technology. 1995;31:251–254. [Google Scholar]
- Ostrolenk M, Kramer N, Cleverdon RC. Comparative studies of enterococci and Escherichia coli as indices of pollution. Journal of Bacteriology. 1947;53:197–203. doi: 10.1128/jb.53.2.197-203.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M. Bacterial loadings from resuspended sediments in recreational beaches. Canadian Journal of Civil Engineering. 1988;15:450–455. [Google Scholar]
- Papadakis JA, Mavridou A, Richardson SC, Lampiri M, Marcelou U. Bather-related microbial and yeast populations in sand and seawater. Water Research. 1997;31:799–804. [Google Scholar]
- Perkins S, Gyr P, James G. The influence of biofilm on the mechanical behavior of sand. ASTM Geotechnical Testing Journal. 2000;23:300–312. [Google Scholar]
- Phillips MC, Solo-Gabriele HM, Piggot AM, Klaus JS, Zhang YJ. Relationships between sand and water quality at recreational beaches. Water Research. 2011a;45:6763–6769. doi: 10.1016/j.watres.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MC, Solo-Gabriele HM, Reniers AJ, Wang JD, Kiger RT, Abdel-Mottaleb N. Pore water transport of enterococci out of beach sediments. Marine Pollution Bulletin. 2011b;62:2293–2298. doi: 10.1016/j.marpolbul.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianetti A, Bruscolini F, Sabatini L, Colantoni P. Microbial characteristics of marine sediments in bathing area along Pesaro-Gabicce coast (Italy): a preliminary study. Journal of Applied Microbiology. 2004;97:682–689. doi: 10.1111/j.1365-2672.2004.02352.x. [DOI] [PubMed] [Google Scholar]
- Piggot AM, Klaus JS, Johnson S, Phillips MC, Solo-Gabriele HM. Relationship between enterococcal levels and sediment biofilms at recreational beaches in South Florida. Applied and Environmental Microbiology. 2012;78:5973–5982. doi: 10.1128/AEM.00603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano LRW, Garza AC, Shibata T, et al. Shedding of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from adult and pediatric bathers in marine waters. BMC Microbiology. 2011;11:5. doi: 10.1186/1471-2180-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommepuy M, Butin M, Derrien A, Gourmelon M, Colwell R, Cormier M. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Applied and Environmental Microbiology. 1996;62:4621–4626. doi: 10.1128/aem.62.12.4621-4626.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environmental Microbiology. 2005;7:631–640. doi: 10.1111/j.1462-2920.2005.00729.x. [DOI] [PubMed] [Google Scholar]
- Prado B, Bernal P, Contreras M, Saavedra M, Del Moral A, Joyas A. Numerical taxonomy of staphylococci isolated from water and beach sand from Valparaíso and viña del Mar, Chile. Revista Latinoamericana de Microbiología. 1994;36:71. [PubMed] [Google Scholar]
- Price W, Burchell M, II, Hunt W, Chescheir G. Long-term study of dune infiltration systems to treat coastal stormwater runoff for fecal bacteria. Ecological Engineering. 2013;52:1–11. [Google Scholar]
- Rijnaarts HHM, Norde W, Bouwer EJ, Lyklema J, Zehnder AJB. Bacterial deposition in porous media related to the clean bed collision efficiency and to substratum blocking by attached cells. Environmental Science and Technology. 1996;30:2869–2876. [Google Scholar]
- Ripp S, Nivens DE, Werner C, Sayler GS. Vertical transport of a field-released genetically engineered microorganism through soil. Soil Biology and Biochemistry. 2001;33:1873–1877. [Google Scholar]
- Rivera S, Hazen T, Toranzos G. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Applied and Environmental Microbiology. 1988;54:513–517. doi: 10.1128/aem.54.2.513-517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC, Soge OO, Giardino MA, Mazengia E, Ma G, Meschke JS. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. Journal of Applied Microbiology. 2009;107:300–307. doi: 10.1111/j.1365-2672.2009.04207.x. [DOI] [PubMed] [Google Scholar]
- Robertson JB, Edberg SC. Natural protection of spring and well drinking water against surface microbial contamination. I. Hydrogeological parameters. Critical Reviews in Microbiology. 1997;23:143–178. doi: 10.3109/10408419709115134. [DOI] [PubMed] [Google Scholar]
- Romero OC, Straub AP, Kohn T, Nguyen TH. Role of temperature and Suwannee river natural organic matter on inactivation kinetics of rotavirus and bacteriophage MS2 by solar irradiation. Environmental Science and Technology. 2011;45:10385–10393. doi: 10.1021/es202067f. [DOI] [PubMed] [Google Scholar]
- Roses Codinachs M, Isern Vins AM, Ferrer Escobar MD, Fernandez Perez F. Microbiological contamination of the sand from the Barcelona city beaches. Revista de Sanidad e Higiene Publica. 1988;62:1537–1544. [PubMed] [Google Scholar]
- Ross N, Villemur R, Deschênes L, Samson R. Clogging of a limestone fracture by stimulating groundwater microbes. Water Research. 2001;35:2029–2037. doi: 10.1016/s0043-1354(00)00476-0. [DOI] [PubMed] [Google Scholar]
- Rusch A, Huettel M, Reimers CE, Taghon GL, Fuller CM. Activity and distribution of bacterial populations in Middle Atlantic Bight shelf sands. FEMS Microbiology Ecology. 2003;44:89–100. doi: 10.1111/j.1574-6941.2003.tb01093.x. [DOI] [PubMed] [Google Scholar]
- Russell TL, Sassoubre LM, Wang D, et al. A Coupled Modeling and Molecular Biology Approach to Microbial Source Tracking at Cowell Beach, Santa Cruz, CA, United States. Environmental Science and Technology. 2013;47:10231–10239. doi: 10.1021/es402303w. [DOI] [PubMed] [Google Scholar]
- Russell TL, Yamahara KM, Boehm AB. Mobilization and transport of naturally occurring enterococci in beach sands subject to transient infiltration of seawater. Environmental Science and Technology. 2012;46:5988–5996. doi: 10.1021/es300408z. [DOI] [PubMed] [Google Scholar]
- Sabino R, Sampaio P, Carneiro C, Rosado L, Pais C. Isolates from hospital environments are the most virulent of the Candida parapsilosis complex. BMC Microbiology. 2011a;11:180. doi: 10.1186/1471-2180-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino R, Veríssimo C, Cunha MA, et al. Pathogenic fungi: An unacknowledged risk at coastal resorts? New insights on microbiological sand quality in Portugal. Marine Pollution Bulletin. 2011b;62:1506–1511. doi: 10.1016/j.marpolbul.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Salmore A, Hollis E, McLellan S. Delineation of a chemical and biological signature for stormwater pollution in an urban river. Journal of Water and Health. 2006;4:247–262. [PubMed] [Google Scholar]
- Sampson RW, Swiatnicki SA, Osinga VL, Supita JL, McDermott CM, Kleinheinz GT. Effects of temperature and sand on E. coli survival in a northern lake water microcosm. Journal of Water and Health. 2006;4:389–393. doi: 10.2166/wh.2006.524. [DOI] [PubMed] [Google Scholar]
- Sanchez PS, Agudo EG, Castro FG, Alves MN, Martins MT. Evaluation of the sanitary quality of marine recreational waters and sands from beaches of the São Paulo State, Brazil. Water Science and Technology. 1986;18:61–72. [Google Scholar]
- Sato MIZ, Di Bari M, Lamparelli CC, Truzzi AC, Coelho MCLS, Hachich EM. Sanitary quality of sands from marine recreational beaches of São Paulo, Brazil. Brazilian Journal of Microbiology. 2005;36:321–326. [Google Scholar]
- Sauer EP, VandeWalle JL, Bootsma MJ, McLellan SL. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Research. 2011;45:4081–4091. doi: 10.1016/j.watres.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Schuch AP, Menck CFM. The genotoxic effects of DNA lesions induced by artificial UV-radiation and sunlight. Journal of Photochemistry and Photobiology B: Biology. 2010;99:111–116. doi: 10.1016/j.jphotobiol.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Seyfried PL, Tobin RS, Brown NE, Ness PF. A prospective study of swimming-related illness. II. Morbidity and microbiologial quality of water. American Journal of Public Health. 1985;75:1071–1075. doi: 10.2105/ajph.75.9.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AH, Abdelzaher AM, Phillips M, et al. Indicator microbes correlate with pathogenic bacteria, yeasts and helminthes in sand at a subtropical recreational beach site. Journal of Applied Microbiology. 2011;110:1571–1583. doi: 10.1111/j.1365-2672.2011.05013.x. [DOI] [PubMed] [Google Scholar]
- Shatti JA, Abdullah THA. Marine pollution due to wastewater discharge in Kuwait. Water Science and Technology. 1999;40:33–39. [Google Scholar]
- Shibata T, Solo-Gabriele HM. Quantitative Microbial Risk Assessment of Human Illness from Exposure to Marine Beach Sand. Environmental Science and Technology. 2012;46:2799–2805. doi: 10.1021/es203638x. doi:10.1021/es203638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigalliano CD, Gidley M, Shibata T, et al. Impacts of Hurricanes Katrina and Rita on the microbial landscape of the New Orleans area. Proceedings of the National Academy of Sciences. 2007;104:9029–9034. doi: 10.1073/pnas.0610552104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalbeck JD, Kinzelman JL, Mayer GC. Fecal indicator organism density in beach sands: impact of sediment grain size, uniformity, and hydrologic factors on surface water loading. Journal of Great Lakes Research. 2010;36:707–714. [Google Scholar]
- Soge OO, Meschke JS, No DB, Roberts MC. Characterization of methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococcus spp. isolated from US West Coast public marine beaches. Journal of Antimicrobial Chemotherapy. 2009;64:1148–1155. doi: 10.1093/jac/dkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. Sources of Escherichia coli in a coastal subtropical environment. Applied and Environmental Microbiology. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MLR. Micoses. [Fungi.] National Institute of Health (INSA), Centre of Epidemiologic Surveillance of Transmissible Diseases (Epidemiology Bulletin No. 5) Lisbon. 1990 [Google Scholar]
- Staff M, Musto J, Hogg G, Janssen M, Rose K. Salmonellosis outbreak traced to playground sand, Australia, 2007–2009. Emerging Infectious Diseases. 2012;18:1159–1161. doi: 10.3201/eid1807.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JL, Evans GE, Aguirre KM. Human beach use affects abundance and identity of fungi present in sand. Journal of Coastal Research. 2012;28:787–792. [Google Scholar]
- Stocker R. Marine microbes see a sea of gradients. Science. 2012;338:628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- Suter E, Juhl AR, O'Mullan GD. Particle association of Enterococcus and total bacteria in the lower Hudson River estuary, USA. Journal of Water Resource and Protection. 2011;3:715–725. [Google Scholar]
- Sutherland BM. Photoreactivation. BioScience. 1981;31:439–444. [Google Scholar]
- Tiefenthaler L, Stein E, Schiff K. Levels and patterns of fecal indicator bacteria in stormwater runoff from homogenous land use sites and urban watersheds. Journal of Water and Health. 2011;9:279–290. doi: 10.2166/wh.2010.056. [DOI] [PubMed] [Google Scholar]
- Twinning TL, Whitman RL, Hoff BE. Occurrence of E. coli in open beach groundwaters of Indiana Dunes National Lakeshore.. 109th Annual Meeting of the Indiana Academy of Science; West Lafayette, Indiana. 1993. [Google Scholar]
- US EPA. Cruise Ship Discharge Assessment Report. EPA 842-R-07-005. U.S. Environmental Protection Agency, Office of Water; Washington, DC: 2008. [Google Scholar]
- Vally H, Whittle A, Cameron S, Dowse GK, Watson T. Outbreak of Aeromonas hydrophila wound infections associated with mud football. Clinical Infectious Diseases. 2004;38:1084–1089. doi: 10.1086/382876. [DOI] [PubMed] [Google Scholar]
- Van Donsel DJ, Geldreich EE, Clarke NA. Seasonal variations in survival of indicator bacteria in soil and their contribution to storm-water pollution. Applied Microbiology. 1967;15:1362–1370. doi: 10.1128/am.15.6.1362-1370.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Heuvel A, McDermott C, Pillsbury R, et al. The green alga, Cladophora, promotes E. coli growth and contamination of recreational waters in Lake Michigan. Journal of Environmental Quality. 2010;39:333–344. doi: 10.2134/jeq2009.0152. [DOI] [PubMed] [Google Scholar]
- Vieira RHSF Rodrigues DP, Menezes EA Evangelista NSS, Reis EMF Barreto LM, Gonçalves FA. Microbial contamination of sand from major beaches in Fortaleza, Ceará State, Brazil. Brazilian Journal of Microbiology. 2001;32:77–80. [Google Scholar]
- Vijayavel K, Fujioka R, Ebdon J, Taylor H. Isolation and characterization of the Bacteroides host strain HB-73 used to detect sewage specific phases in Hawaii. Water Research. 2010;44:3714–3724. doi: 10.1016/j.watres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Vijayavel K, Sadowsky MJ, Ferguson JA, Kashian DR. The establishment of the nuisance cyanobacteria Lyngbya wollei in Lake St. Clair and its potential to harbor fecal indicator bacteria. Journal of Great Lakes Research. 2013;39:560–568. [Google Scholar]
- Vogel C, Rogerson A, Schatz S, Laubach H, Tallman A, Fell J. Prevalence of yeasts in beach sand at three bathing beaches in South Florida. Water Research. 2007;41:1915–1920. doi: 10.1016/j.watres.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittamn TS. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environmental Microbiology. 2007;9:2274–2288. doi: 10.1111/j.1462-2920.2007.01341.x. [DOI] [PubMed] [Google Scholar]
- Wang A, Lin B, Sleep BE, Liss SN. The impact of biofilm growth on transport of Escherichia coli O157: H7 in sand. Ground Water. 2011;49:20–31. doi: 10.1111/j.1745-6584.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- Wanjugi P, Harwood VJ. The influence of predation and competition on the survival of commensal and pathogenic fecal bacteria in aquatic habitats. Environmental Microbiology. 2013;15:517–526. doi: 10.1111/j.1462-2920.2012.02877.x. [DOI] [PubMed] [Google Scholar]
- Weinzirl J, Newton EB. The fate of bacteria in frozen meat held in cold storage, and its bearing on a bacteriological standard for condemnation. American Journal of Public Health. 1915;5:833–835. doi: 10.2105/ajph.5.9.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JE, Jones DT, Harwood VJ. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Research. 2002;36:4273–4282. doi: 10.1016/s0043-1354(02)00139-2. [DOI] [PubMed] [Google Scholar]
- Whitman RL, Byers SE, Shively DA, Ferguson DM, Byappanahalli MN. Occurrence and growth characteristics of Escherichia coli and enterococci within the accumulated fluid of the northern pitcher plant (Sarracenia purpurea L.). Canadian Journal of Microbiology. 2005;51:1027–1037. doi: 10.1139/w05-091. [DOI] [PubMed] [Google Scholar]
- Whitman RL, Horvath TG, Goodrich ML, Nevers MB, Wolcott MJ, Haack SK. Characterization of E. coli levels at 63rd Street Beach. Report to the City of Chicago. Department of the Environment and the Chicago Park District; Chicago, Illinois: 2001. [Google Scholar]
- Whitman RL, Nevers MB. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Applied and Environmental Microbiology. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB, Byappanahalli MN. Examination of the watershed-wide distribution of Escherichia coli along southern Lake Michigan: An integrated approach. Applied and Environmental Microbiology. 2006;72:7301–7310. doi: 10.1128/AEM.00454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB, Korinek GC, Byappanahalli MN. Solar and temporal effects on Escherichia coli concentration at a Great Lakes swimming beach. Applied and Environmental Microbiology. 2004;70:4276–4285. doi: 10.1128/AEM.70.7.4276-4285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Przybyla-Kelly K, Shively DA, Nevers MB, Byappanahalli MN. Hand-mouth transfer and potential for exposure to E. coli and F+ coliphage in beach sand, Chicago, Illinois. Journal of Water and Health. 2009;7:623–629. doi: 10.2166/wh.2009.115. doi:doi:10.2166/wh.2009.115. [DOI] [PubMed] [Google Scholar]
- Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Applied and Environmental Microbiology. 2003;69:4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AP, Avery LM, Killham K, Jones DL. Persistence, dissipation, and activity of Escherichia coli O157:H7 within sand and seawater environments. FEMS Microbiology Ecology. 2007;60:24–32. doi: 10.1111/j.1574-6941.2006.00273.x. [DOI] [PubMed] [Google Scholar]
- Woessner WW, Ball PN, DeBorde DC, Troy TL. Viral transport in a sand and gravel aquifer under field pumping conditions. Ground Water. 2005;39:886–894. doi: 10.1111/j.1745-6584.2001.tb02476.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . Guidelines for safe recreational water environments: Coastal and fresh waters (Vol. 1), vol Volume 1, Coastal and Fresh Waters. World Health Organization; Geneva, Switzerland: 2003. Microbial aspects of beach sand quality. pp. 118–127. [Google Scholar]
- Wright M, Abdelzaher A, Solo-Gabriele H, Elmir S, Fleming L. The inter-tidal zone is the pathway of input of enterococci to a subtropical recreational marine beach. Water Science and Technology. 2011;63:542–549. doi: 10.2166/wst.2011.255. [DOI] [PubMed] [Google Scholar]
- Wright ME, Solo-Gabriele HM, Elmir S, Fleming LE. Microbial load from animal feces at a recreational beach. Marine Pollution Bulletin. 2009;58:1649–1656. doi: 10.1016/j.marpolbul.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin P, Robinson C, Li L, Barry DA, Bakhtyar R. Effects of wave forcing on a subterranean estuary. Water Resources Research. 2010;46:W12505. [Google Scholar]
- Yallop M, Paterson D, Wellsbury P. Interrelationships between rates of microbial production, exopolymer production, microbial biomass, and sediment stability in biofilms of intertidal sediments. Microbial Ecology. 2000;39:116–127. doi: 10.1007/s002489900186. [DOI] [PubMed] [Google Scholar]
- Yamahara KM, Layton BA, Santoro AE, Boehm AB. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environmental Science and Technology. 2007;41:4515–4521. doi: 10.1021/es062822n. [DOI] [PubMed] [Google Scholar]
- Yamahara KM, Sassoubre LM, Goodwin KD, Boehm AB. Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Applied and Environmental Microbiology. 2012;78:1733–1745. doi: 10.1128/AEM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahara KM, Walters SP, Boehm AB. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Applied and Environmental Microbiology. 2009;75:1517–1524. doi: 10.1128/AEM.02278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehms TT, McDermott CM, Kleinheinz GT. Microbial concentrations in sand and their effect on beach water in Door County, Wisconsin. Journal of Great Lakes Research. 2008;34:524–534. [Google Scholar]
- Zhang Q, Yan T. Correlation of intracellular trehalose concentration with desiccation resistance of soil Escherichia coli populations. Applied and Environmental Microbiology. 2012;78:7407–7413. doi: 10.1128/AEM.01904-12. doi:10.1128/aem.01904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wang JD, Solo-Gabriele HM, Fleming LE. A water quality modeling study of non-point sources at recreational marine beaches. Water Research. 2011;45:2985–2995. doi: 10.1016/j.watres.2011.03.015. [DOI] [PubMed] [Google Scholar]