Abstract
The intestinal epithelium is a single cell barrier separating a sterile mucosal tissue from a large microbial community dominated by obligate anaerobic bacteria, which inhabit the gut lumen. To maintain mucosal integrity, any breach in the epithelial barrier needs to be met with an inflammatory host response designed to repel microbial intruders from the tissue, protect the mucosal surface and repair injuries to the epithelium. In addition, inflammation induces mechanisms of nutritional immunity, which limit the availability of metals in the intestinal lumen, thereby imposing new selective forces on microbial growth. However, the inflammatory host response also has important side effects. A by-product of producing reactive oxygen and nitrogen species aimed at eradicating microbial intruders is the luminal generation of exogenous electron acceptors. The presence of these electron acceptors creates a new metabolic niche that is filled by facultative anaerobic bacteria. Here we review the changes in microbial nutrient utilization that accompany intestinal inflammation and the consequent changes in the composition of gut-associated microbial communities.
Keywords: Intestinal inflammation, microbiota, nutritional immunity, anaerobic respiration
Introduction
During intestinal inflammation the epithelium plays an important role in mounting responses that are aimed at clearing the mucosal surface from microbes. For example, production of IFN-γ during inflammation results in the activation of DUOX2 (dual function NADPH oxidase 2) [1], NOX1 (NADPH oxidase 1) [2] and iNOS (inducible nitric oxide synthase) [3] in epithelial cells. Reactive oxygen species (ROS) produced by DUOX2 and NOX1 and reactive nitrogen species (RNS) generated by iNOS create a hostile environment in close proximity to the mucosal surface. Furthermore, the pro-inflammatory cytokine interleukin (IL)-22 induces the luminal release of the antimicrobial proteins lipocalin-2, calprotectin, RegIIIβ (regenerating islet-derived 3 beta) and RegIIIγ from epithelial cells [4-6].
These epithelial defenses can be augmented by the transmigration of neutrophils into the intestinal lumen as the severity of intestinal inflammation increases. Upon transmigration, the phagocyte NADPH oxidase (PHOX), superoxide dismutase (SOD) and myeloperoxidase (MPO) of neutrophils generate additional ROS in the gut lumen. Subsequent lysis of neutrophils in the intestinal lumen releases calprotectin, which constitutes approximately 40% of their cytoplasmic content [7]. As a result, neutrophils are the main sources of luminal calprotectin during severe intestinal inflammation [8].
Some of the antimicrobials released into the intestinal lumen are bacteriocidal, thereby protecting the mucosa from infection. For instance, release of the C-type lectin RegIIIγ contributes to luminal clearance of opportunistic pathogens, such as Listeria monocytogenes or vancomycin-resistant Enterococcus feacium, which are both members of the class Bacilli within the phylum Furmicutes [9,10]. Chronic granulomatous disease, an illness caused by PHOX-deficiency, illustrates that the generation of ROS by phagocytes is essential for preventing recurrent bacterial infections [11-13]. It is thus likely that upon transmigration into the lumen, the respiratory burst of neutrophils aids in clearing bacteria from the vicinity of the mucosal surface. However, recent evidence suggests that in addition to its bacteriocidal effects, the inflammatory host response has also a profound impact on the nutritional environment in the gut lumen, which can lead to alterations in the composition of gut-associated microbial communities (microbiota). Here we review these novel hypothesis and the underlying mechanisms.
Nutritional immunity changes the rules for microbial contestants
One subset of antimicrobial proteins released into the intestinal lumen during inflammation functions in limiting the availability of trace elements required for bacterial growth, such as iron and zinc, a host defense mechanism known as nutritional immunity. Bacteria acquire ferric iron (Fe3+) by releasing high-affinity iron chelators, termed siderophores (reviewed in [14]). Enterobactin, a cyclic trimer of N-(2,3-dihydroxybenzoyl)-L-serine, is the siderophore produced by most members of the Enterobacteriaceae, a family of facultative anaerobic bacteria belonging to the class Gammaproteobacteria within the phylum Proteobacteria [15-17]. After chelating iron, the Fe3+-enterobactin complex is transported actively by an energy-coupled outer membrane receptor protein into the periplasm. The energy required for transporting the Fe3+-enterobactin complex across the outer membrane is provided by the proton motive force of the cytoplasmic membrane, which is transmitted to the outer membrane via the TonB protein (reviewed in [14]).
Lipocalin-2 prevents bacterial iron acquisition by binding and sequestering enterobactin [18-20]. While uptake of Fe3+-enterobactin is a viable strategy for obtaining iron in the non-inflamed intestine, the epithelial release of lipocalin-2 during conditions of inflammation inhibits growth of bacteria relying solely on enterobactin for iron acquisition. Thus, bacteria acquiring iron through mechanisms that are not inhibited by lipocalin-2 gain a relative luminal growth advantage in the inflamed gut. This concept was first described in Salmonella enterica, a member of the Enterobacteriaceae that secretes enterobactin along with a glycosylated derivative of enterobactin, termed salmochelin [21]. Salmochelin is not sequestered by lipocalin-2, thereby conferring resistance against this antimicrobial protein [22,23]. Deletion of the iroN gene, which encodes the TonB-dependent outer membrane siderophore receptor [24], renders S. enterica unable to utilize salmochelin [21]. As a result, an S. enterica iroN mutant solely relies on enterobactin for iron-acquisition. Compared to wild-type bacteria, growth of a S. enterica iroN mutant in the lumen of the mouse gut is reduced in the presence, but not in the absence of intestinal inflammation. Furthermore, S. enterica wild type and iroN mutant grow equally well in the inflamed gut of lipocalin-2-deficient mice [5]. Thus, luminal growth of lipocalin-2 resistant bacteria is favored in the inflamed gut, but not in the absence of intestinal inflammation.
A second metal that is sequestered by the host during inflammation through the release of antimicrobial proteins into the intestinal lumen is zinc. Calprotectin, a heterodimer composed of S100A8 and S100A9, inhibits bacterial growth in tissue by chelating both manganese and zinc [25]. Recent studies suggest that the transepithelial migration of neutrophils and the subsequent release of calprotectin from dead neutrophils reduce the availability of zinc in the intestinal lumen [8]. Zinc is transported across the cytoplasmic membrane of S. enterica by the high-affinity ABC (ATP binding cassette) transporter ZnuABC [26]. Compared to the S. enterica wild type, luminal growth of a znuA mutant is impaired in the inflamed intestine of wild type mice, but not in the inflamed intestine of S100A9-deficient mice [8]. These data support the idea that by overcoming the calprotectin-mediated host zinc sequestration, bacterial high-affinity zinc acquisition confers a luminal fitness advantage during colitis.
Above examples illustrate that the inflammatory host response can influence bacterial growth by changing the nutritional environment in the intestinal lumen. As a result, bacterial metal acquisition strategies that bestow no apparent growth benefit in the healthy gut can confer a luminal fitness advantage in the inflamed intestine. In other words, the host response can alter the contest rules that govern microbial competition for metals.
Interestingly, reducing the availability of metals brings microbes, which rely on similar iron acquisition strategies, into a contest. For example, the commensal Escherichia coli strain Nissle 1917, a member of the family Enterobacteriaceae, elaborates four siderophores, including enterobactin, salmochelin, aerobactin and yersiniabactin [27-29]. Of these siderophores, only enterobactin is sequestered by lipocalin-2. Co-colonization with E. coli Nissle 1917 reduces luminal growth of the pathogenic S. enterica in wild-type mice, but not in lipocalin-2-deficient mice. Furthermore, co-colonization of mice with a siderophore utilization-deficient E. coli Nissle 1917 tonB mutant does not reduce the ability of S. enterica to grow in the intestinal lumen [30]. These data suggest that by lowering the availability of iron in the lumen, the host inflammatory response can alter the outcome of a competition between bacterial species that utilize overlapping siderophore repertoires.
Microbial metabolism in the healthy large intestine
In addition to conferring nutritional immunity by lowering the availability of metals in the intestinal lumen, the host response changes the luminal environment by generating inflammation-derived nutrients as a by-product. The resulting bloom of bacterial species that can utilize inflammation-derived nutrients can alter the composition of gut-associated microbial communities. To understand how inflammation-derived nutrients alter the growth conditions in the large bowel, it is important to first grasp the nutrient acquisition and utilization strategies that characterize a balanced microbiota, which inhabits the healthy gut.
In healthy individuals, obligate anaerobic bacteria belonging to the classes Bacteroidia (phylum Bacteroidetes) and Clostridia (phylum Firmicutes) dominate microbial communities inhabiting the anaerobic environment of the lower gastrointestinal tract [31]. Since simple carbohydrates and proteins are digested and absorbed in the upper gastrointestinal tract, complex carbohydrates (e.g. fiber or mucus carbohydrates) or non-digestible proteins (e.g. gluten) are the main nutrients supporting growth of Bacteroidia and Clostridia in the large bowel. Oxygen or other exogenous electron acceptors are not available in the healthy distal gut to support respiration. Thus, microbes rely largely on fermentation of carbohydrates and amino acids to generate energy via substrate-level phosphorylation and to acquire carbon and nitrogen for the biosynthesis of proteins, carbohydrates, lipids and nucleotides.
To maintain redox balance during fermentation, electrons have to be transferred from NADH onto organic compounds, such as phosphoenolpyruvate, thereby generating metabolic end products that are released. Microbiota-derived fermentation end products that commonly accumulate in the gut lumen include formate, acetate, proprionate, butyrate, lactate and hydrogen (H2) (Fig. 1A). Some bacteria, such as Bacteroides fragilis, maintain redox balance by transferring electrons onto fumarate to generate succinate, a process known as fumarate respiration (reviewed in [32]). During this process, B. fragilis fixes host-derived carbon dioxide (CO2) onto phosphoenolpyruvate to generate oxaloacetate, which is converted by reversing reaction of the tricarboxylic acid (TCA) cycle into the endogenous electron acceptor fumarate [33]. Succinate is released as a metabolic end product of fumarate respiration. Thus fumarate respiration and fermentation have in common that metabolically valuable phosphoenolpyruvate is removed from anabolic reactions and converted into metabolic end products to maintain redox balance.
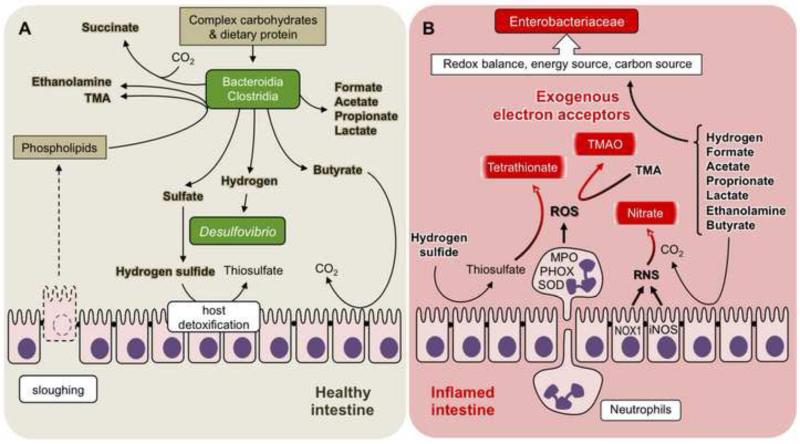
Figure 1.
The inflammatory host response creates a new metabolic niche in the intestine.
(A) The metabolic niche occupied by obligate anaerobic bacteria in the healthy intestine. Clostridia and Bacteroidia degrade complex carbohydrates and proteins in the distal gut to form a variety of metabolic end products, which accumulate in the lumen [32]. Furthermore, the head groups of phospholipids released by sloughing are degraded to TMA and ethanolamine [63]. Hydrogen produced by Clostridia and Bacteroidia fuels the growth of sulfate-reducing bacteria (Desulfovibrio), which produce hydrogen sulfide [35-37], a toxic gas oxidized by colonocytes to form harmless thiosulfate [38,39]. Butyrate is a fermentation product of obligate anaerobic bacteria that serves as nutrient for colonocytes [34]. (B) The metabolic niche occupied by facultative anaerobic Enterobacteriaceae in the healthy intestine. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) generated by the inflammatory host response oxidize luminal compounds (TMA and thiosulfate) to form exogenous electron acceptors (trimethylamine N-oxide [TMAO] and tetrathionate, respectively) [40,48,49]. Some RNS species are converted into the exogenous electron acceptor nitrate in a reaction catalyzed by carbon dioxide (CO2) [45]. The presence of exogenous electron acceptors enables Enterobacteriaceae to utilize microbiota-derived metabolic end products to generate energy, maintain redox balance and acquire carbon for the biosynthesis of primary metabolites.
Metabolic end products generated by Bacteroidia and Clostridia change the nutritional environment for both the host and other intestinal microbes. For example, some metabolic end products of Bacteroidia and Clostridia, such as butyrate, confer benefit to the host by providing nutrition for colonocytes, which mitochondrially oxidize this compound to carbon dioxide [34] (Fig. 1A). Other metabolites, such as hydrogen, are consumed by obligate anaerobic sulfate-reducing bacteria. Sulfate-reducing bacteria of the genus Desulfovibrio (class Gammaproteobacteria, family Desulfovibrionaceae) in turn generate the genotoxic gas hydrogen sulfide (H2S) in the distal gut [35-37]. To avoid toxicity, host colonocytes express sulfide oxidases in their apical membrane that oxidize hydrogen sulfide to generate harmless thiosulfate (S2O32−) [38,39].
By-products of the host response support anaerobic respiration
Inflammation causes a dramatic change in the nutritional environment in the large intestine by generating exogenous electron acceptors. This concept was first established by showing that a by-product of neutrophils mounting a respiratory burst in the gut lumen is the oxidation of thiosulfate to tetrathionate (S4O62−) [40] (Fig. 1B). S. enterica can use tetrathionate as a respiratory electron acceptor, a property that has been used empirically in clinical laboratories since 1923 to enrich for this pathogen in samples containing competing microbes [41]. However, tetrathionate respiration is of biological significance because it confers a luminal fitness advantage upon S. enterica in the inflamed intestine, thereby resulting in a disruption of the microbiota composition, which is characterized by an outgrowth of the pathogen [40]. The uncontrolled growth of S. enterica in the lumen of the inflamed gut enhances its transmission by the fecal oral route, thereby placing tetrathionate respiration under selection [42].
The finding that the inflammatory host response generates an exogenous electron acceptor in the gut lumen suggests that changes in the nutritional environment are a by-product of the antimicrobial activity of ROS and RNS. While ROS and RNS create a hostile environment in close proximity to the mucosal surface, these radicals are short lived and quickly react to form harmless oxidation by-products, such as tetrathionate. In addition to tetrathionate, a number of other oxidation by-products can drastically change bacterial growth conditions in the anaerobic environment of the large bowel. Nitric oxide (NO) generated by iNOS can react with superoxide radicals (O2.−) produced by host NADPH oxidases to yield the bacteriocidal compound peroxynitrite (ONOO−) [43,44]. Peroxynitrite is further converted to nitrate (NO3−) in a reaction catalyzed by carbon dioxide [45]. Through this mechanism, intestinal inflammation generates nitrate in the gut lumen [46,47] (Fig. 1B). Furthermore, ROS and RNS can oxidize organic sulfides, such as methionine, or tertiary amines, such as trimethylamine (TMA), to form the respective S-oxides and N-oxides [48,49]. Nitrate, tetrathionate, S-oxides and N-oxides are harmless oxidation products that can serve as exogenous electron acceptors for anaerobic respiration (reviewed in [50]). Thus, a by-product of releasing bacteriocidal ROS and RNS during inflammation is the generation of a cocktail of host-derived exogenous electron acceptors that enable microbes to perform anaerobic respiration.
The nitrate/nitrite redox couple has a high standard redox potential (E° = 433 mV), which is second only to that of the oxygen/water redox couple (E° = 818 mV) (reviewed in [51]). Under anaerobic conditions, nitrate is therefore the most potent electron acceptor for energy production (reviewed in [50]). Among the phylogenetic groupings that are present within gut-associated microbial communities, genes encoding nitrate reductase activity are found most commonly within genomes of the facultative anaerobic Enterobacteriaceae [52]. In contrast, genes encoding nitrate reductase activity are notably absent in genomes of obligate anaerobic bacteria belonging to the Bacteroidia and Clostridia [52]. The generation of host-derived nitrate during inflammation is thus predicted to favor growth of Enterobacteriaceae, because members of this family happen to be more likely to encode the enzymes required for nitrate respiration.
Direct evidence that inflammation-derived nitrate boosts growth of Enterobacteriaceae comes from studies on two of its representatives, S. enterica and E. coli. Colitis induced by S. enterica infection leads to the production of nitrate in the gut lumen, which in turn increases growth of the pathogen by nitrate respiration [53]. To enhance its growth in the gut lumen, S. enterica uses motility and chemotaxis to actively seek out metabolic niches that contain respiratory electron acceptors, such as nitrate and tetrathionate [54]. Similarly, nitrate generated by iNOS during chemically-induced or genetically-induced colitis markedly increases the luminal abundance of E. coli by supporting growth of this commensal microbe through nitrate respiration [47]. Host-derived nitrate is also generated during low-level intestinal inflammation induced by oral antibiotic treatment, thereby conferring a nitrate respiration-dependent fitness advantage upon commensal E. coli [55]. It has been proposed that the generation of nitrate in the intestinal lumen is one of the mechanisms by which antibiotic treatment reduces colonization resistance against commensal E. coli and other Enterobacteriaceae [56]. Luminal growth of E. coli is likely fueled further by respiration of other inflammation-derived electron acceptors, such as S-oxides and N-oxides [40]. In summary, the generation of exogenous electron acceptors by the inflammatory host response provides Enterobacteriaceae with a decisive luminal fitness advantage, which results in their uncontrolled expansion in the large bowel.
Why inflammation-derived electron acceptors favor Enterobacteriaceae
Exogenous electron acceptors enable Enterobacteriaceae to use strategies for maintaining redox balance, generating energy and acquiring carbon for biosynthesis of primary metabolites that are fundamentally different from those employed by Bacteroidia and Clostridia, thereby creating a new metabolic niche for these facultative anaerobic bacteria (Fig. 1). There are three advantages the metabolic strategy of Enterobacteriaceae has over that employed by Bacteroidia and Clostridia.
First, anaerobic respiration enables Enterobacteriaceae to balance their redox sheet by transferring electrons from NADH onto respiratory electron acceptors, such as nitrate, thereby preserving phosphoenolpyruvate for anabolic reactions. In contrast, fumarate respiration and fermentation performed by Bacteroidia and Clostridia are accompanied by the removal of metabolically valuable phosphoenolpyruvate to form metabolic end products to maintain redox balance.
Second, Enterobacteriaceae can use metabolic end products of Bacteroidia and Clostridia, such as formate or hydrogen, as electron donors to produce energy by anaerobic respiration (reviewed in [50]). This process can be performed solely for the purpose of energy production. For example, the transfer of electrons from hydrogen onto nitrate is independent of both carbon acquisition and maintaining redox balance. The finding that hydrogen enhances luminal growth of S. enterica in a mouse model [57], suggests that the ability to use metabolic end products of Bacteroidia and Clostridia for the sole purpose of producing energy provides a fitness advantage.
Third, anaerobic respiration broadens the spectrum of compounds that can serve as carbon sources in the anaerobic environment of the distal gut. During S. enterica infection, intestinal contents are removed by the flushing action of diarrhea, thereby limiting microbial nutrition to compounds available in the mucus layer. Phosphatidylethanolamine, the most abundant phospholipid of enterocytes [58], is released into the mucus by sloughing and subsequently fermented by the microbiota, which produces ethanolamine as one of the end products (Fig. 1A). In the presence of an exogenous electron acceptor, such as tetrathionate, S. enterica can grow anaerobically on ethanolamine as the sole carbon source [59]. The presence of tetrathionate in the inflamed gut enables S. enterica to utilize ethanolamine to boost its luminal growth [60]. Collectively, these data suggest that the ability to utilize fermentation end products of Bacteroidia and Clostridia as a carbon source for the biosynthesis of proteins, carbohydrates, lipids and nucleotides confers a growth advantage upon S. enterica.
Conclusions
Intestinal inflammation has an impact on microbial metabolism through two different mechanisms. The first is related to the activity of antimicrobial proteins, which are released into the intestinal lumen during inflammation. The presence of these antimicrobial proteins favors growth of bacteria that are resistant against host nutrient withholding mechanisms [5,8]. As a result, nutritional immunity can alter the outcome of a competition between individual microbes [30]. A mechanistic understanding of these processes might facilitate the rational design of probiotics with increased capacity to exclude enteric pathogens.
The second mechanism influencing microbial metabolism is the generation of exogenous electron acceptors as a by-product of the inflammatory host response. In the anaerobic environment of the healthy gut, nutrient acquisition strategies of Enterobacteriaceae are inferior to the metabolic tactics employed by Bacteroidia and Clostridia (reviewed in [61]). However, the generation of exogenous electron acceptors by the inflammatory host response creates a new metabolic niche in the gut lumen. Unlike the obligate anaerobic Bacteroidia and Clostridia, the facultative anaerobic Enterbacteriaceae possess the enzymes to take advantage of the novel opportunities for generating energy, maintaining redox balance and acquiring carbon that become available in this new niche. As Enterobatceriaceae fill the metabolic niche created by intestinal inflammation, their relative abundance within the community increases and the resulting disruption of a balanced microbiota composition is known as dysbiosis. A dysbiosis characterized by a bloom of Enterobacteriaceae in the lower gastrointestinal tract is the ecological pattern observed most consistently in studies describing the changes in microbial communities that accompany gut inflammation (reviewed in [62]). A mechanistic understanding of the mechanisms responsible for dysbiosis identifies anaerobic respiration as a potential target for intervention strategies aimed at restoring a balanced community structure to improve health.
Acknowledgements
Work in A.J.B.’s laboratory is supported by Public Health Service grants AI107393 and AI096528.
Abbreviations
- DUOX2
dual function NADPH oxidase 2
- IFN-γ
gamma interferon
- IL
interleukin
- iNOS
inducible nitric oxide synthase
- MPO
myeloperoxidase
- NADH
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NOX1
NADPH oxidase 1
- PHOX
phagocyte NADPH oxidase
- RegIIIβ
regenerating islet-derived 3 beta
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- TCA
tricarboxylic acid
- TMA
trimethylamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature
- [1].Harper RW, Xu C, Eiserich JP, Chen Y, Kao CY, Thai P, Setiadi H, Wu R. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS letters. 2005;579:4911–4917. doi: 10.1016/j.febslet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- [2].Kuwano Y, Kawahara T, Yamamoto H, Teshima-Kondo S, Tominaga K, Masuda K, Kishi K, Morita K, Rokutan K. Interferon-gamma activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. American journal of physiology Cell physiology. 2006;290:C433–443. doi: 10.1152/ajpcell.00135.2005. [DOI] [PubMed] [Google Scholar]
- [3].Salzman A, Denenberg AG, Ueta I, O’Connor M, Linn SC, Szabo C. Induction and activity of nitric oxide synthase in cultured human intestinal epithelial monolayers. The American journal of physiology. 1996;270:G565–573. doi: 10.1152/ajpgi.1996.270.4.G565. [DOI] [PubMed] [Google Scholar]
- [4].Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- [5].Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Teigelkamp S, Bhardwaj RS, Roth J, Meinardus-Hager G, Karas M, Sorg C. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J Biol Chem. 1991;266:13462–13467. [PubMed] [Google Scholar]
- [8].Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. The Journal of experimental medicine. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hohn DC, Lehrer RI. NADPH oxidase deficiency in X-linked chronic granulomatous disease. The Journal of clinical investigation. 1975;55:707–713. doi: 10.1172/JCI107980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McPhail LC, DeChatelet LR, Shirley PS, Wilfert C, Johnston RB, Jr., McCall CE. Deficiency of NADPH oxidase activity in chronic granulomatous disease. The Journal of pediatrics. 1977;90:213–217. doi: 10.1016/s0022-3476(77)80632-x. [DOI] [PubMed] [Google Scholar]
- [13].Seger RA, Tiefenauer L, Matsunaga T, Wildfeuer A, Newburger PE. Chronic granulomatous disease due to granulocytes with abnormal NADPH oxidase activity and deficient cytochrome-b. Blood. 1983;61:423–428. [PubMed] [Google Scholar]
- [14].Braun V. Iron uptake by Escherichia coli. Frontiers in bioscience : a journal and virtual library. 2003;8:s1409–1421. doi: 10.2741/1232. [DOI] [PubMed] [Google Scholar]
- [15].Reissbrodt R, Rabsch W. Further differentiation of Enterobacteriaceae by means of siderophore-pattern analysis. Zentralbl Bakteriol Mikrobiol Hyg A. 1988;268:306–317. doi: 10.1016/s0176-6724(88)80015-4. [DOI] [PubMed] [Google Scholar]
- [16].Young IG. Preparation of enterochelin from Escherichia coli. Prep Biochem. 1976;6:123–131. doi: 10.1080/00327487608061607. [DOI] [PubMed] [Google Scholar]
- [17].Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- [18].Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- [19].Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- [20].Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Molecular microbiology. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- [23].Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT, Aderem A, Smith KD. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baumler AJ, Norris TL, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- [26].Campoy S, Jara M, Busquets N, Perez De, Rozas AM, Badiola I, Barbe J. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect Immun. 2002;70:4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cress BF, Linhardt RJ, Koffas MA. Draft Genome Sequence of Escherichia coli Strain Nissle 1917 (Serovar O6:K5:H1) Genome Announc. 2013;1:e0004713. doi: 10.1128/genomeA.00047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sun J, Gunzer F, Westendorf AM, Buer J, Scharfe M, Jarek M, Gossling F, Blocker H, Zeng AP. Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J Biotechnol. 2005;117:147–161. doi: 10.1016/j.jbiotec.2005.01.008. [DOI] [PubMed] [Google Scholar]
- [30].Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E, Macfarlane S. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, Mackie RI, Gaskins HR. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zinkevich VV, Beech IB. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol Ecol. 2000;34:147–155. doi: 10.1111/j.1574-6941.2000.tb00764.x. [DOI] [PubMed] [Google Scholar]
- [38].Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharmacol. 2001;62:255–259. doi: 10.1016/s0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- [39].Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest. 1999;104:1107–1114. doi: 10.1172/JCI7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Muller L. Un nouveau milieu d’enrichissement pour la recherche du Bacille Typhique at Paratyphique. Comptes Rendus des Seances de la Societe de Biologie et de ses Filiales. 1923:434–437. [Google Scholar]
- [42].Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhu L, Gunn C, Beckman JS. Bactericidal activity of peroxynitrite. Archives of biochemistry and biophysics. 1992;298:452–457. doi: 10.1016/0003-9861(92)90434-x. [DOI] [PubMed] [Google Scholar]
- [44].De Groote MA, Granger D, Xu Y, Campbell G, Prince R, Fang FC. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- [46].Dudhgaonkar SP, Tandan SK, Kumar D, Raviprakash V, Kataria M. Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology. 2007;15:188–195. doi: 10.1007/s10787-007-1603-3. [DOI] [PubMed] [Google Scholar]
- [47].Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova I, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Balagam B, Richardson DE. The mechanism of carbon dioxide catalysis in the hydrogen peroxide N-oxidation of amines. Inorg Chem. 2008;47:1173–1178. doi: 10.1021/ic701402h. [DOI] [PubMed] [Google Scholar]
- [49].Schoneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer’s disease. Biochim Biophys Acta. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- [50].Gennis RB, Stewart V. Respiration. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and molecular biology. ASM Press; Washington, D.C.: 1996. pp. 217–261. [Google Scholar]
- [51].Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriological reviews. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Winter SE, Bäumler AJ. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes. 2014 doi: 10.4161/gmic.27129. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio. 2012:3. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rivera-Chavez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Baumler AJ. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Baumler AJ. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013:4. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Spees AM, Lopez CA, Kingsbury DD, Winter SE, Baumler AJ. Colonization resistance: battle of the bugs or Menage a Trois with the host? PLoS Pathog. 2013;9:e1003730. doi: 10.1371/journal.ppat.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, von Mering C, Robinson MD, Stecher B, Hardt WD. Microbiota-derived hydrogen fuels salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe. 2013;14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- [58].Kawai K, Fujita M, Nakao M. Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim Biophys Acta. 1974;369:222–233. [PubMed] [Google Scholar]
- [59].Price-Carter M, Tingey J, Bobik TA, Roth JR. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Winter SE, Baumler AJ. Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘Like will to like’ concept. Cell Microbiol. 2013 doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].de la Huerga J, Popper H. Urinary excretion of choline metabolites following choline administration in normals and patients with hepatobiliary diseases. The Journal of clinical investigation. 1951;30:463–470. doi: 10.1172/JCI102463. [DOI] [PMC free article] [PubMed] [Google Scholar]