Abstract
Bile acid synthesis is the major pathway for catabolism of cholesterol. Cholesterol 7α-hydroxylase (CYP7A1) is the rate-limiting enzyme in the bile acid biosynthetic pathway in the liver and plays an important role in regulating lipid, glucose and energy metabolism. Transgenic mice overexpressing CYP7A1 (CYP7A1-tg mice) were resistant to high-fat diet (HFD)-induced obesity, fatty liver, and diabetes. However the mechanism of resistance to HFD-induced obesity of CYP7A1-tg mice has not been determined. In this study, metabolomic and lipidomic profiles of CYP7A1-tg mice were analyzed to explore the metabolic alterations in CYP7A1-tg mice that govern the protection against obesity and insulin resistance by using ultra-performance liquid chromatography-coupled with electrospray ionization quadrupole time-of-flight mass spectrometry combined with multivariate analyses. Lipidomics analysis identified seven lipid markers including lysophosphatidylcholines, phosphatidylcholines, sphingomyelins and ceramides that were significantly decreased in serum of HFD-fed CYP7A1-tg mice. Metabolomics analysis identified 13 metabolites in bile acid synthesis including taurochenodeoxycholic acid, taurodeoxycholic acid, tauroursodeoxycholic acid, taurocholic acid, and tauro-β-muricholic acid (T-β-MCA) that differed between CYP7A1-tg and wild-type mice. Notably, T-β-MCA, an antagonist of the farnesoid X receptor (FXR) was significantly increased in intestine of CYP7A1-tg mice. This study suggests that reducing 12α-hydroxylated bile acids and increasing intestinal T-β-MCA may reduce high fat diet-induced increase of phospholipids, sphingomyelins and ceramides, and ameliorate diabetes and obesity.
Keywords: CYP7A1, lipidomics, tauro-β-muricholic acid, farnesoid X receptor (FXR), bile acid metabolism
1. Introduction
1.1. Bile acid synthesis
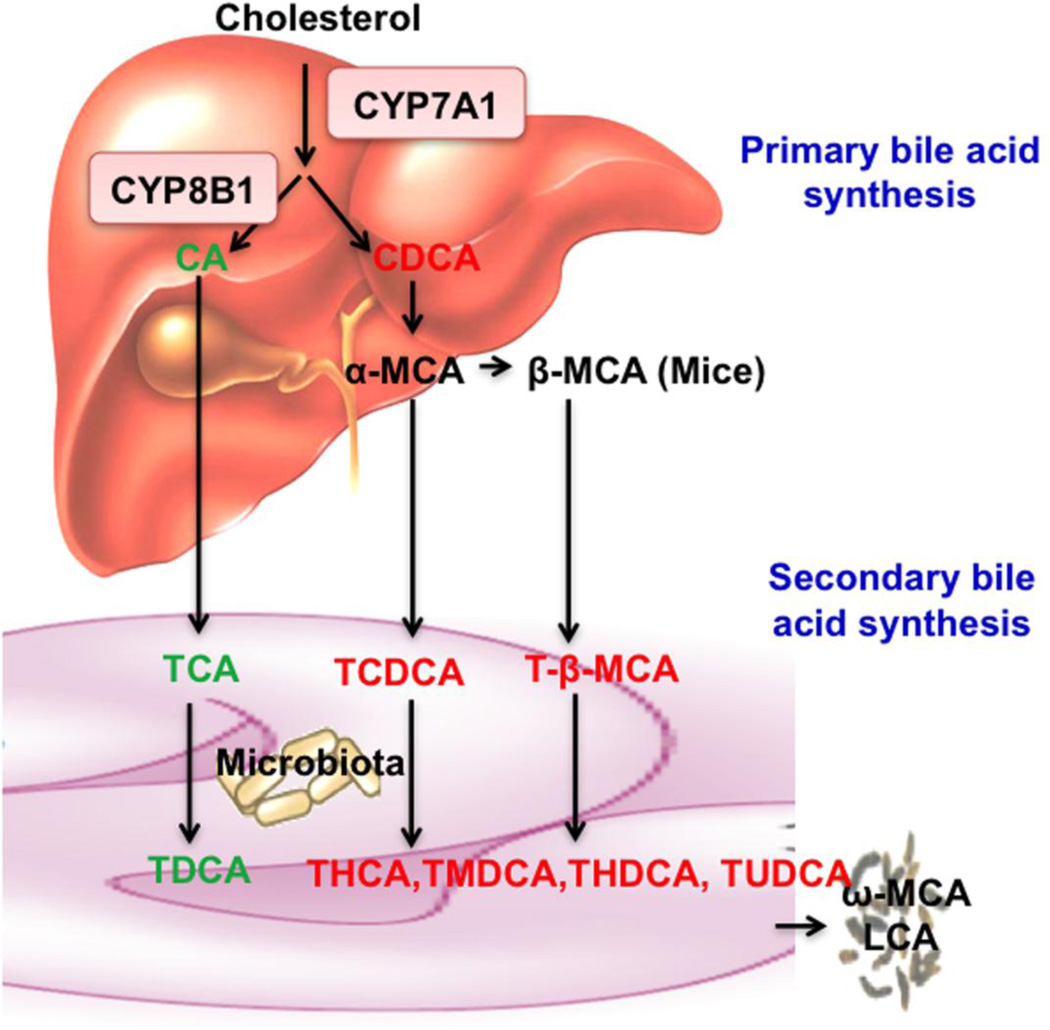
Bile acid synthesis is the major pathway for catabolism of cholesterol to bile acids. In the liver, cholesterol 7α-hydroxylase (CYP7A1) is the first and rate-limiting enzyme of the bile acid biosynthetic pathway producing two primary bile acids, cholic acid (CA, 3α, 7α, 12α-OH) and chenodeoxycholic acid (CDCA, 3α, 7α-OH) in humans (Fig. 1) [1]. Sterol-12α hydroxylase (CYP8B1) catalyzes the synthesis of CA. In mice, CDCA is converted to α-muricholic acid (α-MCA: 3α, 6β, 7α-OH) β-muricholic acid (β-MCA: 3α, 6β, 7β-OH). Bile acids are conjugated to taurine or glycine, secreted into the bile and stored in the gallbladder. After a meal, bile acids are released into the gastrointestinal tract. In the intestine, conjugated bile acids are first de-conjugated and then 7α-dehydroxylase activity in the gut flora converts CA to deoxycholic acid (DCA: 3α, 12α), and CDCA to lithocholic acid (LCA: 3α), two major secondary bile acids in humans. In mice, CDCA is converted to ursodeoxycholic acid (UDCA: 3α, 7β), α-MCA to hyocholic acid (HCA: 3α, 6α, 7α) and murideoxycholic acid (MDCA: 3α, 6β), and β-MCA t ω-MCA (3α, 6α, 7β) and hyodeoxycholic acid (HDCA: 3α, 6α). The number of hydroxyl groups, their location and α- or β-orientation, and conjugation determine their hydrophobicity and detergent properties. In humans, most bile acids are glycine or taurine-conjugated and CA, CDCA and DCA are the most abundant bile acids. In mice, most bile acids are taurine-conjugated and CA and α- and β-MCAs are the most abundant bile acids. Bile acids facilitate absorption of dietary fats, steroids, and lipid soluble vitamins into enterocytes and are transported via portal circulation to the liver for metabolism and distribution to other tissues and organs. About 95% of bile acids are reabsorbed in the ileum and transported to the liver to inhibit CYP7A1 and bile acid synthesis. Enterohepatic circulation of bile acids provides a negative feedback mechanism to maintain bile acid homeostasis. Alteration of bile acid synthesis, secretion and transport causes cholestatic liver diseases, gallstone diseases, fatty liver disease, diabetes and obesity [1]. Deficiency of CYP7A1 in humans is associated with hypercholesterolemia and premature atherosclerosis [2].
Fig 1. Bile acid synthesis.
In the classic bile acid synthesis pathway, cholesterol is converted to cholic acid (CA, 3α, 7α, 12α) and chenodeoxycholic acid (CDCA, 3α, 7α) CYP7A1 is the rate-limiting enzyme and CYP8B1 catalyzes the synthesis of CA. In mouse liver, CDCA is converted to α-muricholic acid (α-MCA, 3α, 6β, 7α) and β-MCA (3α, 6β, 7β) Most bile acids in mice are taurine (T)-conjugated and secreted into bile. In the intestine, gut bacteria de-conjugate bile acids and then remove the 7α-hydroxyl group from CA and CDCA to form secondary bile acids deoxycholic acid (DCA, 3α, 12α) and lithocholic acid (LCA, 3α), respectively. T-α-MCA and T-β-MCA are converted to T-hyodeoxycholic acid (THDCA, 3α, 6α), T-ursodeoxycholic acid (TUDCA, 3α, 7β), T-hyocholic acid (THCA, 3α, 6α, 7α) and T-murideoxycholic acid (TMDCA, 3α, 6β). These secondary bile acids are reabsorbed and circulated to liver to contribute to the bile acid pool. Secondary bile acids ω-MCA (3α, 6α, 7β) and LCA are excreted into feces.
1.2 Bile acid signaling
Bile acids are signaling molecules that activate several intracellular signaling pathways [1, 3]. Bile acids are known to activate the farnesoid X receptor (FXR) [4–6] and a membrane G protein coupled-receptor TGR5 (Gpbar1) [7, 8]. CA and CDCA are potent endogenous ligands of FXR, whereas LCA and DCA and their taurine conjugates are more potent agonists of TGR5. FXR plays a critical role in regulation of bile acid synthesis and secretion, and lipid and glucose metabolism in the liver. TGR5 is a Gαs protein coupled receptor (GPCR) that activates cAMP signaling in many cells and plays a role in energy metabolism in brown adipose tissue, relaxing and refilling gallbladder, secreting glucagon-like peptide 1 (GLP-1) in intestinal endocrine cells and controlling GI motility [9, 10]. TGR5 is expressed in the epithelial cells of the gastrointestinal system, including intestine, spleen, cholangiocytes, gallbladder, hepatic sinusoidal endothelial cells and hepatic macrophages, and Kupffer cells [7, 8, 11, 12]. A recent report shows that conjugated bile acids are potential agonists of sphingosine-1-phosphate receptor 2 (S1PR2) in hepatocytes [13] S1PR2 is a Gαi class of GPCR activated by sphingosine-1-phosphate (S1P).
1.3. Mechanisms of bile acid feedback regulation of bile acid synthesis
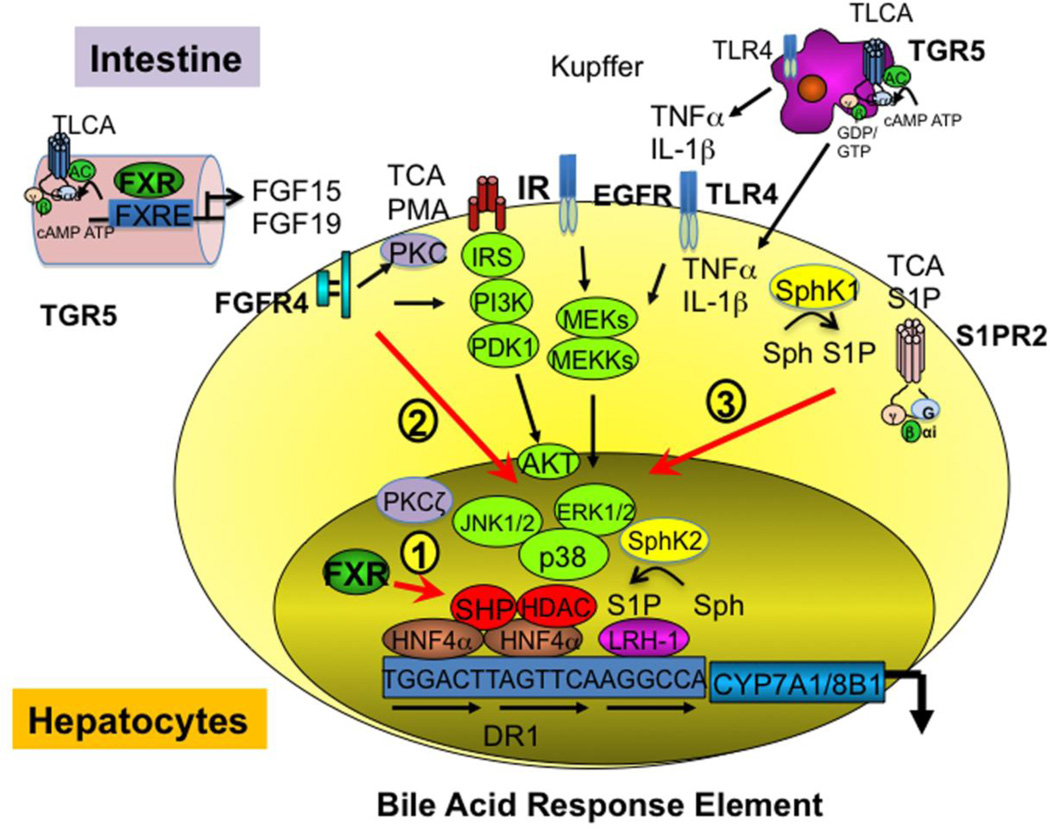
Two FXR-dependent mechanisms are known to inhibit bile acid synthesis. In the liver bile acid-activated FXR induces a negative receptor small heterodimer partner (SHP) to inhibit trans-activation activity of hepatic nuclear factor 4α (HNF4α) and liver receptor homologue-1 (LRH-1) that bind to the bile acid response element in the CYP7A1 and CYP8B1 gene promoters (Fig 2, Pathway 1). In the intestine, bile acids activate FXR to induce fibroblast growth factor (mouse FGF15, or human FGF19), which activates hepatic FGF receptor 4 (FGFR4) and cJun N-terminal kinase 1/2 (JNK1/2) and extracellular-regulated kinase 1/2 (ERK1/2) signaling of mitogen-activated protein kinase (MAPK) pathways to inhibit trans-activation of CYP7A1/CYP8B1 gene by HNF4α [14] (Pathway 2). Several FXR-independent cell-signaling pathways have been reported and are shown as Pathway 3 (Fig 2). Conjugated bile acids are known to activate several protein kinase Cs (PKC) and growth factor receptors, epidermal growth factor receptor (EGFR), and insulin receptor (IR) signaling to inhibit CYP7A1/CYP8B1 and bile acid synthesis via activating the ERK1/2, p38 and JNK1/2 pathways [15–18]. Bile acids and pro-inflammatory cytokines TNFα and IL-1β produced in hepatocytes or secreted from Kupffer cells activated ERK1/2 JNK1/2 to phosphorylate and inactivated HNF4α, and resulted in inhibiting CYP7A1 and CYP8B1 gene transcription [19, 20]. Conjugated bile acids activate S1PR2 in hepatocytes [13]. Extracellular signaling activates sphingosine kinase 1 (SphK1) to phosphorylate membrane sphingosine to S1P, which activates S1PR2 and downstream ERK1/2 and AKT signaling and may inhibit CYP7A1/CYP8B1 gene transcription. Interestingly, nuclear SphK2 is associated with histone 3 and S1P inhibits histone deacetylase 1/2 (HDAC1/2) [21]. It is possible that bile acids may activate SphK2 to stimulate CYP7A1 gene transcription by an epigenetic mechanism.
Fig 2. Bile acid signaling pathways.
Bile acids activate FXR, TGR5 and cell signaling pathways to inhibit CYP7A1 and CYP8B1 gene transcription. 1) Hepatic FXR/SHP pathway: bile acid activated-FXR induces SHP, which inhibits HNF4α and LRH-1 trans-activation of CYP7A1 and CYP8B1 gene transcription in hepatocytes. Bile acid response element binds HNF4α and LRH-1. 2) Intestinal FXR/FGF19/FGFR4 pathway: in the intestine, FXR induces FGF15 (mouse) /FGF19 (human), which is secreted into portal circulation to activate FGF receptor 4 (FGFR4) in hepatocytes. FGFR4 signaling stimulates JNK1/2 and ERK1/2 pathways of MAPK signaling to inhibit CYP7A1 gene transcription by phosphorylation and inhibition of HNF4α binding activity. 3) FXR-independent signaling pathways: Conjugated bile acids activate PKCs, which activate the MAPK pathways to inhibit CYP7A1. Bile acids also activate insulin receptor (IR) signaling IRS/PI3K/PDK1/AKT, possibly via activation of epidermal growth factor receptor (EGFR) signaling, MAPKs (MEK, MEKK), to inhibit CYP7A1 gene transcription. The secondary bile acid TLCA activates TGR5 signaling in Kupffer cells. TGR5 signaling may regulate CYP7A1 by an unknown mechanism. TCA activates sphingosine-1-phosphate (S1P) receptor 2 (S1PR2), which may activate AKT and ERK1/2 to inhibit CYP7A1. S1P kinase 1 (Sphk1) phosphates sphingosine (Sph) to S-1-P, which activates S1PR2. On the other hand, nuclear SphK2 interacts with and inhibits histone deacetylase (HDAC1/2) and may induce CYP7A1. The role of S1P, SphK2, and S1PR2 signaling in regulation of bile acid synthesis is not known.
1.4. Bile acids regulation of lipid and glucose metabolism
Recent studies have unveiled that bile acids play a critical role in maintaining lipid, glucose and energy homeostasis through activation of FXR and TGR5 [1, 9]. It has been known for a long time that treating human gallstone patients with CDCA decreases hepatic VLDL production and plasma triglyceride levels [22], while treating hypercholesterolemic patients with bile acid binding resins increases plasma triglyceride levels [23, 24]. Hepatic lipids and circulating cholesterol and triglycerides are accumulated in FXR−/−mice, whereas activation of FXR by bile acids or FXR agonists decreases plasma cholesterol and triglycerides in wild type mice [25]. It was suggested that the FXR/SHP pathway inhibits steroid response element binding protein 1c (SREBP-1c)-mediated hepatic lipogenesis [26]. Activation of FXR by bile acid feeding or administration of the FXR agonist GW4064 lowered fasting plasma glucose and improved insulin sensitivity in obese and diabetic db/db mice [25, 27], while FXR-deficient mice had insulin resistance and hyperglycemia [27]. Plasma FGF19 increases during the postprandial period in humans, presumably due to increased bile acid signaling [28]. FGF19 transgenic mice were resistant to diet-induced obesity and insulin resistance [29, 30]. In addition, FGF19 has been shown to repress hepatic glucose production [31], promote glycogen synthesis [32], repress lipogenesis [33, 34] and increase metabolic rate [29, 30].
1.5. Bile acids protect against high fat diet induced obesity and diabetes
Transgenic mice overexpressing CYP7A1 (CYP7A1-tg) have been used to demonstrate that increasing conversion of cholesterol to bile acids can protect against lithogenic diet -induced atherosclerosis in mice [35]. We used CYP7A1-tg mice as a model to demonstrate that increasing bile acid synthesis and pool size may protect against high-fat diet (HFD)-induced obesity, fatty liver, and insulin resistance [36]. These mice had lower body fat mass and higher lean mass when fed HFD than did strain-matched wild-type (WT) mice. In CYP7A1-tg mice, bile acid pool size increased 2.5-fold and CYP8B1 expression was abolished. The gallbladder bile acid composition was significantly altered. The major bile acids in CYP7A1-tg mice are CDCA (55%), α- and β-MCA (27%) and UDCA (15%). Microarray gene profiling analysis showed markedly increased expression of key genes in de novo cholesterol synthesis in CYP7A1-tg mice [37]. Hepatic cholesterol synthesis was increased 10-fold but fatty acid synthesis was reduced 60%. Biliary cholesterol, bile acid and phospholipid secretion and fecal cholesterol and bile acid excretion were increased, so that hepatic cholesterol homeostasis is maintained [38]. However, the specific metabolic profiles characterizing the resistance to HFD-induced obesity of CYP7A1-tg mice have not been determined. This novel mouse model provides a unique in vivo system to study the underlying molecular mechanism of the anti-diabetic and anti-obesity functions of bile acids.
1.6. Metabolomics and lipidomics
To investigate the metabolic changes in CYP7A1-tg mice that govern protection against obesity, metabolomics and lipidomics profiling were employed. Metabolomics aims to reveal various metabolic characteristics of external or internal perturbations to biological systems by profiling low-molecular-weight metabolites in bio-samples [39–41]. Lipidomics is a sub-metabolomics platform that provides a comprehensive analysis of lipid species within a cell or tissue, which plays an essential role in defining the biochemical mechanisms of lipid-related disease processes through identifying alterations in cellular lipid metabolism, trafficking and homeostasis [42, 43]. The current study of metabolomics and lipidomics profiling of CYP7A1-tg mice using ultra-performance liquid chromatography-coupled with electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOFMS) detected and characterized small organic molecules in biological materials [44–47]. This study explores the involvement of metabolites such as phospholipids and bile acids in the regulation of metabolic homeostasis, and provides insights into the mechanism for the anti-obesity and anti-diabetic effect of increased bile acid pool and altered bile acid composition.
2. Materials and Methods
2.1. Chemicals and reagents
The lipid standards1-palmitoyl-sn-glycero-3-phosphocholine (LPC 16:0), 1-stearoyl-sn-glycero-3-phosphocholine (LPC 18:0), 1,2-diarachidonoyl-sn-glycero-3-phosphocholine (PC 20:4), N-palmitoyl-D-erythro-sphingosylphosphorylcholine (SM 18:1/16:0), and N-stearoyl-D-erythro-sphingosine (C18 ceramide), etc., were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). Bile acid standards cholic acid (CA), taurocholic acid (TCA), chenodeoxycholic acid (CDCA), tauro-β-muricholic acid (T-β-MCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA) and the internal standard chlorpropamide were purchased from Sigma-Aldrich (St. Louis, MO). All solvents and organic reagents were of the highest obtainable grade.
2.2. Animals
CYP7A1-tg mice overexpressing rat CYP7A1 under the control of the apolipoprotein E3 (Apoe3) promoter (B6.Cg-Tg (APOE-CYP7A1) 1Rjd/Mmcd) were obtained as described previously [48]. The mice were further bred with wild-type C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME). Transgenic mice and wild-type littermates were backcrossed between 8–9 generations to yield ~99% C57BL/6J background. Mice were maintained on a standard chow diet and water ad libitum and housed in a room with a 12-hour light (6 am to 6 pm)/12-hour dark (6 pm to 6 am) cycle. To induce obesity and insulin resistance, 16–20-week-old male CYP7A1-tg and wild type mice were fed a HFD (Harlan-Teklad, #88137, 42% fat calories and 0.2% cholesterol) or chow diet for 4 months. For other experimenets, 16–20-week-old female CYP7A1-tg and female wild type mice were used. Food intake was determined for a two-day period. All animal manipulations and treatments were performed under protocols approved by the Institutional Animal Care and Use Committees at Northeast Ohio Medical University. Serum and harvested tissues were frozen at −80°C for further analysis at the end of the study.
2.3. RNA Analysis
RNA was extracted using TRIzol reagent (Invitrogen). Quantitative real-time PCR (qPCR) was performed using cDNA generated from 1 µg total RNA with the SuperScript II Reverse Transcriptase kit (Invitrogen). Primers were designed for qPCR using Primer Express software (Applied Biosystems, Foster City, CA), and sequences are available upon request. qPCRs were carried out using SYBR green PCR master mix (Applied Biosystems) in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Values were quantified using the comparative threshold cycle method, and samples were normalized to β-actin.
2.4 Metabolomics analysis
2.4.1. Sample preparation
For serum lipidomics analysis 25 µl serum was extracted by 4-fold cold chloroform: methanol (2:1) solution containing 2 µM LPC (17:0), PC (17:0), SM (17:0) and CER (17:0) (Avanti Polar Lipids, Alabaster, AL) as internal standards. The samples were vortexed for 30 s and then allowed to stand for 5 min at room temperature. The mixture was centrifuged at 13,000 rpm for 5 min and then the lower organic phase was collected and evaporated at room temperature under vacuum and the residue was dissolved in chloroform: methanol (1:1), followed by diluting with isopropanol: acetonitrile: H2O (2:1:1) prior to UPLC-MS analysis. For tissue lipidomics analysis, about 50 mg accurately weighted tissues were homogenized with 700 µL methanol: H2O (4:3) solution and then extracted using 800 µL chloroform containing 2 µM LPC (17:0), PC (17:0), SM (17:0) and CER (17:0) as internal standards. The homogenate was incubated at 37°C for 20 min followed by centrifugation for 20 min at 13,000 rpm. The lower organic phase was transferred to a new tube and dried under vacuum. The residue was suspended with 100 µL chloroform: methanol (1:1) solution and then diluted with isopropanol: acetonitrile: H2O (2:1:1) solution before injection.
2.4.2. Metabolomics analysis
For metabolomics analysis, 50 mg tissue samples were homogenized in 500 mL 50% aqueous acetonitrile containing 5 µM of chlorpropamide (internal standard). The samples were vortexed and centrifuged at 13,000 rpm for 20 min at 4°C to remove particulates and precipitate protein. The supernatant was transferred to an autosampler vial for analysis.
For metabolomics discovery, a 5 µl aliquot of supernatant samples was injected into the UPLC-ESI-QTOFMS system (Waters, Milford, MA) with a Waters Acquity BEH 1.7um C18 (2.1×50mm) column. The gradient mobile phase comprises 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The gradient was maintained at initial 95% A for 0.5 min, to 40% A at 4 min, and then to 1% A at 8 min. Flush for 1 min, then equilibrate at initial conditions for 1.5 min. Flow rate was 0.5 ml/min. Column temperature was maintained at 60°C. Waters Synapt HDMS Q-TOF was operated in both positive and negative modes, scanning 50–850amu, at a rate of 0.3 scans/sec. The following instrument conditions were used: capillary 3kV, source temp 120°C, sampling cove 30V, desolvation gas flow 850 L/h at 400°C.
2.4.3. Lipidomics analysis
For lipidomics discovery, samples were analyzed by UPLC-ESI-QTOF MS using a Water Acquity CSH 1.7um C18 column (2.1×100 mm) under the following conditions: UPLC: A-acetonitrile/water (60/40), B-isopropanol/acetonitrile (90/10), both A and B contain 10 mM Ammonium acetate and 0.1% formic acid. Gradient: initial 60% A to 57% A at 2 min, to 50% A at 2.1 min*, to 46%A at 12 min, to 30% A at 12.1 min*, to 1% A at 18 min before returning to initial conditions at 18.5 min with equilibration for 2 additional minutes (an *indicates ballistic gradient). Flow rate was 0.4 ml/min. Column temperature was maintained at 55°C. MS was run under the same conditions as above, except run time is 18 min. A typical chromatogram using the lipidomics method is shown in Suppl. Fig. S1.
2.4.4. Biomarker identification and quantitation
Biomarkers were screened by analyzing ions in the loading scatter plot, and metabolomics databases (METLIN and Madison Metabolomics Consortium Database) were searched to find potential candidates. To confirm the identities of the putative markers, the authentic standards were compared with the metabolites based on MS/MS fragmentation pattern and retention time.
Concentrations of the metabolites were determined by multiple reaction-monitoring mass spectrometry based on standard curves using authentic standards.
Quantification of lipid markers was performed by MRM and/or parent ion scanning using a Waters UPLC Acquity system coupled to a Waters Xevo TQ mass spectrometer. A Waters Acquity BEH C18 column (2.1×100mm) was used. UPLC: A-water, B-acetonitrile/IPA (5/2), both A and B contain 10 mM Ammonium acetate and 0.1% formic acid. Gradient: initial 70% A for 1 min, then linear gradient to 50% A at 3 min, to 1% A at 8 min, held until 15 min, then returning to initial conditions over 1 min, and held for an additional 2 min for column equilibration. Flow rate was 0.4 ml/min. Column temperature was maintained at 50°C. Waters Xevo TQ was operated in MRM mode. The following instrument conditions were used: Capillary 2.2kV, source temp 150°C, desolvation gas flow 850 L/h at 450°C. Total run time is 18 min. The cone voltage and collision energy for the MRM transitions was optimally determined for each transition by the instrument’s IntelliStart software.
Quantification of bile acids was performed on a Waters Acquity H-class UPLC system using a Waters Acquity BEH C18 column (2.1×100mm) coupled to a Waters Xevo G2 Q-TOF mass spectrometer. UPLC: A-0.1% formic acid in water, B-0.1% formic acid in acetonitrile. Gradient: initial 80% A for 4 min, to 60% A at 15 min, to 40% A at 20 min, to 10% A at 21 min. Flush for 1 min, then equilibrate at initial conditions for 4 min. Flow rate 0.4 ml/min. Column temperature was maintained at 45°C. Waters Xevo G2 Q-TOF was operated in negative mode, scanning 50–850amu, at a rate of 0.3 scans/sec. The following instrument conditions were used: Capillary 1.5kV, source temp 150°C, sampling cove 30V, desolvation gas flow 850 L/h at 500°C.
2.4.5. Data Processing and Multivariate Data Analysis
Chromatographic and spectral data were de-convoluted by MarkerLynx software (Waters). A multivariate data matrix containing information on sample identity, ion identity (retention time and m/z), and ion abundance was generated through centroiding, deisotoping, filtering, peak recognition, and integration. The intensity of each ion was calculated by normalizing the single ion counts vs. the total ion counts in the whole chromatogram. The data matrix was further exported into SIMCA-P software (Umetrics, Kinnelon, NJ) and transformed by mean centering and pareto scaling, the technique that increases the importance of low abundance ions without significant amplification of noise. Statistical models including principal components analysis (PCA), partial least squares-discriminant analysis (PLS-DA), orthogonal projections to latent structures-discriminant analysis (OPLS-DA) were established to represent the major latent variables in the data matrix.
2.4.6. Data Analysis
Experimental values were expressed as mean±S.D. Statistical analysis was performed with two-tailed Student’s t-test or Mann-Whitney test using Prism 6.0 (GraphPad Software, Inc., San Diego, CA). A p-value < 0.05 was considered statistically significant.
3. Results
3.1. CYP7A1-tg and wild-type mice challenged with HFD had distinct lipidomic profiles
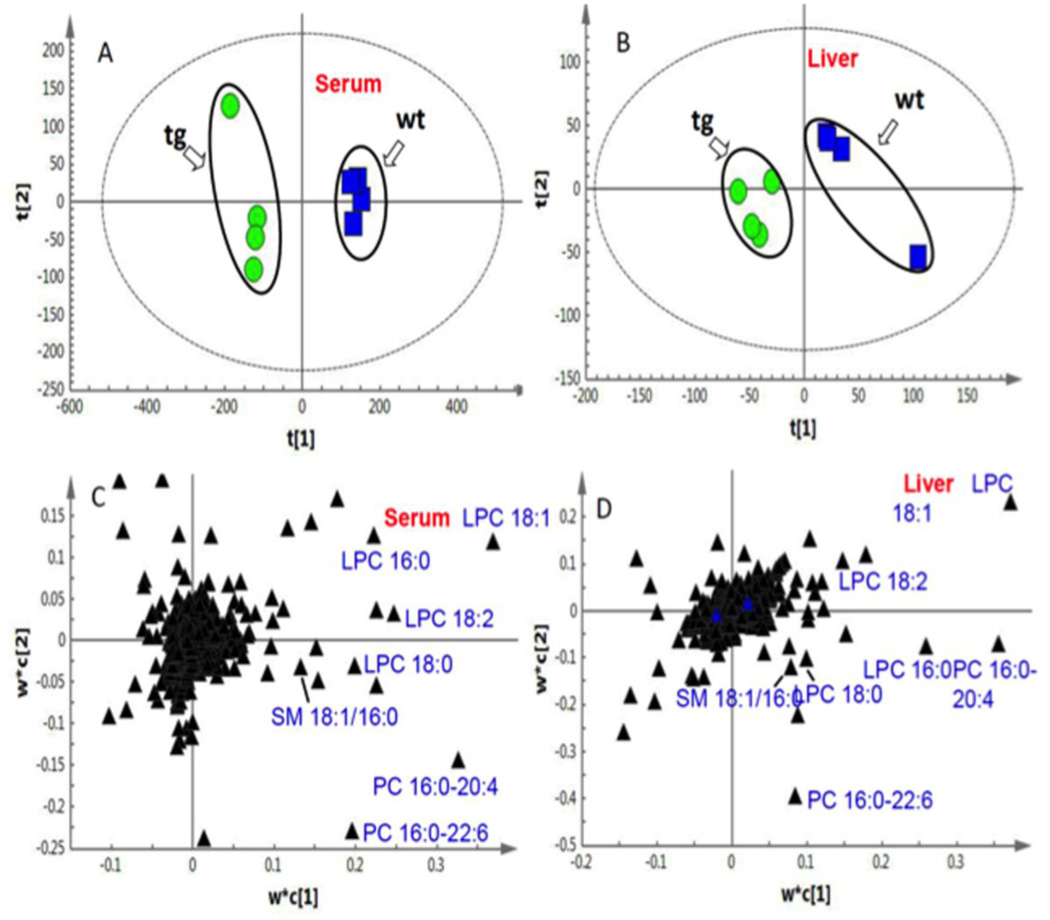
When challenged with a HFD, CYP7A1-tg mice had lower body fat mass and higher lean mass compared to wild-type mice [38]. As a platform for comprehensive and quantitative description of the set of lipid species, lipidomics was used to investigate the mechanism of this phenotype. By use of an unsupervised PCA model with the cumulative R2X 0.677 for serum and 0.593 for liver, CYP7A1-tg and wild-type mice were clearly separated based on the scores plot (Suppl. Fig.S2), indicating that these two groups have distinct lipidomic profiles. Supervised PLS-DA models were then established to maximize the difference of metabolic profiles between CYP7A1-tg and wild-type groups as well as to facilitate the screening of lipid marker metabolites (Fig.3). For serum samples, two latent variables (LV) were calculated, and the cumulative R2Y and Q2 were 0.993 and 0.878, respectively. For liver samples, two LV were calculated with the cumulative R2Y and Q2 0.937 and 0.495, respectively. No over-fitting of the data was observed according to results of the permutation test. Based on the PLS-DA model, the most significant ions that led to the separation between CYP7A1-tg and WT groups in serum (Fig. 3A) and liver (Fig.3B) were obtained from the loading plots shown in Fig.3C (serum) and Fig. 3D (liver), in which the ions furthest away from the origin contribute significantly to the clustering of the two groups. The statistically significant thresholds of variable influence on projection (VIP) values larger than 1.5 and p values less than 0.05 were used [49]. Finally, seven ions were identified (Table 1) by tandem mass spectrometry and retention time comparisons with authentic standards (Suppl. Fig.S3). These lipid markers (Table 1) include LPC 16:0, LPC 18:0, LPC 18:1, LPC 18:2, PC 16:0–20:4, PC 16:0–22:6, and SM 16:0.
Fig 3. PLS-DA analysis of CYP7A1-tg and wild-type (WT) mice challenged with HFD.
Based on the score plots, distinct lipidomic profiles of male CYP7A1-tg and wild-type groups were shown for serum (A) and liver samples (B). Based on the loading plots (C for serum and D for liver) the most significant ions that led to the separation between CYP7A1-tg and wild-type groups were obtained and identified as follows: 1. LPC16:0; 2. LPC18:0; 3. LPC18: 1; 4. LPC 18:2; 5. PC16:0-20:4; 6. PC16:0-22:6; 7. SM16:0.
Table 1.
List of identified lipidomics markers
| No. | Metabolites Name | MF | Retention time, M+H |
|---|---|---|---|
| 1 | LPC 16:0 | C24H50NO7P | 1.4873, 496.3403 |
| 2 | LPC 18:0 | C26H54NO7P | 2.1969, 524.3711 |
| 3 | LPC 18:1 | C26H52NO7P | 1.5914, 522.3554 |
| 4 | LPC 18:2 | C26H50NO7P | 1.2159, 520.3404 |
| 5 | PC 16:0–20:4 | C44H80NO8P | 9.7434, 782.5695 |
| 6 | PC 16:0–22:6 | C46H80NO8P | 9.0373, 806.5699 |
| 7 | SM 16:0 | C39H79N2O6P | 8.584, 703.5749 |
The most significant seven ions based on PLS-DA models were identified by tandem mass spectrometry and retention time comparisons with authentic standards.
Abbreviations: MF, molecular formula.
3.2. Lipidomics marker levels decreased in HFD-fed CYP 7A1-tg mice
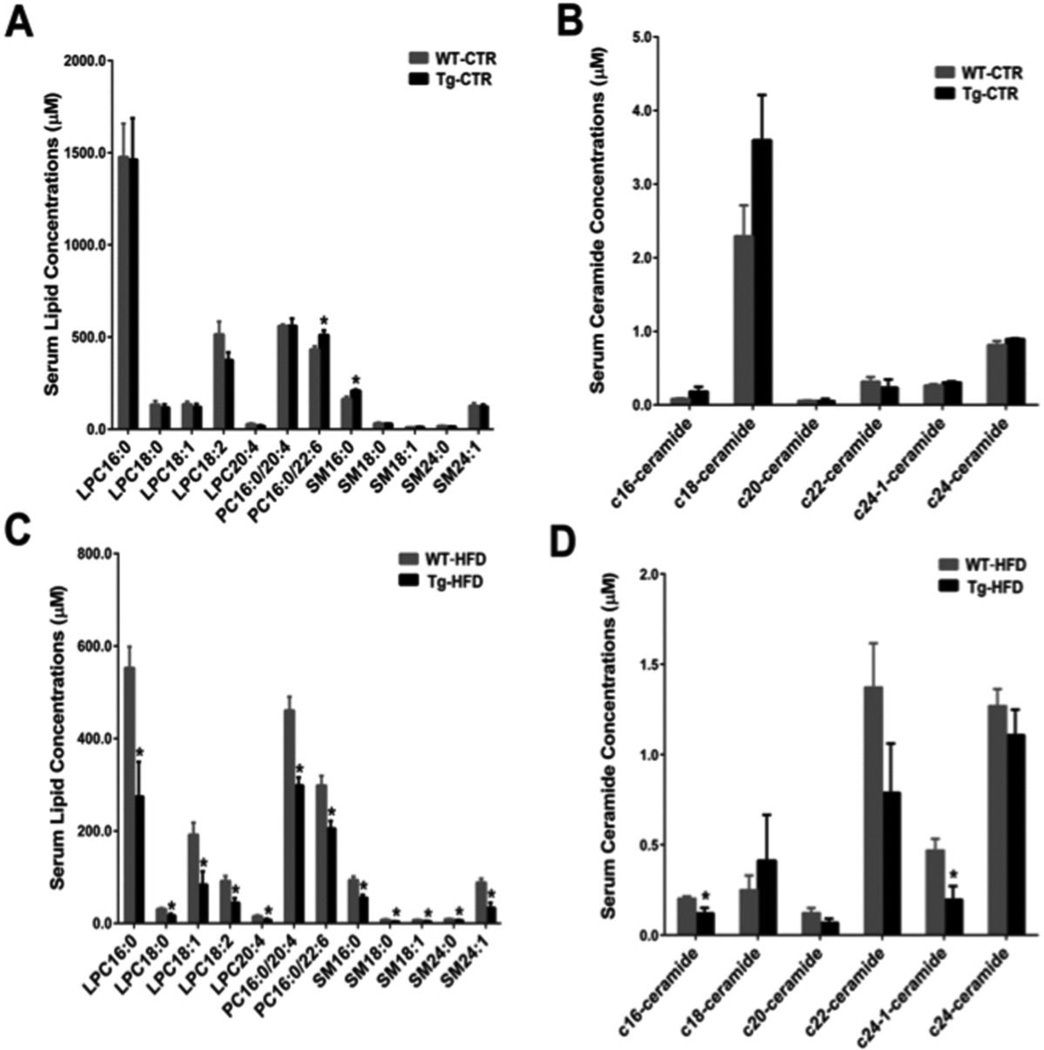
A number of endogenous lysophosphatidylcholine (LPC), phosphatidylcholine (PC), sphingomyelin (SM) and ceramide (CER) were determined and quantified. In control diet-treated mice, except PC 16:0/22:6 and SM 16:0, no clear differences were observed between the two groups (Fig.4A and Fig.4B). In the HFD-fed groups, all the lipid markers listed in Table 1 were significantly decreased in CYP7A1-tg mice compared to wild-type mice. Notably, all the detected SMs, and some CER (C16, C24: 1) were significantly decreased in serum of the CYP7A1-tg group compared to the wild-type group (Fig.4C and Fig. 4D). CER is produced by hydrolysis of SM and synthesized from serine and palmitoyl-CoA [50]. CERs are typically found at trace levels in tissues [51, 52]. Hence, CER may fail to be screened out as metabolite markers using statistical methods. Since both SM and CER were implicated in the development of atherosclerosis, insulin resistance, diabetes, and obesity [53], reduced SM and CER may reduce inflammation and contribute to the phenotypes of resistant to diabetes and obesity of CYP7A1-tg mice [37]. Interestingly, in ileum of control diet-fed CYP7A1-tg mice, these detected lipid markers were decreased (Suppl. Fig.S4A), whereas this is not the case for liver (Suppl. Fig.S4B).
Fig 4. Serum lipid marker quantitation results by multiple reaction-monitoring mass spectrometry based on standard curves using authentic standards.
Data were expressed as mean ± SD. Significant comparison was based on two-tailed Student’s t-test or Mann-Whitney test. An * indicates p< 0.05 (with respect to the wild-type group). Abbreviations: WTCTR, control diet-treated male wild-type mice; Tg-CTR, control diet-treated male CYP7A1 transgenic mice; WT-HFD, high-fat diet-treated male wild-type mice; Tg-HFD, high-fat diet-treated male CYP7A1 transgenic mice.
3.3. Metabolomics profiling of CYP7A1-tg and wild-type mice highlighted markers in bile acid pathway
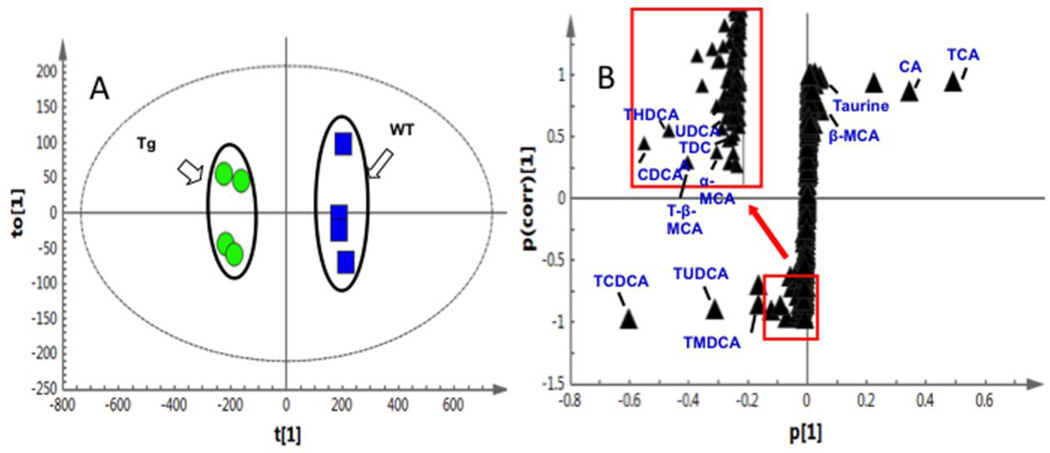
PCA of the UPLC-ESI-QTOFMS-negative mode data from mouse gallbladder, liver, ileum and colon samples revealed distinct metabolic profiles of the CYP7A1-tg and wild-type mice (Suppl. Fig.S5). Subsequently, OPLS-DA was performed to screen for potential markers that contribute to clustering. The S-plot of OPLS-DA data revealed covariance and correlation between the variables and the model, and decreases the risk of false positives in the selection of potential biomarkers [54]. Using a statistically significant threshold of variable confidence approximately 0.75 in the S-plot, a number of ions were selected as potential markers. For example, ions m/z 514.2838 at 11.02 min and m/z 124.0073 at 0.30 min had significantly reduced levels, whereas m/z 498.2883 at 8.80 min and m/z 498.2907 at 13.83 min were markedly enriched in the CYP7A1-tg mice. Ion identification was then performed as described above and identified 13 metabolites (Fig. 5 and Table 2), including taurine, CDCA, UDCA, α-MCA, β-MCA, CA, tauro-murideoxycholic acid (TMDCA), tauro-ursodeoxycholic acid (TUDCA), tauro-hyodeoxycholic acid (THDCA), TCDCA, TDCA, T-β-MCA, and TCA.
Fig 5. OPLS-DA highlighted thirteen markers in bile acid pathway that contribute significantly to the clustering of CYP7A1-tg and wild-type (WT) mice.
Ileum bile acids are shown. (A) In the score plot, female CYP7A1-tg and WT mice were well separated; (B) using a statistically significant thresholds of variable confidence approximately 0.75 in the S-plot, a number of ions were screened out as potential markers, which were later identified as 13 bile acid metabolites including α-MCA, TCA, CDCA, and TCDCA etc.
Table 2.
Metabolomics analysis highlighted 13 markers in bile acid pathway
| No. | Metabolites Name | MF | Retention time, M−H |
|---|---|---|---|
| 1 | Taurine | C2H7NO3S | 0.30, 124.0073 |
| 2 | CDCA | C24H40O4 | 19.07, 391.2858 |
| 3 | UDCA | C24H40O4 | 16.16, 391.2848 |
| 4 | α-MCA | C24H40O5 | 13.23, 407.2809 |
| 5 | β-MCA | C24H40O5 | 13.64, 407.2794 |
| 6 | CA | C24H40O5 | 15.96, 407.2815 |
| 7 | TMDCA | C26H45NO6S | 8.80, 498.2883 |
| 8 | TUDCA | C26H45NO6S | 10.57, 498.2904 |
| 9 | THDCA | C26H45NO6S | 10.77, 498.2911 |
| 10 | TCDCA | C26H45NO6S | 13.83, 498.2907 |
| 11 | TDCA | C26H45NO6S | 14.46, 498.2914 |
| 12 | T-β-MCA | C26H45NO7S | 7.73, 514.2848 |
| 13 | TCA | C26H45NO7S | 11.02, 514.2838 |
The most significant thirteen ions based on OPLS-DA models were identified by tandem mass spectrometry and retention time comparisons with authentic standards.
Abbreviations: MF, molecular formula.
3.4. Metabolomics marker quantitation revealed altered bile-acid pattern in CYP7A1-tg mice
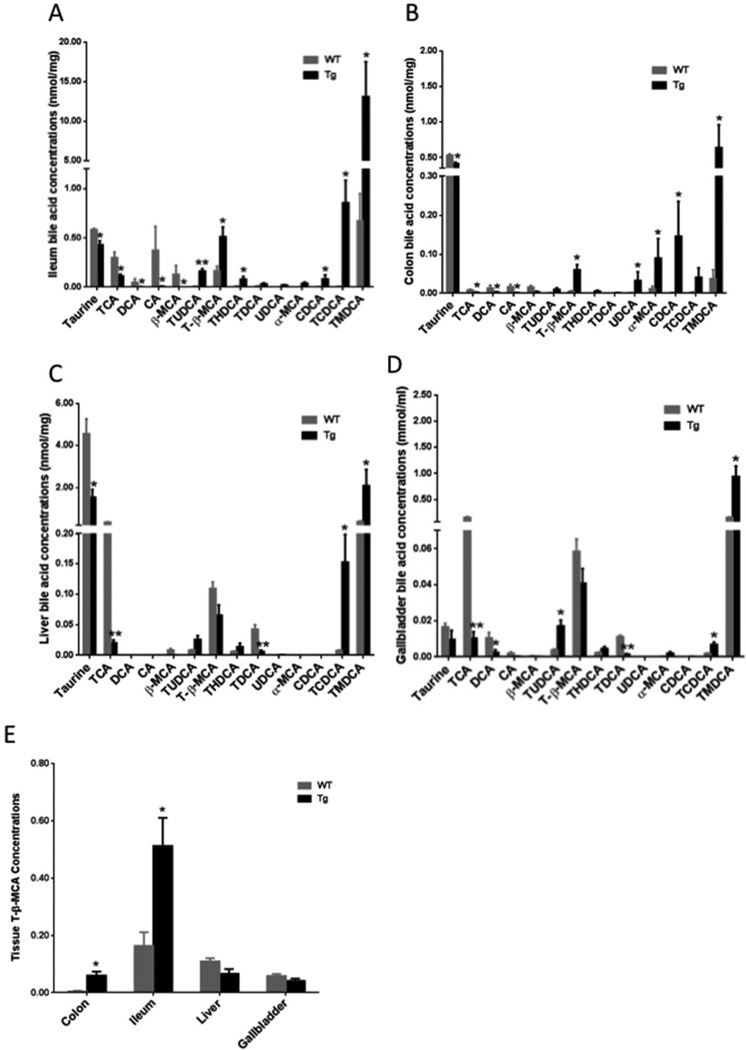
To examine the bile-acid composition and their levels in the tested tissues, 13 bile acid markers in gallbladder, liver, ileum and colon samples were quantified. A representative heat map of ileum bile acid markers of wild type and CYP7A1-tg mice is shown in Suppl. Fig. S6. Fig. 6 shows quantities of identified bile acid metabolites in ileum (Fig. 6A), colon (Fig. 6B), liver (Fig. 6C) and gallbladder (Fig. 6D) in wild type and CYP7A1-tg mice. Consistent with and apart from the previous findings in gallbladders of CYP7A1-tg mice [38], these data show that in most of the tested tissues, 12α-hydroxylated bile acids (CA, TCA, DCA and TDCA) were decreased due to inhibition of CYP8B1 by increased bile acid pool in CYP7A1-tg mice [55]. On the other hand, TCDCA and TMDCA were increased in all tissues. Taurine was markedly decreased in all tissues due to increased taurine-conjugation of the enlarged bile acid pool. These data indicate that the enlarged bile acid pool in CYP7A1-tg mice contains mostly taurine-conjugated CDCA and muricholic acids and their derivatives. The most notable finding in the bile acid metabolomics is the significant increase of T-β-MCA, a potent FXR antagonist [56] in ileum and colon, but not in liver or gallbladder of CYP7A1-tg mice compared to wild-type mice (Fig. 6E).
Fig 6. Quantitation analysis of the bile acid markers.
Bile acid marker levels in the ileum (A), colon (B), liver (C), and gallbladder (D) of female CYP7A1-tg and wild-type (WT) mice. (E) T-β-MCA levels in colon, ileum, liver and gallbladder samples (nmol/mg tissue for colon, ileum and liver, and mmol/ml for gallbladder). Data were expressed as mean ± SD. Significant comparison was based on two tailed Student’s t-test or Mann-Whitney test. An * indicates p < 0.05, and a ** indicates p<0.01 (with respect to the WT group).
3.5. Intestinal mRNA expression revealed inhibition of FXR signaling and induced fatty acid β-oxidation in CYP7A1-tg mice
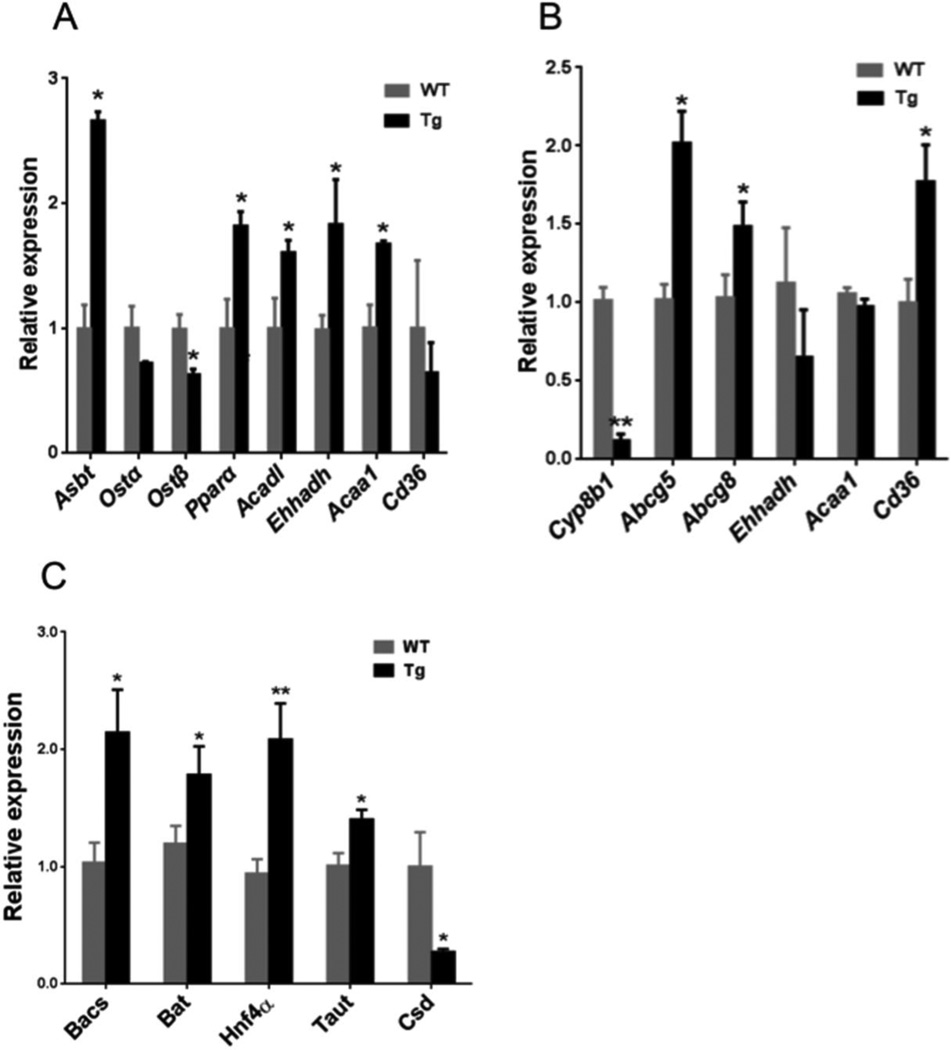
Analysis of intestinal mRNA expression revealed that in chow-fed CYP7A1-tg mice, apical sodium dependent bile salt transporter (ASBT), an intestinal bile acid reabsorption transporter was significantly induced, while sinusoidal bile acid efflux transporters (OSTα) and OSTβ were somewhat decreased (Fig.7A). Increasing ASBT indicates reduced fecal bile acid excretion. OSTα and OSTβ are induced by FXR. Intestinal mRNA levels of the genes involved in mitochondrial fatty acid β-oxidation, such as peroxisome proliferator-activated receptor α (PPPARα), long-chain specific acyl-CoA dehydrogenase (ACADL), enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (EHHADH), and acetyl-CoA acyltransferase 1 (ACAA1), and a fatty acid uptake transporter CD36 were up-regulated (Fig. 7A), indicating increased fatty acid β-oxidation.
Fig 7. Quantitative real time PCR analysis of ileum and liver mRNA expression in CYP7A1-tg and wild-type mice.
(A) Ileum mRNA expression of genes involved in bile acid metabolism and transport (Asbt, Ostα, Ostβ) and fatty acid β-oxidation (Pparα, Acad1, Ehhadh, Acaa1), and fatty acid transporter Cd36 in female chow-fed mice. (B) Hepatic mRNA expression of genes in bile acid metabolism (Cyp8b1), cholesterol efflux transporters (Abcg5, Abcg8) and fatty acid oxidation (Ehhadh, Acaa1) and fatty acid transport (Cd36) in female chow-fed mice. (C) Hepatic mRNA expression of enzymes in bile acid conjugation (Bacs and Bat), HNF4α, taurine biosynthesis (Taut and Csd) in chow fed female CYP7A1-tg mice. Data were expressed as mean ± SD. Significance comparison was based on two-tailed Student’s t-test or Mann-Whitney test. An *Indicates p<0.05 and an **indicates p<0.01 with respect to the wild-type (WT) group. Abbreviations: Tg, transgenic mice overexpressing CYP7A1 in the liver; WT, wild-type mice; Asbt, apical sodium-dependent bile acid transporter; Ostα, organic solute transporter-α; Ostβ, organic solute transporter-β; Pparα, peroxisome proliferator-activated receptor α; Acadl, long-chain specific acyl-CoA dehydrogenase; Ehhadh, enoyl-CoA hydratase/liter-3-hydroxyacyl-CoA dehydrogenase; Acaa1, acetyl-CoA acyltransferase 1; Abcg5, ATP-binding cassette G5; Abcg8, ATP-binding cassette G8; Cd36, fatty acid transporter; Taut, taurine transporter; Csd, cysteine sulfonic acid decarboxylase.
3.6. Hepatic mRNA expression in CYP7A1-tg mice
CYP8B1 was markedly suppressed in liver of CYP7A1-tg mice (Fig 7B), consistent with the perturbed bile acid profiles as revealed in metabolomics (Fig. 6C). Still, hepatic expression of mRNAs encoding the cholesterol transporters ATP-binding cassette G5 (ABCG5) and ABCG8 was significantly induced in CYP7A1-tg mice, in accordance with previous results [38]. Although the intestinal expression levels of fatty acid β-oxidation genes were significantly increased in CYP7A1-tg mice, their hepatic expression was not changed (Fig.7B).
Newly synthesized bile acids are conjugated with taurine in a two-step process, an initial step catalyzed by bile acid: CoA synthase (BACS) and a second step requiring bile acid transferase (BAT) [57]; both are expressed from FXR target genes. Expression of BACS and BAT and their regulator, hepatocyte nuclear factor 4α (HNF4α) were up regulated in CYP7A1-tg mice. Also, significant increase of the taurine transporter (TAUT) mRNA and decrease of cysteine sulfonic acid decarboxylase (CSD) mRNA encoding the rate-limiting enzyme for synthesis of taurine were observed (Fig.7C). These results suggest that the reduced taurine is due to decreased taurine synthesis and increased taurine transport as well as increased conjugation of bile acids.
4. Discussion
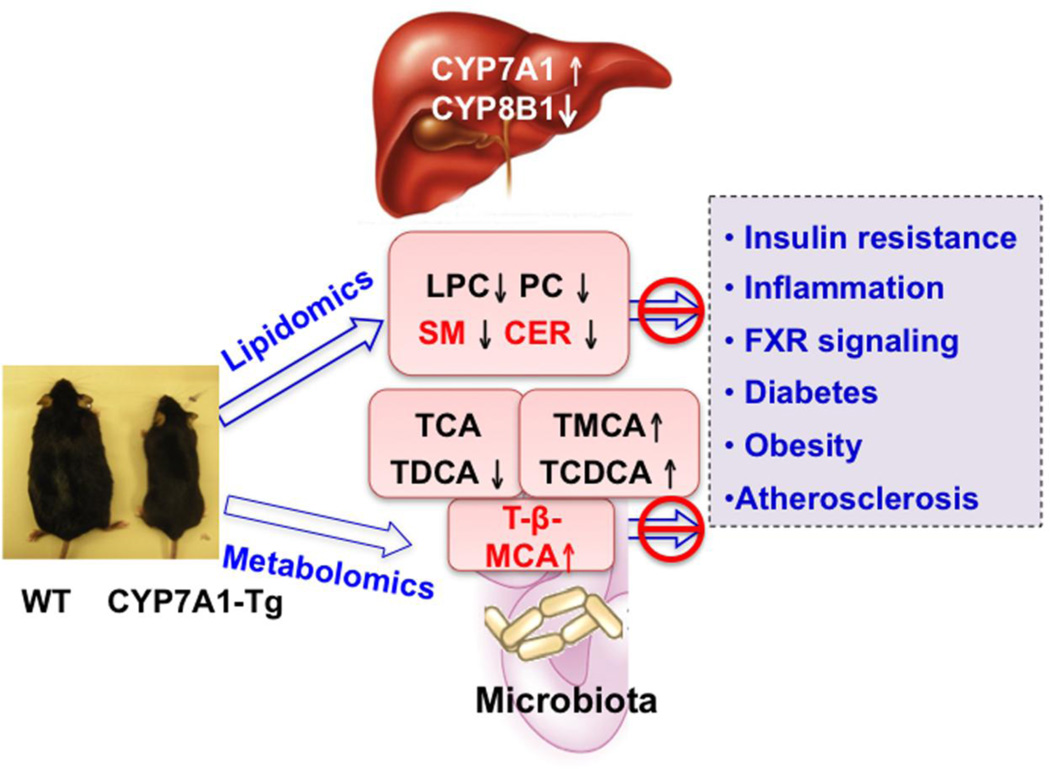
Our recent study of CYP7A1-tg mice revealed that increased CYP7A1 expression and enlarged bile acid pool resulted in significant improvement of lipid homeostasis and resistance to high-fed diet-induced hepatic steatosis, insulin resistance, and obesity in CYP7A1-tg mice [36]. In this study, metabolomics and lipidomics were employed to characterize the metabolic profiles of CYP7A1-tg mice and to provide new insights into the critical role of bile acids in regulation of lipid metabolism and metabolic diseases. Lipidomics analysis of serum lipid profiles of high fat diet-fed CYP7A1-tg identified 7 lipidomic markers that were reduced in CYP7A1-tg mice compared to wild type mice. Metabolomics analysis identified 13 bile acid metabolites that were altered in CYP7A1-tg mice. In CYP7A1-tg mice, TCA and TDCA were reduced, whereas T-β-MCA was increased in the intestine compared to that of wild type mice. The decrease of serum PLC, PC, SM and CER, and 12α-hydroxylated bile acids, and increase of T-β-MCA may contribute to the resistance to diet-induced obesity and diabetes in CYP7A1-tg mice (Fig. 8).
Fig 8. Mechanisms of anti-diabetic and anti-obesity function of bile acids in CYP7A1-tg mice.
In CYP7A1-tg mice, overexpressing CYP7A1 increases bile acid pool size and reduces cholic acid by inhibiting CYP8B1. Lipidomics analysis revealed decreased serum LPC, PC, SM and CER. These lipidomic markers are increased in hepatic steatosis and NAFLD. Bile acids may reduce LPC, PC, SM and CER levels and protect against high fat diet-induced insulin resistance and obesity in CYP7A1-tg mice. Metabolomics analysis showed decreased intestinal TCA and TDCA and increased intestinal T-β-MCA In CYP7A1-tg mice. High fat diets are known to increase CA synthesis and intestinal inflammation. It is proposed that decreasing CA and DCA synthesis may increase intestinal T-β-MCA, which antagonizes FXR signaling to increase bile acid synthesis and prevent high fat diet-induced insulin resistance and obesity.
The present metabolomics and lipidomics analysis revealed that even upon challenging with HFD, CYP7A1-tg mice had reduced lipid levels including LPC, PC, SM and CER. Metabolomics studies of human steatotic liver tissues and HFD-fed mice showed that serum and liver LPC and PC and other lipids levels were increased compared with non-steatotic livers, suggesting altered lipid metabolism contributes to non-alcoholic fatty liver disease (NAFLD) [58, 59]. In HFD-fed CYP7A1-tg mice, reduced serum PC, LPC, SM and CER levels suggest a role for bile acids in maintaining phospholipid homeostasis to prevent NAFLD. SMs are important membrane phospholipids that interact with cholesterol in membrane rafts and regulate cholesterol distribution and homeostasis. A role for SM and CER in the pathogenesis of insulin resistance, diabetes and obesity [60] and development of atherosclerosis [53] has been reported. CER has a wide range of biological functions in cellular signaling such as activating protein kinase C and c-Jun N-terminal kinase (JNK), induction of β-cell apoptosis and insulin resistance [61, 62]. CER increases reactive oxidizing species and activates the NF-κB pathway, which induces pro-inflammatory cytokines, diabetes and insulin resistance [63]. CER is synthesized from serine and palmitoyl-CoA or hydrolysis of SM by acid sphingomyelinase (ASM). HFD is known to increase CER and SM in liver [64]. The present observation of decreased SM and CER levels in HFD-fed CYP7A1-tg mice indicated that bile acids might reduce HFD-induced increase of SM and CER. A previous report revealed that DCA activates an ASM to convert SM to CER, and Asm−/− hepatocytes are resistant to DCA induction of CER and activation of the JNK pathway [65]. In CYP7A1-tg mice, enlarged bile acid pool inhibits CYP8B1 and reduces CA and DCA levels. Thus, decreasing DCA may reduce ASM activity and SM and CER levels, and contribute to reducing inflammation and improving insulin sensitivity in CYP7A1-tg mice. It has been reported recently that in diabetic patients, serun 12α-hydroxylated bile acids are increased and correlated to insulin resistance [66]. Reduction of CA is known to reduce intestinal cholesterol and fat absorption, and improve insulin resistance and obesity in CYP7A1-tg mice [38].
Several recent studies show that gut microbiota are altered in obesity and NAFLD [67]. Gut microbiota regulate bile acid metabolism and reduce T-β-MCA [56, 68]. In germ free mice and antibiotic-treated mice, tauro-conjugated bile acids, especially T-β-MCA, are increased as compared to conventionally raised mice [68]. Ampicillin increased hepatic primary bile acid synthesis and suppressed ileal FGF15 expression [69]. It has been suggested that T-β-MCA and T-α-MCA are FXR antagonists that reduce FXR induction of FGF15 resulting in increased hepatic CYP7A1 expression and bile acid synthesis [56]. A study of Cyp8b 1−/− mice showed increased bile acid synthesis and pool size, reduced fecal bile acid excretion and increased bile acid reabsorption in the intestine [70]. The present metabolomics analysis revealed a significant increase of T-β-MCA in ileum and colon of CYP7A1-tg mice, which had increased taurine synthesis and bile acid conjugation. Bile acids also have been implicated in modulation of gut microbiota [71]. The lack of CA and DCA in the intestine may alter gut microbiota to decrease bile acid de-conjugation and increase T-β-MCA in the intestine. Inhibition of CYP8B1 increases synthesis of CDCA, which is converted to α- and β-MCAs in the liver. Thus, reducing CA synthesis may increase T-β-MCA and resulting in increasing bile acid synthesis, intestinal bile acid reabsorption and bile acid pool size [70]. On the other hand, high saturated fats increase TCA to promote the expansion of the low abundant Bilophila wadsworthia, which is associated with increased pro-inflammatory cytokines and colitis in IL 10−/− mice [72].
In conclusion, metabolomics and lipidomics were employed to characterize the metabolic profiles of CYP7A1-tg mice, aiming to provide new insights into the mechanism of bile acid signaling in regulation of lipid metabolism and maintain lipid homeostasis. A number of lipid and bile acid markers were unveiled in this study. Decreasing of lipid markers, especially SM and CER may explain the improved insulin sensitivity and obesity in CYP7A1-tg mice. Furthermore, this study uncovered that enlarged bile acid pool size and altered bile acid composition may reduce de-conjugation by gut microbiota and increase tauro-conjugated muricholic acids, which partially inhibit intestinal FXR signaling without affecting hepatic FXR signaling. This study is significant in applying metabolomics for diagnosis of lipid biomarkers for fatty liver diseases, obesity and diabetes. Increasing CYP7A1 activity and bile acid synthesis coupled to decreasing CYP8B1 and 12α-hydroxylated-bile acids may be a therapeutic strategy for treating diabetes and obesity.
Supplementary Material
Highlights.
Lipidomics identified 7 lipid markers decreased in HFD-fed CYP7A1-tg mice.
Metabolomics identified 13 bile acid metabolites that were altered in CYP7A1-tg mice.
Bile acids protect against HDF-induced obesity and insulin resistance.
Acknowledgment
This study was supported by the National Cancer Institute Intramural Research Program and by R37DK058379 and R01DK044442 to JYLC from NIDDK, NIH.
Abbreviations
- CYP7A1
cholesterol 7α-hydroxylase
- CYP7A1-tg mice
transgenic mice overexpressing CYP7A1 in the liver
- HFD
high-fat diet
- UPLC-ESI-QTOFMS
Ultra performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry
- FXR
farnesoid X receptor
- LPC
lysophosphatidylcholine
- PC
phosphatidylcholine
- SM
sphingomyelin
- CER
ceramide
- CA
cholic acid
- TCA
taurocholic acid
- CDCA
chenodeoxycholate
- T-β-MCA
tauro-β-muricholic acid
- TCDCA
taurochenodeoxycholate
- TDCA
taurodeoxycholic acid
- PCA
Principal Components Analysis
- PLS-DA
Partial Least Squares-Discriminant Analysis
- OPLS-DA
Orthogonal Projections to Latent Structures-Discriminant Analysis
- α-MCA
α-muricholic acid
- UDCA
ursodeoxycholic acid
- β-MCA
β-muricholic acid
- DCA
deoxycholic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiang JY. Bile acids: regulation of synthesis. Journal of lipid research. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, et al. Human cholesterol 7α-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. The Journal of clinical investigation. 2002;110:109–117. doi: 10.1172/JCI15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. Journal of lipid research. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 5.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 7.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. The Journal of biological chemistry. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 9.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell metabolism. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alemi F, Poole DP, Chiu J, Schoonjans K, Cattaruzza F, Grider JR, et al. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology. 2013;144:145–154. doi: 10.1053/j.gastro.2012.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keitel V, Cupisti K, Ullmer C, Knoefel WT, Kubitz R, Haussinger D. The membrane-bound bile acid receptor TGR5 is localized in the epithelium of human gallbladders. Hepatology. 2009 doi: 10.1002/hep.23032. [DOI] [PubMed] [Google Scholar]
- 12.Keitel V, Ullmer C, Haussinger D. The membrane-bound bile acid receptor TGR5 (Gpbar-1) is localized in the primary cilium of cholangiocytes. Biological chemistry. 2010;391:785–789. doi: 10.1515/BC.2010.077. [DOI] [PubMed] [Google Scholar]
- 13.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell metabolism. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, et al. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 2005;42:1291–1299. doi: 10.1002/hep.20942. [DOI] [PubMed] [Google Scholar]
- 16.Stravitz RT, Rao YP, Vlahcevic ZR, Gurley EC, Jarvis WD, Hylemon PB. Hepatocellular protein kinase C activation by bile acids: implications for regulation of cholesterol 7α-hydroxylase. Am J Physiol. 1996;34:G293–G303. doi: 10.1152/ajpgi.1996.271.2.G293. [DOI] [PubMed] [Google Scholar]
- 17.Song KH, Ellis E, Strom S, Chiang JY. Hepatocyte growth factor signaling pathway inhibits cholesterol 7α-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 2007;46:1993–2002. doi: 10.1002/hep.21878. [DOI] [PubMed] [Google Scholar]
- 18.Seok S, Kanamaluru D, Xiao Z, Ryerson D, Choi SE, Suino-Powell K, et al. Bile Acid Signal-induced Phosphorylation of Small Heterodimer Partner by Protein Kinase Czeta Is Critical for Epigenomic Regulation of Liver Metabolic Genes. The Journal of biological chemistry. 2013;288:23252–23263. doi: 10.1074/jbc.M113.452037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahan A, Chiang JY. Cytokine regulation of human sterol 12α-hydroxylase (CYP8B1) gene. American journal of physiology Gastrointestinal and liver physiology. 2005;288:G685–G695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoenfield LJ, Lachin JM. Chenodiol (chenodeoxycholic acid) for dissolution of gallstones: the National Cooperative Gallstone Study. A controlled trial of efficacy and safety. Annals of internal medicine. 1981;95:257–282. doi: 10.7326/0003-4819-95-3-257. [DOI] [PubMed] [Google Scholar]
- 23.Garg A, Grundy SM. Cholestyramine therapy for dyslipidemia in non-insulin-dependent diabetes mellitus. A short-term, double-blind, crossover trial. Annals of internal medicine. 1994;121:416–422. doi: 10.7326/0003-4819-121-6-199409150-00004. [DOI] [PubMed] [Google Scholar]
- 24.Angelin B, Einarsson K, Hellstrom K, Leijd B. Effects of cholestyramine and chenodeoxycholic acid on the metabolism of endogenous triglyceride in hyperlipoproteinemia. Journal of lipid research. 1978;19:1017–1024. [PubMed] [Google Scholar]
- 25.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. The Journal of clinical investigation. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. The Journal of clinical investigation. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundasen T, Galman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. Journal of internal medicine. 2006;260:530–536. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- 29.Tomlinson E, Fu L, John L, Hultgren B, Huang X, Renz M, et al. Transgenic mice expressing human fibroblast growth factor-19 display increased metabolic rate and decreased adiposity. Endocrinology. 2002;143:1741–1747. doi: 10.1210/endo.143.5.8850. [DOI] [PubMed] [Google Scholar]
- 30.Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, et al. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology. 2004;145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 31.Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell metabolism. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatnagar S, Damron HA, Hillgartner FB. Fibroblast growth factor-19, a novel factor that inhibits hepatic fatty acid synthesis. The Journal of biological chemistry. 2009;284:10023–10033. doi: 10.1074/jbc.M808818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata M, Sakaida Y, Matsuzawa H, Yoshinari K, Yamazoe Y. Fibroblast growth factor 19 treatment ameliorates disruption of hepatic lipid metabolism in farnesoid X receptor (Fxr)-null mice. Biological & pharmaceutical bulletin. 2011;34:1885–1889. doi: 10.1248/bpb.34.1885. [DOI] [PubMed] [Google Scholar]
- 35.Miyake JH, Duong-Polk XT, Taylor JM, Du EZ, Castellani LW, Lusis AJ, et al. Transgenic Expression of Cholesterol-7α-Hydroxylase Prevents Atherosclerosis in C57BL/6J Mice. Arteriosclerosis, thrombosis, and vascular biology. 2002;22:121–126. doi: 10.1161/hq0102.102588. [DOI] [PubMed] [Google Scholar]
- 36.Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7α-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013;58:1111–1121. doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, et al. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica; the fate of foreign compounds in biological systems. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nature reviews Drug discovery. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 41.Oresic M, Vidal-Puig A, Hanninen V. Metabolomic approaches to phenotype characterization and applications to complex diseases. Expert review of molecular diagnostics. 2006;6:575–585. doi: 10.1586/14737159.6.4.575. [DOI] [PubMed] [Google Scholar]
- 42.Hartler J, Tharakan R, Kofeler HC, Graham DR, Thallinger GG. Bioinformatics tools and challenges in structural analysis of lipidomics MS/MS data. Briefings in bioinformatics. 2013;14:375–390. doi: 10.1093/bib/bbs030. [DOI] [PubMed] [Google Scholar]
- 43.Murphy SA, Nicolaou A. Lipidomics applications in health, disease and nutrition research. Molecular nutrition & food research. 2013;57:1336–1346. doi: 10.1002/mnfr.201200863. [DOI] [PubMed] [Google Scholar]
- 44.Li F, Patterson AD, Krausz KW, Jiang C, Bi H, Sowers AL, et al. Metabolomics reveals that tumor xenografts induce liver dysfunction. Molecular & cellular proteomics : MCP. 2013;12:2126–2135. doi: 10.1074/mcp.M113.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson CH, Slanar O, Krausz KW, Kang DW, Patterson AD, Kim JH, et al. Novel metabolites and roles for alpha-tocopherol in humans and mice discovered by mass spectrometry-based metabolomics. The American journal of clinical nutrition. 2012;96:818–830. doi: 10.3945/ajcn.112.042929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsubara T, Tanaka N, Krausz KW, Manna SK, Kang DW, Anderson ER, et al. Metabolomics identifies an inflammatory cascade involved in dioxin- and diet-induced steatohepatitis. Cell metabolism. 2012;16:634–644. doi: 10.1016/j.cmet.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, et al. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer research. 2011;71:6590–6600. doi: 10.1158/0008-5472.CAN-11-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyake JH, Doung XD, Strauss W, Moore GL, Castellani LW, Curtiss LK, et al. Increased production of Apo B100-containing lipoproteins in the absence of hyperlipidemia in transgenic mice expressing cholesterol 7α- hydroxylase. The Journal of biological chemistry. 2001;276:23304–23311. doi: 10.1074/jbc.M101853200. [DOI] [PubMed] [Google Scholar]
- 49.Jonsson P, Bruce SJ, Moritz T, Trygg J, Sjostrom M, Plumb R, et al. Extraction, interpretation and validation of information for comparing samples in metabolic LC/MS data sets. The Analyst. 2005;130:701–707. doi: 10.1039/b501890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hannun YA. The sphingomyelin cycle and the second messenger function of ceramide. The Journal of biological chemistry. 1994;269:3125–3128. [PubMed] [Google Scholar]
- 51.Hammad SM, Pierce JS, Soodavar F, Smith KJ, Al Gadban MM, Rembiesa B, et al. Blood sphingolipidomics in healthy humans: impact of sample collection methodology. Journal of lipid research. 2010;51:3074–3087. doi: 10.1194/jlr.D008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Hu C, Zhao X, Dai W, Chen S, Lu X, et al. Large-scaled human serum sphingolipid profiling by using reversed-phase liquid chromatography coupled with dynamic multiple reaction monitoring of mass spectrometry: method development and application in hepatocellular carcinoma. Journal of chromatography A. 2013;1320:103–110. doi: 10.1016/j.chroma.2013.10.064. [DOI] [PubMed] [Google Scholar]
- 53.Alewijnse AE, Peters SL. Sphingolipid signalling in the cardiovascular system: good, bad or both? European journal of pharmacology. 2008;585:292–302. doi: 10.1016/j.ejphar.2008.02.089. [DOI] [PubMed] [Google Scholar]
- 54.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical chemistry. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 55.Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissuespecific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Chanda D, Lee CH, Kim YH, Noh JR, Kim DK, Park JH, et al. Fenofibrate differentially regulates plasminogen activator inhibitor-1 gene expression via adenosine monophosphate-activated protein kinase-dependent induction of orphan nuclear receptor small heterodimer partner. Hepatology. 2009;50:880–892. doi: 10.1002/hep.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Canaveras JC, Donato MT, Castell JV, Lahoz A. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. Journal of proteome research. 2011;10:4825–4834. doi: 10.1021/pr200629p. [DOI] [PubMed] [Google Scholar]
- 59.Beyoglu D, Idle JR. The metabolomic window into hepatobiliary disease. Journal of hepatology. 2013;59:842–858. doi: 10.1016/j.jhep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chavez JA, Summers SA. A ceramide-centric view of insulin resistance. Cell metabolism. 2012;15:585–594. doi: 10.1016/j.cmet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Blachnio-Zabielska AU, Pulka M, Baranowski M, Nikolajuk A, Zabielski P, Gorska M, et al. Ceramide metabolism is affected by obesity and diabetes in human adipose tissue. Journal of cellular physiology. 2012;227:550–557. doi: 10.1002/jcp.22745. [DOI] [PubMed] [Google Scholar]
- 62.Galadari S, Rahman A, Pallichankandy S, Galadari A, Thayyullathil F. Role of ceramide in diabetes mellitus: evidence and mechanisms. Lipids in health and disease. 2013;12:98. doi: 10.1186/1476-511X-12-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin J, Zhang X, Lu Z, Perry DM, Li Y, Russo SB, et al. Acid sphingomyelinase plays a key role in palmitic acid-amplified inflammatory signaling triggered by lipopolysaccharide at low concentrations in macrophages. American journal of physiology Endocrinology and metabolism. 2013;305:E853–E867. doi: 10.1152/ajpendo.00251.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chocian G, Chabowski A, Zendzian-Piotrowska M, Harasim E, Lukaszuk B, Gorski J. High fat diet induces ceramide and sphingomyelin formation in rat's liver nuclei. Molecular and cellular biochemistry. 2010;340:125–131. doi: 10.1007/s11010-010-0409-6. [DOI] [PubMed] [Google Scholar]
- 65.Gupta S, Natarajan R, Payne SG, Studer EJ, Spiegel S, Dent P, et al. Deoxycholic Acid Activates the c-Jun N-terminal Kinase Pathway via FAS Receptor Activation in Primary Hepatocytes: ROLE OF ACIDIC SPHINGOMYELINASE-MEDIATED CERAMIDE GENERATION IN FAS RECEPTOR ACTIVATION. The Journal of biological chemistry. 2004;279:5821–5828. doi: 10.1074/jbc.M310979200. [DOI] [PubMed] [Google Scholar]
- 66.Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013;62:4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aron-Wisnewsky J, Gaborit B, Dutour A, Clement K. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2013;19:338–348. doi: 10.1111/1469-0691.12140. [DOI] [PubMed] [Google Scholar]
- 68.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyata M, Takamatsu Y, Kuribayashi H, Yamazoe Y. Administration of ampicillin elevates hepatic primary bile acid synthesis through suppression of ileal fibroblast growth factor 15 expression. The Journal of pharmacology and experimental therapeutics. 2009;331:1079–1085. doi: 10.1124/jpet.109.160093. [DOI] [PubMed] [Google Scholar]
- 70.Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. Journal of internal medicine. 2014;275:27–38. doi: 10.1111/joim.12140. [DOI] [PubMed] [Google Scholar]
- 71.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.