Abstract
To understand the immunomodulatory effects of Lactobacillus acidophilus L-92 cells suggested from our previous study of in vivo anti-allergy and anti-virus effects, host immune responses in macrophage-like THP-1 cells after 4 h (the early phase) and 24 h (the late phase) of cocultivation with L-92 cells were investigated by transcriptome analysis. In the early phase of L-92 treatment, various transcription regulator genes, such as, NFkB1, NFkB2, JUN, HIVEP2 and RELB, and genes encoding chemokines and cytokines, such as CCL4, CXCL11, CCL3 and TNF, were upregulated. Two transmembrane receptor genes, TLR7 and ICAM1, were also upregulated in the early phase of treatment. In contrast, many transmembrane receptor genes, such as IL7R, CD80, CRLF2, CD86, CD5, HLA-DQA1, IL2RA, IL15RA and CSF2RA, and some cytokine genes, including IL6, IL23A and CCL22, were significantly upregulated in the late phase after L-92 exposure. Some genes encoding cytokines, such as IL1A, IL1B and IL8, and the enzyme IDO1 were upregulated at both the early and the late phases of treatment. These results suggest that probiotic L-92 might promote Th1 and regulatory T-cell responses by activation of the MAPK signaling pathway, followed by the NOD-like receptor signaling pathway in THP-1 cells.
Keywords: Lactobacillus acidophilus L-92, transcriptome analysis, transcription regulator, cytokine, THP-1
INTRODUCTION
A variety of lactic acid bacteria (LAB) have been isolated and used in mainly dairy foods and supplements for their health benefits in humans. One of the most important probiotic effects of LAB in a host would be its action on the gastrointestinal immune system, which may be beneficial in the prevention of gastrointestinal infections [1, 2], and inflammatory bowel diseases [3] and the alleviation of allergic diseases [4]. Among many probiotic strains that have been developed, Lactobacillus strains have been characterized as strong inducers of pro-inflammatory cytokines, such as IL-12 and tumor necrosis factor (TNF-α) [5, 6], and they seem to be different from the weak IL-12 inducer bifidobacteria.
Lactobacillus acidophilus L-92 has been developed as a strain with anti-allergy effects for pollen allergy [7], perennial allergy [8], and atopic dermatitis [9, 10], and it also has the ability to control gastrointestinal disorders [11]. To explain these in vivo anti-allergy effects, various mechanisms have been proposed, including an improvement in the balance of T helper 1 (Th1) cells and T helper 2 (Th2) cells, induction of apoptosis, induction or activation of regulatory T cells, and activation of the innate immune system [9, 12,13,14]. Our recent study demonstrated that surface layer protein A (SlpA) on the cell wall of the L. acidophilus L-92 strain is important for enhancement of the release of IL-12, known as a Th1-type cytokine, from splenocytes [15]. However, the detailed mechanisms of such immune modulation events are unknown.
The Peyer’s patch, lamina propria and mesenteric lymph nodes of the gastrointestinal tract play a crucial role in controlling the gastrointestinal immune system. In the gastrointestinal immune system, contact between LAB and antigen-presenting cells (APCs), such as dendritic cells (DCs) and macrophages, is likely to be a particularly important process for initiation of host immune responses by priming innate and adaptive immunity [16]. Probiotic microbial signals are thought to be transmitted to DCs via Toll-like receptors [17]; however, few reports have addressed the action of probiotics on cells of the immune system. To improve understanding of the events in DCs, transcriptome analyses have been conducted both in animal studies and in human trials [18,19,20]. Macrophage-like THP-1 cells represent a well responding and physiologically meaningful model system for investigation of the context of innate immunity.
We herein report a detailed analysis of cell metabolism of macrophages in response to L-92 by using transcriptomic analysis on THP-1 cells, which show a response to L-92 cells similar to that of splenocytes by releasing IL-12. The present study suggests that probiotic L-92 might activate Th1 and regulatory T-cell responses by engaging the MAPK and NOD-like receptor signaling pathways.
MATERIALS AND METHODS
THP-1 cell culture
The human monocyte cell line THP-1 [21, 22] was obtained from the Health Science Research Resources Bank (Tokyo, Japan) and maintained in RPMI-1640 (Sigma, St. Louis, MO, USA) containing 10% (V/V) of heat-inactivated fetal bovine serum (FBS; Hana-Nesco Bio, Tokyo, Japan), streptomycin (100 µg/ml) and penicillin (100 U/ml) (GIBCO, Grand Island, NY, USA) at 37°C in 5% CO2/95% air.
Preparation of heat-killed L-92 cells
Lactobacillus acidophilus L-92 from our stock culture collection was cultured anaerobically at 37°C for 20 hr in de Man, Rogosa and Sharpe (MRS) broth (Difco Laboratories, Detroit, MI, USA), and washed with phosphate-buffered saline (PBS). Bacteria were suspended in PBS and killed by heating at 100°C for 10 min.
L-92 treatment of THP-1 cells
For transcriptome analysis of THP-1 metabolism, THP-1 cells were grown in RPMI-1640 containing 10% (V/V) of heat-inactivated FBS, streptomycin (100 µg/ml) and penicillin (100 U/ml) in a 250 mL flask (Sumitomo Bakelite Co., Ltd., Tokyo, Japan) at 37°C in 5% CO2/95% air and stimulated to macrophage-like cells by a method described previously, with some modifications [23]. Briefly, THP-1 cells were suspended in 2 mL of RPMI-1640 at a cell density of 5 × 106 cells/ml and cultivated in a 6-well plate (Asahi Glass Co., Ltd., Tokyo, Japan). The cells were differentiated by cultivation in 1.2% (V/V) Me2SO containing RPMI-1640 for 24 hr and stimulated with IFN-γ (5000 U/ml) for 16 hr. Unstained cells were counted as viable cell after treatment with Trypan Blue Solution (0.4%) (Sigma, St. Louis, MO, USA). Next, 1 × 109 cells of heat-killed L-92, suspended in 0.2 mL of culture buffer, were added to stimulate the THP-1 cells. The THP-1 cells were then harvested after 4 and 24 hr of cocultivation with L-92 in the culture medium. The THP-1 cells were collected by centrifugation at 4°C for 10 min at 430 × g, and cell pellets were washed twice with cold D-PBS (Sigma) for total RNA extraction. Cells untreated with L-92 cells and stimulated with IFN-γ (5000 U/ml) for 16 hr were used as controls. Three samples were prepared from three independent experiments for each group.
Microarray analysis
Total RNA was extracted by using an RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). Double-stranded cDNA was synthesized from 5 µg of total RNA, and the cDNA was subjected to in vitro transcription in the presence of biotinylated nucleotide triphosphates. Human genome-wide gene expression was examined by using the GeneChip® Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, USA), which contains oligonucleotide probe sets for approximately 28,869 full-length genes and expressed sequence tags. The biotinylated cDNA was hybridized with a probe array for 16 hr at 45°C. The hybridized products were stained with streptavidin-phycoerythrin, and then scanned with a Hewlett-Packard Gene Array Scanner (Palo Alto, CA, USA). The fluorescence intensity of each probe was quantified by using the GeneChip Analysis Suite 5.0 software (Affymetrix). The level of gene expression was determined as the average difference by using the GeneChip software.
Statistical and functional analysis of microarray data
Data analysis was performed with GeneSpring software version 6.1 (Silicon Genetics, San Carlos, CA, USA). Expression data were considered significant when they differed at least twofold between L-92-treated and PBS-treated cells, and were statistically analyzed by the unpaired t-test with the Benjamini-Hochberg post test. P-values of less than 0.05 were considered significant. Biological interaction networks among regulated genes activated in response to L-92 were identified using Ingenuity Pathways Analysis (IPA). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used for deeper understanding of the metabolism of the altered gene expression.
Quantitative real-time (qRT)-PCR
A two-step RT-PCR reaction was employed. cDNA was synthesized according to the protocol of the Superscript III First-Strand cDNA Synthesis System (Invitrogen, Carlsbad, CA, USA) using 1 µg of total RNA. cDNA was diluted, and frozen aliquots were stored at −20°C. The cDNA generated was amplified using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara Bio, Inc., Shiga, Japan). All assays were performed on a TP800 Thermal Cycler Dice® Real Time System (Takara Bio, Inc.). Primers were prepared for amplification of specific genes, TNF, CRLF2, GAPDH, CD86, STAT4 and IL-8 genes. The sequences of the primers (forward and reverse primers) used in PCR reactions were 5′-AGATGATCTGACTGCCTGGG-3′ and 5′-CTGCTGCACTTTGGAGTGAT-3′ for TNF, 5′-CTGATGCCACGAAAATCTCA-3′ and 5′-TTCTCCATCAGGAATGGGAC-3′ for CRLF25’, 5′-GCACCGTCAAGGCTGAGAAC-3′ and 5′-TGGTGAAGACGCCAGTGGA -3′ for GAPDH, 5′-AGAGGAGCAGCACCAGAGAG-3′ and 5′-CAGAAGCAGCCAAAATGGAT-3′ for CD86, 5′-CACAGCTACATGCATTGGATT-3′and 5′-CGTGTTTCCAAAGAGAAAAACC-3′ for STAT4 and 5′-CTGGCCGTGGCTCTCTTG-3′ and 5′-CCTTGGCAAAACTGCACCTT-3′ for and IL8. A cycle threshold (Ct) was assigned at the beginning of the logarithmic phase of PCR amplification. Data were analyzed by ABI software, and the gene expression was quantified using the 2−ΔΔCT method and normalized to the constitutively expressed housekeeping gene GAPDH.
RESULTS
Altered gene pathways in THP-1 cells
To understand the host immunomodulatory effects of L. acidophilus L-92 cells suggested from previous studies of in vivo anti-allergy and anti-virus effects, responses in THP-1 cells after 4 hr (the early phase) and 24 hr (the late phase) of cocultivation with L-92 cells were investigated by transcriptome analysis. For cultivation of THP-1 cells in RPMI-1640 medium, a high concentration of IFN-γ was added to enhance IL-12 release for the Th1 type cell response based on our previous study. Viability of the THP-1 cells after inoculation in RPMI-1640 medium with IFN-γ at 16 hr was 96.3%. Differential expression analysis showed a significant (more than 2-fold) up- or downregulation of 282 genes in the early phase and 2,411 genes in the late phase in THP-1 cells caused by treatment with L-92 (data not shown).
Next, IPA was used to integrate the transcriptional networks for a more detailed understanding of the main networks linked to immunomodulatory responses in THP-1 cells, as described in the Materials and Methods. In the IPA analysis, the most significant differences in biological functions, referred to as “hematological process”, were found after 4 hr (66 molecules; p=1.34 × 10−28) and 24 hr (253 molecules; p=1.42 × 10−37) of treatment with L-92 cells. At the early phase of the treatment (4 hr), transcription regulators, cytokines and transmembrane receptors in particular showed highly altered expression (Table 1). In the late phase of the treatment with L-92, transcription regulators, enzymes, transmembrane receptors and cytokines were mostly upregulated (Table 1). Over the course of the treatment, the majority of genes with altered expression were transcription regulators, enzymes, cytokines and transmembrane receptors (Table 1). These results revealed that the genes showing changes in expression in THP-1 cells caused by L-92 treatment were mainly associated with immune response and transcriptional regulation.
Table 1. Ingenuity pathway analysis for altered gene expression in THP-1 cells treated with heat-killed Lactobacillus acidophilus L-92 cells.
| Category | 4 hr | 24 hr |
| Transcription regulator | 19 | 53 |
| Enzyme | 5 | 31 |
| Transmembrane receptor | 7 | 28 |
| Cytokine | 14 | 24 |
| Kinase | 4 | 22 |
| G-protein coupled receptor | 2 | 10 |
| Phosphatase | 2 | 7 |
| Growth factor | 1 | 5 |
| Peptidase | 0 | 4 |
| Ion channel | 0 | 3 |
| Ligand-dependent nuclear receptor | 0 | 3 |
| Transporter | 0 | 1 |
| Other | 12 | 62 |
| 66 | 253 |
Altered genes in THP-1 cells
For a deeper understanding of each cellular response in THP-1 cells caused by L-92 treatment, genes that showed significant changes, especially those in the transcription regulator, enzyme, transmembrane receptor and cytokine categories that showed over a 4-fold change after 4 hr of treatment and less than a 4-fold change from 4 hr to 24 hr of L-92 treatment, are listed in Table 2 as “early responding genes”. Among these genes, 6 categorized as “cytokine” were markedly upregulated after 4 hr of treatment but were moderately upregulated after 24 hr of treatment. Moreover, 2 genes in the “transmembrane receptor” category, 2, 2 genes in the “enzyme” category and 5 genes in the “transcription regulator” category were upregulated in THP-1 cells by L-92 treatment at 4 hr of treatment but showed moderate upregulation at 24 hr of treatment (Table 2). In contrast, genes that showed less than a 4-fold change after 4 hr, but over a 4-fold increase in expression after 24 hr of treatment compared with that after 4 hr were categorized as “slowly responding genes” (Table 2). Among these genes, 9 categorized as “transmembranes” were markedly upregulated after 24 hr but showed mild changes after 4 hr of treatment. Moreover, 6 genes in the “cytokine” category and 6 genes in “transcription regulator” category also showed a significant increase after 24 hr but only mild changes after 4 hr of treatment with L-92 (Table 2). Finally, four genes categorized as “cytokine” and 1 gene categorized as an “enzyme” were constitutively expressed after 4 and 24 hr of treatment (“constitutively responding genes” in Table 2). These results suggest that L-92 treatment in THP-1 cells might cause a rapid change in expression among genes in the “cytokine” category after 4 hr of treatment and subsequently among genes involved in cell signaling categorized here as “transmembrane receptors” and “cytokine” (Table 2). For these cell responses, the activation of “transcription regulator” genes observed in the early and late phases of responses seems to be important for subsequent gene expression.
Table 2. Major altered genes categorized as transcription regulators, enzymes, transmembrane receptors and cytokines in THP-1 cells after L-92 treatment for 4 hr and 24 hr.
| Category | Entrez Gene Name | Symbol | Fold change |
|
| 0–4 hr | 4–24 hr | |||
| Early responding genes | > 4.0 | 4.0 > | ||
| Cytokine | ||||
| chemokine (C-C motif) ligand 4 | CCL4 | 58.29 | 2.02 | |
| chemokine (C-X-C motif) ligand 11 | CXCL11 | 27.97 | 2.28 | |
| chemokine (C-C motif) ligand 3 | CCL3 | 20.04 | 2.77 | |
| tumor necrosis factor | TNF | 14.57 | 1.15 | |
| interleukin 1 receptor antagonist | IL1RN | 5.11 | 1.95 | |
| Transmembrane receptor | ||||
| intercellular adhesion molecule 1 | ICAM1 | 10.01 | 2.33 | |
| toll-like receptor 7 | TLR7 | 6.63 | 0.32 | |
| Enzyme | ||||
| superoxide dismutase 2, mitochondrial | SOD2 | 10.04 | 2.50 | |
| heme oxygenase (decycling) 1 | HMOX1 | 4.26 | 2.21 | |
| Transcription regulator | ||||
| jun proto-oncogene | JUN | 4.99 | 2.72 | |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | NFKB1 | 5.18 | 2.21 | |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | NFKB2 | 6.10 | 1.85 | |
| human immunodeficiency virus type I enhancer binding protein 2 | HIVEP2 | 4.18 | 2.29 | |
| v-rel reticuloendotheliosis viral oncogene homolog B | RELB | 4.34 | 1.04 | |
| Slowly responding genes | 0–4 hr | 4–24 hr | ||
| Transmembrane receptor | 4.0 > | > 4.0 | ||
| interleukin 7 receptor | IL7R | 1.83 | 63.99 | |
| CD80 molecule | CD80 | 1.74 | 61.40 | |
| cytokine receptor-like factor 2 | CRLF2 | 2.02 | 52.00 | |
| CD86 molecule | CD86 | 0.86 | 30.10 | |
| CD5 molecule | CD5 | 1.10 | 16.20 | |
| interleukin 15 receptor, alpha | IL15RA | 2.06 | 4.37 | |
| major histocompatibility complex, class II, DQ alpha 1 | HLA-DQA1 | 0.94 | 5.93 | |
| interleukin 2 receptor, alpha | IL2RA | 1.00 | 4.98 | |
| colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | CSF2RA | 0.85 | 4.15 | |
| Cytokine | ||||
| interleukin 12B (natural killer cell stimulatory factor 2, p40) | IL12B | 3.39 | 18.10 | |
| interleukin 6 (interferon, beta 2) | IL6 | 1.08 | 54.45 | |
| interleukin 23, alpha subunit p19 | IL23A | 1.29 | 28.01 | |
| secreted phosphoprotein 1 | SPP1 | 2.73 | 11.06 | |
| chemokine (C-C motif) ligand 22 | CCL22 | 1.07 | 17.16 | |
| chemokine (C-C motif) ligand 19 | CCL19 | 1.13 | 9.65 | |
| Transcription regulator | ||||
| early B-cell factor 1 | EBF1 | 1.12 | 41.89 | |
| signal transducer and activator of transcription 4 | STAT4 | 1.42 | 23.28 | |
| runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | RUNX1T1 | 0.97 | 8.18 | |
| zinc finger protein, multitype 2 | ZFPM2 | 1.05 | 5.65 | |
| B-cell CLL/lymphoma 11A (zinc finger protein) | BCL11A | 0.85 | 6.92 | |
| growth factor independent 1 transcription repressor | GFI1 | –2.21 | –5.00 | |
| Enzyme | ||||
| prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | PTGS2 | 2.58 | 14.29 | |
| adenosine deaminase | ADA | 1.07 | 9.62 | |
| sphingosine-1-phosphate lyase 1 | SGPL1 | 1.06 | 5.90 | |
| 3’-phosphoadenosine 5’-phosphosulfate synthase 2 | PAPSS2 | 1.05 | 5.47 | |
| fibronectin 1 | FN1 | 0.95 | 5.84 | |
| coagulation factor XIII, A1 polypeptide | F13A1 | –1.33 | –12.62 | |
| Constitutively responding genes | 0–4 hr | 4–24 hr | ||
| Cytokine | > 4.0 | > 4.0 | ||
| interleukin 1, beta | IL1B | 26.00 | 4.09 | |
| interleukin 8 | IL8 | 16.28 | 4.84 | |
| interleukin 1, alpha | IL1A | 4.32 | 16.48 | |
| Epstein-Barr virus induced 3 | EBI3 | 6.64 | 7.44 | |
| Enzyme | ||||
| indoleamine 2,3-dioxygenase 1 | IDO1 | 5.67 | 30.67 | |
Early responding genes were those altered at 4 hr; slowly responding genes were those altered at 24 hr.
Quantitative analysis of the gene expressions in THP-1 cells
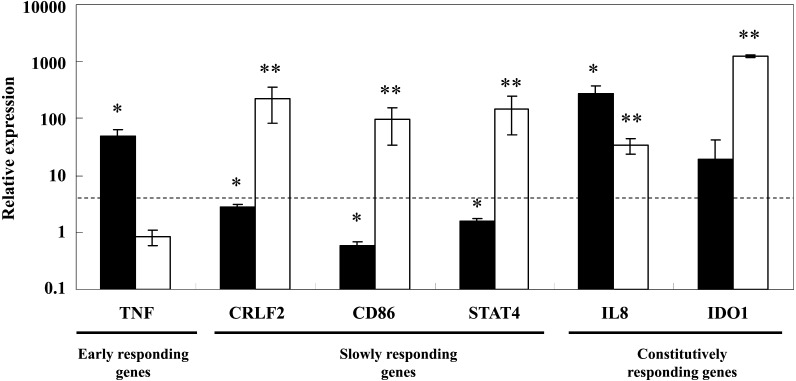
To validate the gene expression changes observed in THP-1 cells in response to L-92 cell treatment (Table 2), qRT-PCR analysis was performed for 6 randomly selected genes with specific primer pairs: TNF (early responding genes), CRLF2, CD86 and STAT4 (slowly responding genes); and IL8 and IDO1 (constitutively responding genes). As shown in Fig. 1, statistical changes were confirmed for most of the quantified gene expressions in L-92-treated THP-1 cells. The TNF gene categorized as an early responding gene was significantly upregulated at 4 hr after treatment (black) but not at 24 hr after treatment (white) (Fig. 1). In contrast, in the genes categorized as slowly responding genes, CRLF2, CD86 and STAT4, the change in expression of each upregulated gene was bigger at 24 hr (white) than that at 4 hr (black) after treatment (Fig. 1). Genes categorized as constitutively responding genes, IL8 and IDO1, were upregulated both at 4 hr and 24hr (Fig. 1). As shown in Fig. 1, the gene expressions randomly quantified by qRT-PCR in THP-1 cells after L-92 treatment showed changes similar to those quantified by microarray analysis (Table 2).
Fig. 1.
Changes in gene expressions from 0 to 4 hr (black bar) and 4 to 24 hr (white bar) observed in THP-1 cells after treatment with L-92 cells as shown by qRT-PCR analysis. The dotted line represents a 4-fold change in expression. The mean ± SD levels of expression of differentially expressed genes and statistical significance of each gene’s expression between 0 and 4 hr (black) and 4 and 24 hr (white) are shown. *p<0.05; **p<0.01 (determined by Student’s t-test).
Highlighted responses in THP-1 cells
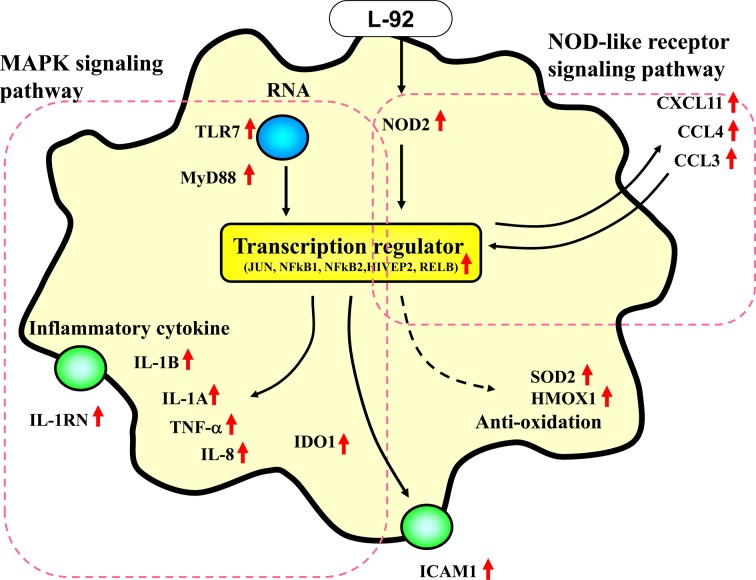
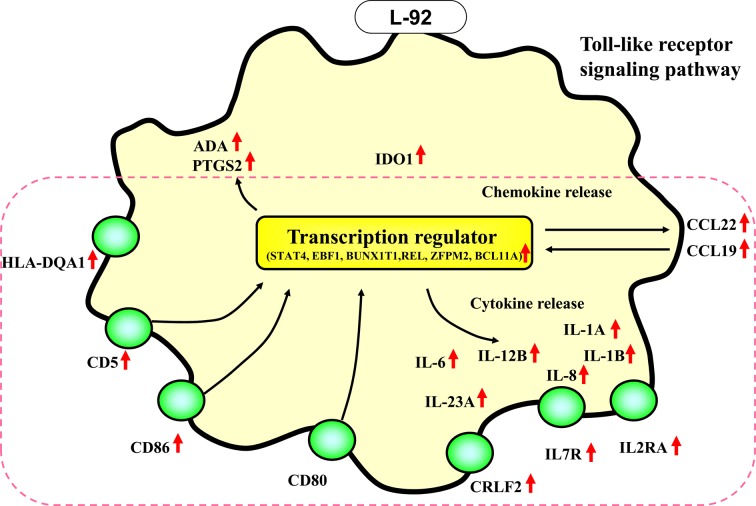
These cell responses at 4 hr and 24 hr are illustrated in Fig. 2 as the early response and in Fig. 3 as the slow response. As summarized in Fig. 2, in the early phase of the cellular response (4 hr of treatment), various transcription regulator genes, such as JUN, NFkB1, NFkB2, HIVEP2 and RELB, as well as genes encoding chemokines and cytokines, such as CXCL11, CCL4,, CCL3, IL1B, IL8 and TNF, were upregulated by L-92 treatment. Two transmembrane receptor genes, TLR7 and ICAM1, were also upregulated at the early phase of the treatment (Fig. 2). Moreover, the NOD2 and MyD88 genes, which were not involved in the hematological processes in the shown in Table 2, were selected as important genes in the MAPK signal pathway and NOD-like receptor signal pathway by KEGG pathway analysis, with 2.99- fold and 1.25-fold upregulated gene expressions at 4 hr (p<0.05 and p<0.05, respectively) in response to L-92 treatment (Fig. 2). During the slow response of the THP-1 cells (24 hr of treatment), mainly transmembrane receptors, such as IL2RA, IL7R, CD80, CRLF2, CD86, CD5, HLA-DQA1, IL15RA and CSF2RA, were activated (Fig. 3). Some cytokine genes, namely IL6, IL23A and CCL22, were also significantly upregulated.
Fig. 2.
Changes in gene expression in THP-1 cells after 4 hr of treatment with Lactobacillus acidophilus L-92 cells. Genes that were significantly upregulated by L-92 treatment were categorized by the IPA method as described in the Materials and Methods. Upregulated genes are indicated by red arrows. Black thin arrows represent the predicted flow of the pathway. Genes grouped by KEGG analysis are indicated by the red dotted line.
Fig. 3.
Changes in gene expression in THP-1 cells after 24 hr of treatment with Lactobacillus acidophilus L-92 cells. Genes that were significantly upregulated genes by L-92 treatment were categorized by the IPA method as described in the Materials and Methods. Upregulated genes are indicated by red arrows. Black thin arrows represent the predicted flow of the pathway. Genes grouped by KEGG analysis are indicated by the red dotted line.
As listed in Table 2 and Figs. 2 and 3, in the early phase of the response in THP-1 cells, mainly transcription regulator genes and cytokines/chemokines were activated. Subsequently, many transmembrane receptor genes were mainly upregulated in THP-1 cells in the late phase of treatment.
DISCUSSION
Increases in some Th1-type cytokines and chemokines have been observed in previous in vitro and in vivo studies with L-92 [12, 13], although those studies did not include detailed analyses of the underlying mechanisms. In the present study, we first investigated the immunomodulatory responses that occurred in THP-1 cells as a model of APCs in response to probiotic L-92 cell exposure by using microarray analysis for a detailed understanding of the host gastrointestinal immune response. As expected from a previous study showing the probiotic anti-allergy effects of L-92 in vivo, many Th1-type cytokines and chemokines that would be expected to increase during anti-allergic and anti-inflammatory events were elevated in THP-1 cells treated with L-92 in the present study, as summarized in Figs. 2 and 3. The THP-1 response in the early phase involved mainly the activation of transcription regulator genes and cytokine/chemokine genes categorized as the MAPK signaling pathway and the NOD-like receptor signaling pathway (Fig. 2). Subsequently, many transmembrane receptor and transcription regulator genes in the Toll-like receptor signaling pathway were upregulated in the late phase of the treatment (Fig. 3). Taking into account the above results, the transcriptional upregulation observed in THP-1 cells co-cultured with L-92 seems to well explain the previous in vivo immunomodulatory effects of L-92 treatment [7,8,9,10, 13, 15].
Regarding the activation of the Th1-type immune response by L-92 in THP-1 cells, one of the first events would be the induction of transcription regulator genes involved in TLR-7 signaling; this would trigger the release of some of Th1-type cytokines and inflammatory cytokines in the MAPK signaling pathway (Fig. 2). These molecules, together with the upregulation of surface markers involved in the maturation of APCs such as CD86, CD80 and CD40, stimulate the activation of Th1 cells [5, 23]. These changes in gene expression observed in THP-1 cells strongly suggest that the in vivo anti-allergy effect of L-92 might be linked to a Th1-type host response in a phase-dependent manner in the gastrointestinal immune system.
Considering the constitutive upregulation of the indoleamine 2,3-dioxygenase (IDO) gene observed in the present study (Table 2), other upregulated genes encoding cytokines, such as IL8, TNF, IL1B and IL12, seem to be closely linked to IDO enhancement. IDO has been identified as a key enzyme with immunomodulatory effects, resulting from its enzymatic activity that leads to catabolism of the essential amino acid L-tryptophane [24, 25]. This enzyme is expressed in epithelial cells, macrophages, and DCs induced by proinflammatory cytokines, including type I and type II IFN [26]. Recent studies suggested the involvement of IL-1β, TNF-α and IL-12 in the induction of IDO in DCs [27, 28] and engagement of CTLA-4 with CD80/CD86 on the membrane of DCs in the stimulation of IDO transcriptional expression and activity [29, 30]. These observations suggest that the upregulation of CD80 and CD86 on the membrane of APCs caused by treatment with L-92 might be closely linked to activation of the IDO gene. IDO1 is known to induce regulatory T cells (Tregs) [31, 32] and to inhibit T-cell proliferation and promote T-cell apoptosis by degradation of L-tryptophan to convert into toxic metabolites such as quinolinic acid and 3-hydroxy-anthranilic acid [31, 33].
In conclusion, key molecules are induced in cultured THP-1 cells by L-92 stimulation that are suggested to play a central role in skewing toward Th1 cells and also in inducing T cell suppression networks. The present study on cultured THP-1 cell opens the way for understanding the immunomodulatory networks modulated by L-92 in the gastrointestinal immune system in vivo. The primary information on both gene profiling and the key genes altered in THP-1 cells after L-92 exposure obtained in the present study will be very important for a detailed understanding of the strain-dependent immune responses to other Lactobacillus species in future studies.
Acknowledgments
This study was supported in part by Grants-in-Aid for Scientific research (A) (H.O.) and Scientific Research on Innovative Areas (H.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Gabryszewski SJ, Bachar O, Dyer KD, Percopo CM, Killoran KE, Domachowske JB, Rosenberg HF, Rosenberg FH. 2011. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J Immunol 186: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youn HN, Lee DH, Lee YN, Park JK, Yuk SS, Yang SY, Lee HJ, Woo SH, Kim HM, Lee JB, Park SY, Choi IS, Song CS. 2012. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res 93: 138–143. [DOI] [PubMed] [Google Scholar]
- 3.Mack DR. 2011. Probiotics in inflammatory bowel diseases and associated conditions. Nutrients 3: 245–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME. 2009. Probiotics and immunity. J Gastroenterol 44: 26–46. [DOI] [PubMed] [Google Scholar]
- 5.Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frøkiaer H, Ferlazzo G. 2007. Distinct gut-derived lactic acid bacteria elicit divergent dendritic cell-mediated NK cell responses. Int Immunol 19: 1319–1327. [DOI] [PubMed] [Google Scholar]
- 6.Zeuthen LH, Christensen HR, Frøkiaer H. 2006. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin Vaccine Immunol 13: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishida Y, Nakamura F, Kanzato H, Sawada D, Yamamoto N, Kagata H, Oh-Ida M, Takeuchi H, Fujiwara S. 2005. Effect of milk fermented with Lactobacillus acidophilus strain L-92 on symptoms of Japanese cedar pollen allergy: a randomized placebo-controlled trial. Biosci Biotechnol Biochem 69: 1652–1660. [DOI] [PubMed] [Google Scholar]
- 8.Ishida Y, Nakamura F, Kanzato H, Sawada D, Hirata H, Nishimura A, Kajimoto O, Fujiwara S. 2005. Clinical effects of Lactobacillus acidophilus strain L-92 on perennial allergic rhinitis: a double-blind, placebo-controlled study. J Dairy Sci 88: 527–533. [DOI] [PubMed] [Google Scholar]
- 9.Shah MM, Miyamoto Y, Yamada Y, Yamashita H, Tanaka H, Ezaki T, Nagai H, Inagaki N. 2010. Orally supplemented Lactobacillus acidophilus strain L-92 inhibits passive and active cutaneous anaphylaxis as well as 2,4-dinitroflurobenzene and mite fecal antigen induced atopic dermatitis-like skin lesions in mice. Microbiol Immunol 54: 523–533. [DOI] [PubMed] [Google Scholar]
- 10.Torii S, Torii A, Itoh K, Urisu A, Terada A, Fujisawa T, Yamada K, Suzuki H, Ishida Y, Nakamura F, Kanzato H, Sawada D, Nonaka A, Hatanaka M, Fujiwara S. 2011. Effects of oral administration of Lactobacillus acidophilus L-92 on the symptoms and serum markers of atopic dermatitis in children. Int Arch Allergy Immunol 154: 236–245. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda N, Kaneko K, Ejiri M, Mizutani J, Ohshima K, Kajimoto O, Takahashi T, Yamamoto N. 2000. Improvement of the constipation of healthy volunteers by uptake of lactic acid bacteria fermented milk. J Nutr Food 3: 59–68. [Google Scholar]
- 12.Kanzato H, Fujiwara S, Ise W, Kaminogawa S, Sato R, Hachimura S. 2008. Lactobacillus acidophilus strain L-92 induces apoptosis of antigen-stimulated T cells by modulating dendritic cell function. Immunobiology 213: 399–408. [DOI] [PubMed] [Google Scholar]
- 13.Torii A, Torii S, Fujiwara S, Tanaka H, Inagaki N, Nagai H. 2007. Lactobacillus acidophilus strain L-92 regulates the production of Th1 cytokine as well as Th2 cytokines. Allergol Int 56: 293–301. [DOI] [PubMed] [Google Scholar]
- 14.Sagitani A.Anti-allergic effects of Lactobacillus acidophilus L-92 strain. 2010. Jpn J Lactic Acid Bact 21: 1–7 (in Japanese). [Google Scholar]
- 15.Ashida N, Yanagihara S, Shinoda T, Yamamoto N. 2011. Characterization of adhesive molecule with affinity to Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J Biosci Bioeng 112: 333–337. [DOI] [PubMed] [Google Scholar]
- 16.Sansonetti PJ. 2004. War and peace at mucosal surfaces. Nat Rev Immunol 4: 953–964. [DOI] [PubMed] [Google Scholar]
- 17.Akira S, Takeda K. 2004. Toll-like receptor signalling. Nat Rev Immunol 4: 499–511. [DOI] [PubMed] [Google Scholar]
- 18.Weiss G, Rasmussen S, Nielsen Fink L, Jarmer H, Nøhr Nielsen B, Frøkiaer H. 2010. Bifidobacterium bifidum actively changes the gene expression profile induced by Lactobacillus acidophilus in murine dendritic cells. PLoS ONE 5: e11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss G, Rasmussen S, Zeuthen LH, Nielsen BN, Jarmer H, Jespersen L, Frøkiaer H. 2010. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology 131: 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. 2010. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS ONE 5: e10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HG, Gim MG, Kim JY, Hwang HJ, Ham MS, Lee JM, Hartung T, Park JW, Han SH, Chung DK. 2007. Lipoteichoic acid from Lactobacillus plantarum elicits both the production of interleukin-23p19 and suppression of pathogen-mediated interleukin-10 in THP-1 cells. FEMS Immunol Med Microbiol 49: 205–214. [DOI] [PubMed] [Google Scholar]
- 22.Jin C, Wu L, Li J, Fang M, Cheng L, Wu N. 2012. Multiple signaling pathways are involved in the interleukine-4 regulated expression of DC-SIGN in THP-1 cell line. J Biomed Biotechnol 2012: 357060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Wu QL, Feng YH, Wang YF, Li XY, Zuo JP. 2005. Triptolide suppresses CD80 and CD86 expressions and IL-12 production in THP-1 cells. Acta Pharmacol Sin 26: 223–227. [DOI] [PubMed] [Google Scholar]
- 24.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. 1999. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med 189: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellor AL, Munn DH. 1999. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation? Immunol Today 20: 469–473. [DOI] [PubMed] [Google Scholar]
- 26.Taylor MW, Feng GS. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J 5: 2516–2522. [PubMed] [Google Scholar]
- 27.Robinson CM, Shirey KA, Carlin JM. 2003. Synergistic transcriptional activation of indoleamine dioxygenase by IFN-gamma and tumor necrosis factor-alpha. J Interferon Cytokine Res 23: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson CM, Hale PT, Carlin JM. 2005. The role of IFN-gamma and TNF-alpha-responsive regulatory elements in the synergistic induction of indoleamine dioxygenase. J Interferon Cytokine Res 25: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallarino F, Grohmann U, Hwang KW, Orabona C, Vacca C, Bianchi R, Belladonna ML, Fioretti MC, Alegre ML, Puccetti P. 2003. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol 4: 1206–1212. [DOI] [PubMed] [Google Scholar]
- 30.Fallarino F, Bianchi R, Orabona C, Vacca C, Belladonna ML, Fioretti MC, Serreze DV, Grohmann U, Puccetti P. 2004. CTLA-4-Ig activates forkhead transcription factors and protects dendritic cells from oxidative stress in nonobese diabetic mice. J Exp Med 200: 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellor AL, Munn DH. 2008. Creating immune privilege: active local suppression that benefits friends, but protects foes. Nat Rev Immunol 8: 74–80. [DOI] [PubMed] [Google Scholar]
- 32.Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, Puccetti P, Chieppa M, Rescigno M. 2010. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut 59: 595–604. [DOI] [PubMed] [Google Scholar]
- 33.Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, Fioretti MC, Puccetti P. 2002. T cell apoptosis by tryptophan catabolism. Cell Death Differ 9: 1069–1077. [DOI] [PubMed] [Google Scholar]