Highlights
-
•
We isolated a chronologically long-lived mutant of S. pombe.
-
•
We identified one mutation in pma1+ gene that encoded for an essential P-type proton ATPase.
-
•
The identified Asp-138 to Asn mutation resulted in reduced Pma1 activity.
-
•
This was concomitant with an increase in the chronological lifespan of S. pombe.
Keywords: Chronological lifespan, Fission yeast, Pma1
Abstract
We isolated a chronologically long-lived mutant of Schizosaccharomyces pombe and found a new mutation in pma1+ that encoded for an essential P-type proton ATPase. An Asp-138 to Asn mutation resulted in reduced Pma1 activity, concomitant with an increase in the chronological lifespan of this fission yeast. This study corroborates our previous report indicating Pma1 activity is crucial for the determination of life span of fission yeast, and offers information for better understanding of the enzyme, Pma1.
1. Introduction
The chronological lifespan of yeast cells is defined as the period during which cells can survive in a nondividing state and is determined by their viability after entry into the stationary phase [1]. Studies on Saccharomyces cerevisiae have identified several novel longevity factors, including Ras2, Tor1, and Sch9 [2]. In the fission yeast Schizosaccharomyces pombe, disruptions of pka1+ and sck2+ reportedly increased its chronological lifespan [3]. The lifespan of S. pombe can also be extended by calorie restriction (CR), which is known to extend the life spans of various organisms from yeast to mammals. In S. pombe, this process relies on the Sty1 MAP kinase [4].
Because lifespan is a complex phenomenon, identifying new factors involved in regulating the chronological lifespan is essential for understanding lifespan as a whole. To this end, we have screened for S. pombe short-lived and long-lived mutants and identified lcf1 and pma1 mutants, respectively. lcf1+ encodes for a long-chain fatty acyl-CoA synthetase that is involved in fatty acid utilization and/or metabolism [5]. pma1+ encodes for an essential P-type proton ATPase [6]. We have also determined that deleting php2, which encodes for a subunit of the CCAAT-binding factor complex, results in extending the chronological lifespan of this fission yeast [7].
In this study we identified and characterized another allele of the pma1 mutation that prolonged the chronological lifespan of S. pombe.
2. Materials and methods
2.1. Strains and media
S. pombe strain JY333 (h- leu1-32 ade6-M216) was used for mutant screening. Strains were grown in SD medium [0.67% yeast nitrogen base without amino acids (Difco), and 2% glucose] supplemented with necessary growth requirements in standard amounts at 30 °C. Chronological lifespan analysis was done as described previously [8].
2.2. Linkage analysis
To conduct linkage analysis, a Km-resistant gene was inserted at 1824–1835 bp downstream of pma1+ termination codon using previously described methods [9]. Both the upstream and downstream regions of the desired insertion region were PCR-amplified using F1 and F2 primers and R1 and R2 primers, respectively. After mixing both DNA fragments with pFA6a-kanMX6, a PCR was performed using the F1 and R1 primers. JY336 (h+ leu1-32 ade6-M210) was transformed with the amplified DNA fragment, and stable G418-resisitant transformants were selected. The chromosome construct was then confirmed by PCR using appropriate primers. The primers used were: F1, AGAAGTTATCGTGAGCTACG; F2, TTAATTAACCCGGGGATCCGGAAATCATTGATTTATCTATATAC; R1, GTCTTGGTCTGGTATCAACG; and R2, GTTTAAACGAGCTCGAATTCCATGGATAAGCTGCTAATCCATAAT.
2.3. Preparation of a Pma1 antibody
An antibody directed against Pma1 was prepared by immunizing a rabbit with the peptide MMNGKPKESRNQRSIEDL (Sigma–Aldrich), which corresponded to amino acids 886–903 of the Pma1 protein.
2.4. Pma1 ATPase assay
Pma1 ATPase activity was determined using previously described methods [10]. Total cell lysates were used as the enzyme sources. This assay was conducted with or without 0.1 mM sodium vanadate, and released inorganic phosphate was determined using a Phospha C-Test (Wako Co., Japan). Vanadate-sensitive ATPase activity was determined and expressed as Pma1 activity.
2.5. Assay for glucose concentrations
Cells were grown in SD medium. In addition to monitoring cell growth, 20 μl of culture was sampled to determine the remaining glucose concentrations in medium using a Glucose CII-test kit (Wako Co., Ltd).
3. Results and discussion
3.1. L16 mutant phenotypic characterization
We previously screened for some long-lived mutant S. pombe candidates [6]. In this study, we analyzed one uncharacterized mutant, designated L16. We first analyzed the long-lived phenotype of L16 that was grown in SD medium (Fig. 1). As expected, L16 mutant cells’ viability was maintained for a long period after their entry into the stationary phase as compared with wild type cells (Fig. 1B). Cell morphology was also monitored along with cell growth in SD medium. As shown in Fig. 1C, there were no differences in cell morphology between wild type and L16 mutant cells during the logarithmic growth phase (sample point “a” shown in Fig. 1A). However, after entry into the stationary phase (sample points “b” and “c” in Fig. 1A), wild type cells exhibited abnormal morphologies, such as broken or shrunken figures. By comparison, many of the L16 mutant cells had normal morphologies. This phenotypic difference in morphology may have been due to the differences in viability after entry into the stationary phase.
Fig. 1.
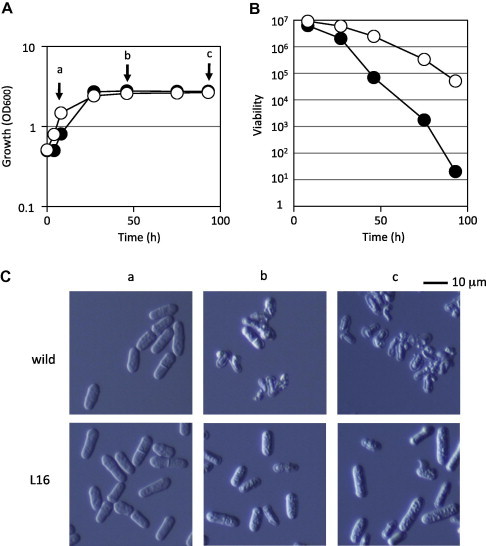
Phenotypes of a mutant with increased cell viability after entry into the stationary phase. Cell growth (A) and cell viability (B) of wild type cells (closed circles) and L16 mutant cells (open circles) in SD medium were monitored. Experiments were repeated twice, with similar results. The representative data are shown. (C) Cell morphologies at various growth phases. Wild type and L16 mutant cells were sampled at each time point (a, b, and c) indicated on the growth curves (A) and observed microscopically.
3.2. Identifying a mutation site in the L16 mutant
We suspected a mutation in pma1+ that encodes for a plasma membrane P-type ATPase, as we previously isolated a long-lived mutant and identified its mutation in pma1+ [6]. Thus, we sequenced the ORF region of pma1+ in the L16 mutant and identified one missense (G to A) mutation that caused an Asp-138 to Asn change in the predicted first extracellular domain of the Pma1 protein (Fig. 2). Next, we confirmed that the identified mutation (designated, pma1-L16 allele) was the causative mutation for the long-lived phenotype of the L16 mutant.
Fig. 2.
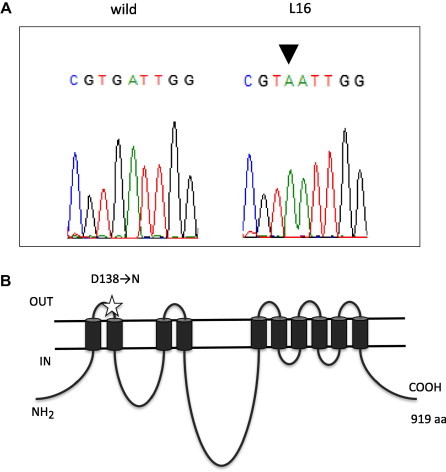
L16 has a mutation lesion in pma1+. (A) pma1 of the L16 mutant was sequenced and an identified mutation (G to A substitution) is shown (arrowhead). (B) Schematic representation for Pma1 protein topology in the plasma membrane. The mutation site (Asp-138 to Asn) is indicated by a star.
First, a Km-resistant cassette was inserted in the downstream region of pma1+, after which the Km-resistant strain was mated with the L16 mutant to assess the linkage between the Km-resistant phenotype and the chronologically long-lived phenotype. After crossing, phenotype analysis of the progeny revealed that 90% of chronologically long-lived cells (n = 20) were Km-sensitive, which indicated that the pma1-L16 mutation was located close to the site at which the Km-resistant cassette was inserted on the chromosome. Next, after crossing the L16 mutant with a wild type strain, we randomly isolated both long-lived cells (n = 4) and non-long-lived cells (n = 4), and sequenced their chromosome regions corresponding to the pma1-L16 mutation. We confirmed that all long-lived cells and non-long-lived cells had a pma1-L16 mutation and a wild type pma1+ allele, respectively. On the basis of these results, we concluded that the pma1-L16 mutation was the causative mutation that conferred the long-lived phenotype.
3.3. L16 mutant ATPase activity
Pma1 is a P-type plasma membrane ATPase that is a hydrogen ion pump [11,12]. H+-ATPase activity of the L16 mutant was analyzed to characterize the effect of the pma1-L16 mutation. Cell lysates were prepared from cells after growth to the mid-logarithmic phase (OD600 = 1) and to the stationary phase (OD600 = 2), after which their H+-ATPase activities were determined. As shown in Fig. 3A, the ATPase activity of the L16 mutant was lower than that of wild type cells at the stationary phase. In S. cerevisiae, it is known that Pma1 H+-ATPase activity is positively regulated by the glucose concentration in the medium; when glucose is added to carbon-starved cells, their ATPase activity increases [13,14]. Thus, we assessed S. pombe Pma1 activity in response to the glucose concentration.
Fig. 3.
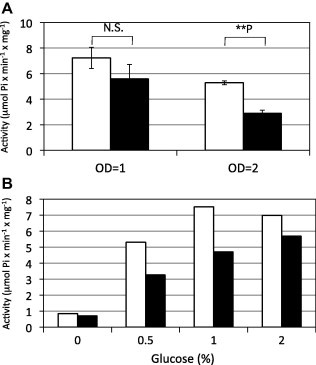
Pma1 activities. (A) Pma1 activities in wild type (open bars) and L16 mutant (filled bars) were assayed using cell lysates prepared from cells grown in SD medium until OD600 = 1 or 2. Results are the means ± s.d.’s of three independent experiments. Statistical analyses were performed using the Student’s t-test and indicated as follows: N.S. nonsignificant; ∗∗P < 0.01. (B) Pma1 activities in wild type (open bars) and L16 mutant (filled bars) were assayed using cell lysates prepared from cells grown in SD medium that contained the indicated glucose concentrations (%). Experiments were repeated twice, with similar results. The representative data are shown.
Wild type and L16 mutant cells were grown in SD medium until the logarithmic growth phase was achieved, and were then transferred to SD media that contained different glucose concentrations. Cells were grown in each medium for 90 min, after which H+-ATPase activity was determined. As shown in Fig. 3B, wild type cells’ H+-ATPase activity was regulated by glucose, as their ATPase activity was low in medium without glucose and increased in response to the glucose concentration. For the L16 mutant, ATPase activity increased in response to glucose. However, the activity was lower than that of wild type cells in medium that contained 0.5–2% glucose. These results suggested that the Pma1-L16 protein had defective ATPase activity but that its regulation by glucose was not affected. Taken together, we concluded that the L16 mutant had low H+-ATPase activity.
3.4. Expression profiles for pma1+ mRNA and Pma1 protein
To determine the expression profiles associated with pma1+, wild-type and L16 mutant cells were grown in SD medium, after which pma1+ mRNA levels were determined by Northern hybridization. As shown in Fig. 4A, similar pma1+ mRNA levels were expressed in both L16 mutant and wild type cells. Next, we determined the amounts of Pma1 protein by Western blotting under the same growth conditions. As shown in Fig. 4B, similar amounts of Pma1 protein were expressed in both cell types. On the basis of these results, we concluded that there were no differences in the amounts and stabilities of the Pma1 protein expressed in L16 and wild type cells. This indicated that the specific H+-ATPase activity of the Pma1-L16 protein was lower than that of the wild type Pma1 protein.
Fig. 4.
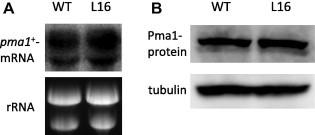
Pma1 RNA and protein expression. (A) Wild type and L16 mutant cells were grown in SD medium at 30 °C. Total RNAs were isolated from cells at the log phase (OD600 = 1.5) and subjected to Northern blotting analysis with a radiolabeled pma1+ probe. An ethidium bromide stained gel shows that total RNA was present as a loading control. (B) Wild type and L16 mutant cells were grown as described above, after which cell lysates were prepared. Pma1 protein expression was assessed by Western blotting using anti-Pma1 serum. Tubulin was used as a loading control.
3.5. The pma1-L16 mutant consumes less glucose
Pma1 H+-ATPase is involved in H+-dependent nutrient uptake [14]. Pma1 ATPase functions physiologically to pump protons out of a cell, thus generating an electrochemical gradient that drives solute uptake by an array of H+-coupled co-transporters. In S. pombe, glucose uptake is energy dependent and is driven by the plasma membrane ATPase-generated electrochemical gradient [15]. We compared glucose consumption during growth between wild-type and L16 mutant cells (Fig. 5). This revealed that the L16 mutant consumed less glucose compared with wild type cells. This difference in glucose consumption was likely due to the differences in Pma1 activity that provides the proton gradient for glucose uptake.
Fig. 5.
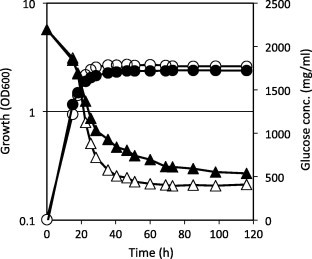
Glucose consumption. Cells were grown in SD medium at 30 °C, and the glucose concentrations in medium with wild type (open triangles) and L16 mutant (closed triangles) cells were determined. Wild type (open circles) and L16 mutant (closed circles) cell growth was monitored at OD600. Experiments were duplicated, with similar results. The representative data are shown.
On the basis of these findings, we propose the following scenario to explain the long-lived phenotype of the L16 mutant. In the L16 mutant, reduced Pma1 activity causes some defect in glucose uptake. This might cause physiological changes that are equivalent to changes caused by calorie restriction.
In summary, we propose that Pma1 ATPase activity is crucial for determining the chronological lifespan of S. pombe. Because Pma1 is conserved among many organisms, their chronological life spans might be manipulated by modulating Pma1 activity. Verification of this possibly novel means to regulate lifespan awaits further experimentation.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (22580079 and 25660290). Part of this work was also supported by the program from The Nagase Science and Technology Foundation and The Asahi Glass Foundation (to HA). HA conceived and designed the project, CN, HI, TO and HO acquired the data, CN, HI, TO, HO, HM and HA analyzed and interpreted the data, HM and HA discussed the data, HA wrote the paper.
References
- 1.Fabrizio P., Longo V.D. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 2.Wei M., Fabrizio P., Hu J., Ge H., Cheng C., Li L., Longo V.D. PLoS Genet. 2008;4:139–149. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux A.E., Quissac A., Chartrand P., Ferbeyre G., Rokeach L.A. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- 4.Zuin A., Carmona M., Morales-Ivorra I., Gabrielli N., Vivancos A.P., Ayté J., Hidalgo E. Lifespan extension by calorie restriction relies on the Sty1 MAP kinase stress pathway. EMBO J. 2010;29:981–991. doi: 10.1038/emboj.2009.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohshiro T., Aiba H., Mizuno T. A defect in a fatty acyl-CoA synthetase gene, lcf1+, results in a decrease in viability after entry into the stationary phase in fission yeast. Mol. Genet. Genomics. 2003;269:437–442. doi: 10.1007/s00438-003-0841-3. [DOI] [PubMed] [Google Scholar]
- 6.Ito H., Oshiro T., Fujita Y., Kubota S., Naito C., Ohtsuka H., Murakami H., Aiba H. Pma1, a P-type proton ATPase, is a determinant of chronological life span in fission yeast. J. Biol. Chem. 2010;285:34616–34620. doi: 10.1074/jbc.M110.175562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takuma K., Ohtsuka H., Azuma K., Murakami H., Aiba H. The fission yeast php2 mutant displays a lengthened chronological lifespan. Biosci. Biotechnol. Biochem. 2013;77:1548–1555. doi: 10.1271/bbb.130223. [DOI] [PubMed] [Google Scholar]
- 8.Ohtsuka H., Mita S., Ogawa Y., Azuma K., Ito H., Aiba H. A novel gene, ecl1+, extends the chronological lifespan in fission yeast. FEMS Yeast Res. 2008;8:520–530. doi: 10.1111/j.1567-1364.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- 9.Krawchuk M.D., Wahls W.P. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briskin D., Leonard R., Hodges T. Isolation of the plasma membrane: membrane markers and general principles. Methods Enzymol. 1987;148:542–558. [Google Scholar]
- 11.Ulaszewski S., Van Herck J.C., Dufour J.P., Kulpa J., Nieuwenhuis B., Goffeau A. A single mutation confers vanadate resistance to the plasma membrane H+-ATPase from the yeast Schizosaccharomyces pombe. J. Biol. Chem. 1987;262:223–228. [PubMed] [Google Scholar]
- 12.Ghislain M., De Sadeleer M., Goffeau A. Altered plasma membrane H+-ATPase from the Dio-9-resistant pmal-2 mutant of Schizosaccharomyces pombe. Eur. J. Biochem. 1992;209:275–279. doi: 10.1111/j.1432-1033.1992.tb17286.x. [DOI] [PubMed] [Google Scholar]
- 13.Serrano R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 1983;156:11–14. doi: 10.1016/0014-5793(83)80237-3. [DOI] [PubMed] [Google Scholar]
- 14.Lecchi S., Nelson C.J., Allen K.E., Swaney D.L., Thompson K.L., Coon J.J., Sussman M.R., Slayman C.W. Tandem phosphorylation of Ser-911 and Thr-912 at the C-terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 2007;282:35471–35481. doi: 10.1074/jbc.M706094200. [DOI] [PubMed] [Google Scholar]
- 15.Höfer M., Nassar F.R. Aerobic and anaerobic uptake of sugars in Schizosaccharomyces pombe. J. Gen. Microbiol. 1987;133:2163–2172. [Google Scholar]


