Abstract
In contrast to FLT3 ITD mutations, in-frame deletions in the FLT3 gene have rarely been described in adult acute leukemia. We report two cases of AML with uncommon in-frame mutations in the juxtamembrane domain of the FLT3 gene: a 3-bp (c.1770_1774delCTACGinsGT; p.F590_V592delinsLF) deletion/insertion and a 12-bp (c.1780_1791delTTCAGAGAATAT; p.F594_Y597del) deletion. We verified by sequencing that the reading frame of the FLT3 gene was preserved and by cDNA analysis that the mRNA of the mutant allele was expressed in both cases. Given the recent development of FLT3 inhibitors, our findings may be of therapeutic value for AML patients harboring similar FLT3 mutations.
Keywords: FLT3, Juxtamembrane domain, Deletion/insertion, Mutation, Acute myeloid leukemia (AML)
1. Introduction
Mutations in the FLT3 gene have been described in about 25% of acute myeloid leukemia (AML). They are somewhat more common in acute promyelocytic leukemia (APL), and have been associated with an increased risk of relapse, decreased disease-free survival, decreased event-free survival, and decreased overall survival [1]. These mutations result in constitutive activation of the FLT3 protein and are of two types: internal tandem duplication (ITD) mutations in exon 14 resulting from the duplication and tandem insertion of a portion of the juxtamembrane (JM) domain of the FLT3 gene and missense mutations in exon 20 which alter the aspartic acid residue at position 835 (D835) within the kinase domain of the FLT3 protein. In the case of ITD mutations, the duplicated segment length ranges in size from 3 to several hundred base pairs and is always in-frame and therefore expected to produce a functional protein [2]. Rare deletion and deletion/insertion mutations affecting the FLT3 juxtamembrane region have been described in childhood acute lymphoblastic leukemia [3,4]. Here, we report two cases of deletion and deletion/insertion mutations in the juxtamembrane domain of FLT3 in adult AML. Proper identification of these mutations may have prognostic and therapeutic significance for AML patients.
2. Methods
2.1. Patients
2.1.1. Patient #1
A 47 year-old man presented with complaints of shortness of breath, fatigue, and weakness over several days. He had WBC of 42.3×109/L and hemoglobin of 4.8 g/dL. Bone marrow morphology showed 95% cellularity with 83% blasts and the case was classified as AML M0 with myelodysplasia-related changes based on the detection of del(5q) by FISH, as the minimal differentiation of the leukemic blasts made the assessment of multilineage dysplasia rather difficult. Molecular diagnostic studies detected wild-type NPM1 gene and atypically mutated FLT3 gene. The patient underwent induction with cytarabine and idarubicin-based chemotherapy, but had evidence of primary refractory FLT3 mutation-positive AML on bone marrow biopsy performed 14 days after initiation of therapy. He then received high-dose cytarabine and mitoxantrone re-induction therapy. Repeat bone marrow evaluation upon count recovery revealed remission with 2% blasts and no evidence of FLT3 mutation. He subsequently underwent allogeneic stem cell transplantation from his sister and remained in remission for five years, after which the AML relapsed with a FLT3 D835 mutation and del(q5). He died shortly afterwards of infectious complications following re-induction chemotherapy.
2.1.2. Patient #2
A 54 year-old man with a prior medical history of coronary artery disease, diabetes, hypercholesterolemia, and hyperlipidemia presented with new onset of widespread bruising and blood in stool. Physical exam demonstrated scattered ecchymoses. Blood work revealed WBC of 8.6×109/L, hemoglobin of 9.8 g/L, and platelet count of 26×109/L. Prothrombin time was slightly elevated at 15.6 s (INR 1.25) with normal activated partial thromboplastin and a reduced fibrinogen level of 163 mg/dL. Hematopathologic evaluation of blood and bone marrow confirmed the diagnosis of acute promyelocytic leukemia (APL) with 91% marrow blasts/abnormal promyelocytes. Cytogenetics revealed a reciprocal translocation between the long arms of chromosomes 15 and 17 in 19/20 cells, t(15;17)(q24;q21). Molecular studies demonstrated a high level of the PMLRARalpha t(15;17) fusion transcript (164% of control) by quantitative RT-PCR. An atypical FLT3 mutation was also identified. The patient was initiated on differentiation therapy with oral retinoic acid (ATRA) 45 mg/m2 and arsenic trioxide 0.15 mg/kg intravenously daily as previously described [5]. Pseudotumor cerebri, scrotal ulcerations, and persistent headaches necessitated ATRA dose reduction. The patient was subsequently found to have CNS involvement by APL and received multiple intrathecal methotrexate injections. He was discharged home with count recovery two months after diagnosis and in complete remission.
2.2. FLT3 ITD and D835 mutation fragment analysis
DNA was extracted from blood or bone marrow samples using the EZ1 DNA Blood Kit (Qiagen, Germantown, MD) on the BioRobot EZ1 system (Qiagen). The FLT3 PCR-based fragment analysis assay was performed as previously described [6].
2.3. FLT3 juxtamembrane domain Sanger sequencing
Mutations detected in the juxtamembrane domain of the FLT3 gene underwent Sanger sequencing in both forward and reverse directions with the Big Dye Terminator v 3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA). Results were analyzed in Sequencing Analysis v5.2 software (Life Technologies) and Lasergene SeqMan Pro v10.0 (DNAStar, Madison, WI), and aligned to the FLT3 reference gene (NCBI RefSeq NM_004119.2).
2.4. FLT3 mRNA analysis
RNA was extracted from patient samples using the miRNeasy Mini Kit (Qiagen) and converted to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Pittsburgh, PA), which was subsequently amplified with primers FLT3F: 5′-6-FAM-GCCAGCTACAGATGGTACAGG-3′ and FLT3R: 5′-TTGCGTTCATCACTTTTCCA-3′. PCR products were analyzed on the ABI 3130xl Genetic Analyzer instrument (Life Technologies).
3. Results and discussion
Upon FLT3 ITD fragment analysis during routine molecular diagnostics work-up at presentation, both patient samples showed an unusual peak in the electropherogram (Fig. 1A). Besides the wild-type allele of 330 bp, a shorter PCR product in the same reaction pointed to the presence of a mutated allele showing a deletion in the PCR-amplified juxtamembrane domain region. Fragments shorter by 3-bp (327-bp) in patient #1 with a mutant allele/wild-type FLT3 ratio of 0.29, and by 12-bp (318-bp) in patient #2 with a ratio of 0.49 were detected. These fragments were further analyzed by Sanger sequencing to elucidate the nature of the deletions. Compared to the wild-type FLT3 sequence, patient #1 had a 5-bp deletion (CTACG) mutation combined with a 2-bp (GT) insertion: c.1770_1774delCTACGinsGT mutation (Fig. 1B), giving an overall 3 bp deletion as detected by FLT3 fragment analysis. The deletion resulted in a p.F590_V592delinsLF amino acid change in the juxtamembrane domain. Patient #2 had a c.1780_1791delTTCAGAGAATAT (12-bp deletion) mutation (Fig. 1B) resulting in p.F594_Y597del amino acid deletion in the juxtamembrane domain. Notably, these deletion and deletion/insertion mutations were in-frame and the reading frame of the FLT3 gene was preserved in both cases. Subsequently, the samples were tested for mutant versus wild type allele expression using cDNA fragment analysis. Both wild-type and mutant alleles were expressed at ratios comparable to the results of the FLT3 ITD assay (Fig. 2).
Fig. 1.
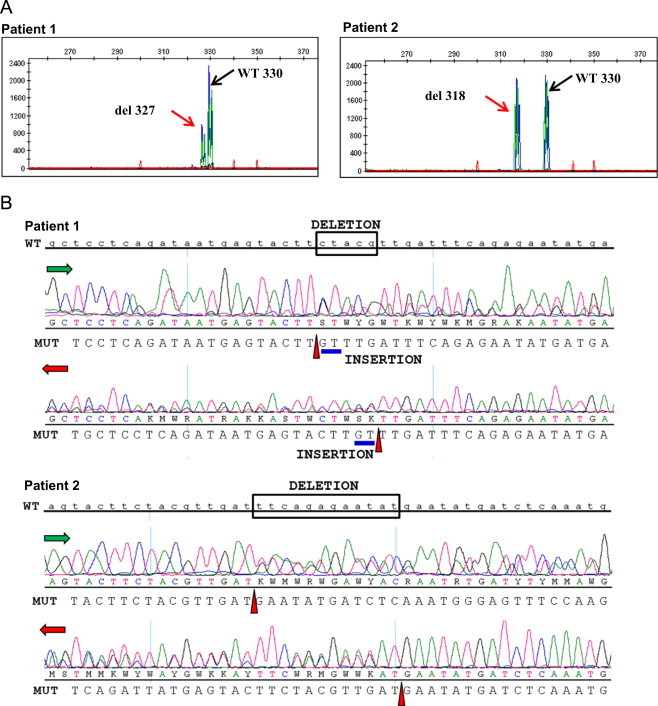
Deletions in the juxtamembrane domain of the FLT3 gene. (A) FLT3 ITD fragment analysis showing wild-type peak (330 bp), and smaller PCR amplification products (327 bp and 318 bp) which correspond to a 3-bp deletion in patient #1 and a 12-bp deletion in patient #2, respectively. (B) Results of Sanger sequencing confirming the deletions. The wild-type (WT) FLT3 sequence is in lowercase. The sequence of the deletion is boxed. The deletion mutation starting point is depicted by ▲(red) in the respective read directions, and the insertion is indicated by a blue underline. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
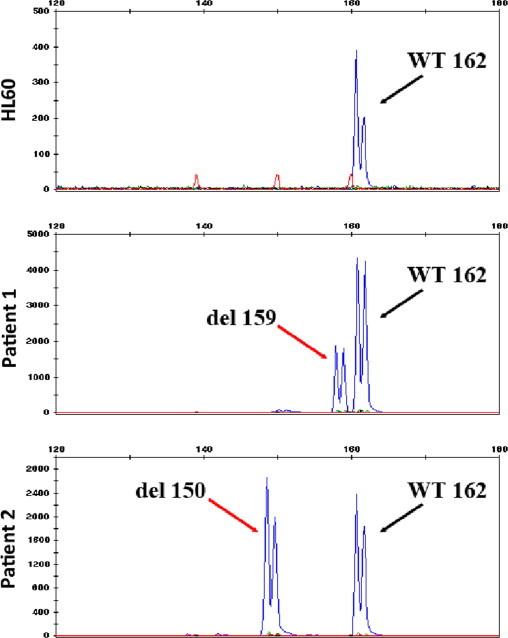
FLT3 mutant allele expression. RT-PCR across the FLT3 juxtamembrane domain containing the deletions shows that all samples (HL60 serves as WT control) express the wild-type allele (162 bp). In addition to that, the patient samples express the mutant alleles (del159 and del150, respectively).
FLT3 deletion and deletion/insertion mutations were previously reported in cases of pediatric acute lymphoblastic leukemia [3,4], but seldom described in adult acute leukemia. While the biological significance of this type of FLT3 mutations is unknown in human disease, a small 10-amino acid (Tyr589 to Tyr599) deletion in the juxtamembrane domain of FLT3 has been previously shown to lead to constitutive activation of the FLT3 protein in transformed murine IL3-dependent myeloid progenitor 32D cell line [7]. Similar deletion mutations are found in receptor tyrosine kinase KIT in gastrointestinal stromal tumors (GIST) [8,9]. An in-frame deletion of 7-amino acids in the juxtamembrane domain of the KIT gene resulted in receptor autophosphorylation and malignant transformation of mast cells [10]. These studies and our findings that both patients showed in-frame deletions with mRNA expressed (unfortunately, the samples did not yield enough material for a Western blot) suggest that deletion and deletion/insertion mutations in FLT3 juxtamembrane domain may lead to receptor activation. Animal models would be a way to prove this hypothesis and show, if inhibition of FLT3 can be therapeutically exploited in such cases. Whether the presence of these mutations in adult acute leukemia has prognostic significance warrants further investigation of a larger patient cohort.
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Kiyoi H., Naoe T. FLT3 in human hematologic malignancies. Leuk Lymphoma. 2002;43(8):1541–1547. doi: 10.1080/1042819021000002866. [DOI] [PubMed] [Google Scholar]
- 2.Schnittger S., Schoch C., Dugas M., Kern W., Staib P., Wuchter C. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100(1):59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong S.A., Mabon M.E., Silverman L.B., Li A., Gribben J.G., Fox E.A. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103(9):3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 4.Chang P., Kang M., Xiao A., Chang J., Feusner J., Buffler P. FLT3 mutation incidence and timing of origin in a population case series of pediatric leukemia. BMC Cancer. 2010;10:513. doi: 10.1186/1471-2407-10-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravandi F., Estey E., Jones D., Faderl S., O׳Brien S., Fiorentino J. Effective treatment of acute promyelocytic leukemia with all-trans-retinoic acid, arsenic trioxide, and gemtuzumab ozogamicin. J Clin Oncol. 2009;27(4):504–510. doi: 10.1200/JCO.2008.18.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy K.M., Levis M., Hafez M.J., Geiger T., Cooper L.C., Smith B.D. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5(2):96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyoi H., Ohno R., Ueda R., Saito H., Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002;21(16):2555–2563. doi: 10.1038/sj.onc.1205332. [DOI] [PubMed] [Google Scholar]
- 8.Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 9.Nishida T., Hirota S., Taniguchi M., Hashimoto K., Isozaki K., Nakamura H. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19(4):323–324. doi: 10.1038/1209. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimura T., Morimoto M., Hashimoto K., Moriyama Y., Kitayama H., Matsuzawa Y. Constitutive activation of c-kit in FMA3 murine mastocytoma cells caused by deletion of seven amino acids at the juxtamembrane domain. Blood. 1996;87(1):273–283. [PubMed] [Google Scholar]


