Abstract
Cell-in-cell structures, also referred to as 'entosis', are frequently found in human malignancies, although their prognostic impact remains to be defined. Two articles recently published in Cell Research report the stimulation of entosis by one prominent oncogene, Kras, as well as by one class of tumor suppressors, namely epithelial cadherins E and P, illustrating the complex regulation of this biological process.
A number of different terms have been used to describe live cell engulfments giving rise to cell-in-cell structures (CICS): entosis, emperipolesis, cannibalism and phagocytosis. Heterotypic live cell engulfment usually involves the ingestion of leukocytes by non-leukocytes (such as epithelial cells or fibroblasts). Homotypic live cell engulfment (among cells of the same type) mostly occurs in cancers, probably reflecting major alterations in cellular physiology that are associated with oncogenesis and tumor progression.
CICS can be visualized by conventional hematoxylin-eosin staining and have been described to occur in many different human cancers1. CICS produced as the result of entosis exhibit β-catenin localization patterns that are indicative of a cell junction-mediated mechanism of engulfment, and this polarized distribution of β-catenin can be taken advantage of to visualize CICS in vivo, in tumors1. However, the prognostic impact of CICS is highly context-dependent. Thus, CICS are particularly frequent in high-grade, aggressive breast cancer with dismal prognosis2. CICS are only found in castration-resistant, not in androgen-dependent, prostate cancer and hence correlate with poor prognosis in this particular malignancy3. In contrast, in pancreas adenocarcinomas, high levels of CICS correlate with a lower incidence of metastases4. These findings point to a complex role of CICS in cancer biology.
Two papers by Sun et al.5,6 recently published in Cell Research characterized one particular mechanism of homotypic live cell engulfment termed entosis. The first paper of this series5 provides evidence that one of the most prominent oncogenes, activated Kras, can stimulate entosis, while the second paper6 demonstrates that a prominent tumor suppressor, epithelial cadherin (E-cadherin), can increase entosis as well.
Cancers are highly complex mixtures of cells in which the malignant population is genetically and epigenetically heterogeneous, reflecting a history of clonal selection. One particular type of competition among distinct cells may consist in the engulfment of one cell (the 'loser') by another (the 'winner'), as demonstrated by Sun et al. in several cell culture models, as well as in human cancers that were xenografted into immunodeficient mice5. Importantly, co-culture of non-transformed cells with their malignant counterparts systematically leads to engulfment of the former by the latter, suggesting that oncogenic transformation is coupled to the 'winner' status5. Indeed, competition by entosis leads to the physical elimination of the 'loser' cells, which usually succumb to non-apoptotic cell death as soon as the phagosome enveloping the engulfed cell is decorated with LC3 and then fuses with lysosomes1,7. What is then the difference between 'loser' and 'winner' cells? Sun et al.5 propose that one cardinal feature of 'winners' is a high degree of mechanic deformability, as demonstrated by biophysical experiments and computer simulations. This is a highly provocative finding because human tumors are known to be more mechanically heterogeneous than normal tissues and that tumor progression is increased with an elevated mechanic deformability of the cancer cells. This reduction in cell stiffness may hence not only increase the metastatic potential of tumor cells8, but may also reflect an increased entotic activity5.
Transfection-enforced expression of active KrasV12 was sufficient to confer winner status onto non-tumorigenic cells, correlating with an increase in mechanic deformability5. This effect of KrasV12 relied on Rac1, as demonstrated by the facts that knockdown of Rac1 suppressed the 'winner' status conferred by KrasV12, expression of constitutively active Rac1 induced a 'winner' phenotype and dominant-negative Rac1N17 imparted a 'loser' status5. However, at this point it remains to be explored whether other pathways downstream of Kras such as the phosphatidylinositide 3-kinases (PI3K)/protein kinase B (PKB, best known as AKT)/mechanistic target of rapamycin (mTOR) pathway may contribute to 'winner' status. Inhibition of mTOR interferes with degradation of engulfed cells9, suggesting that activation of the PI3K/AKT/mTOR axis might favor the manifestation of the 'winner' phenotype as well. Similarly, it remains an open question as to whether other oncogenes than Kras may regulate entosis as well.
Breast cancers cells engineered to express epithelal E- or P-cadherins (but not mesenchymal-type cadherins, such as N-cadherin and cadherin-11) re-establish epithelial junctions and engulf and kill non-transfected parental cells in transformed growth assays6. The induction of entosis by epithelial E- or P-cadherins is associated to the polarized distribution of RhoA and contractile actomyosin dependent on the p190A Rho-GTPase-Activating Protein (p190A RhoGAP) that is recruited to epithelial junctions6. Inhibition of RhoA by overexpression of RhoA-N19 or p190A RhoGAP was sufficient to impart winner status to cells mixed with controls, whereas overexpression of RhoA, ROCKI, or ROCKII had the opposite effect and hence created 'loser' cells5. It has been known that Rho-GTPase and Rho-kinase are not required in engulfing cells but are required in internalizing cells1, underscoring the idea that 'loser' cells are not just passive 'victims' of a cannibalistic attack but somehow contribute to their fatal fate. The 'loser' status was accompanied by the ROCK-dependent accumulation of actomyosin6, and computer simulations suggest that actomyosin contractility within 'loser' cells constitutes a critical driving force of entosis5. The levels of phosphorylated myosin light chain 2 at Ser19 (pMLC2), a readout of contractile myosin downstream of ROCKI/II, were also increased in 'loser' cells as compared to 'winners'6. RhoA, ROCKI/II, MLC2, actin and myosins all accumulated at particularly high levels in 'losers' at the cell cortex oriented away from cell-cell adhesions6.
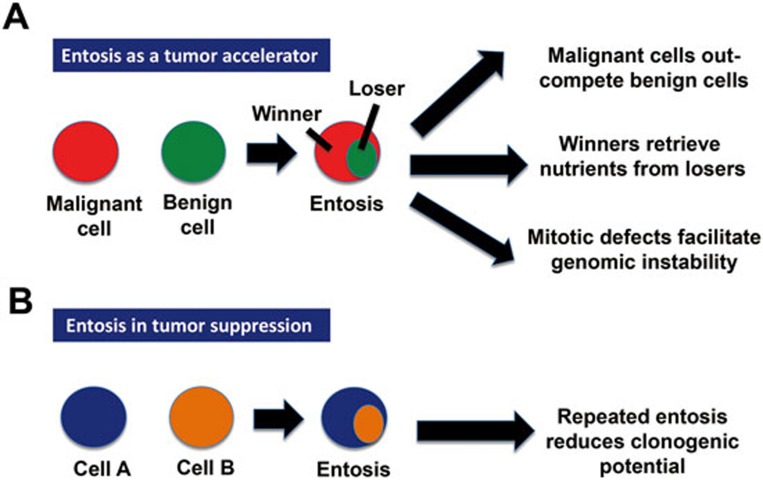
The aforementioned data support a dual implication of entosis in carcinogenesis (Figure 1). On one hand, entosis carried out by 'winner' cells may constitute a competitive advantage of aggressive tumor cells, perhaps allowing the 'winners' to retrieve amino acids and other building blocks for anabolic reaction from their cannibalistic activity9 or increasing their genomic instability subsequent to mitotic aberrations2,10. In this context, pharmacological suppression of entosis by Y27632, a ROCKI/II inhibitor, abolished the competitive advantage of transformed cells over their non-transformed siblings in mixed culture experiments5. On the other hand, stimulation of entosis by re-expression of epithelal E- or P-cadherins reduced the clonogenic potential of breast cancer cells. In this context, Y27632 facilitated tumor cell growth in vitro6. These observations underscore the need of exploring the detailed mechanisms through which entosis may repress or favor oncogenesis and tumor progression.
Figure 1.
A dual role for entosis in cancer. (A) Entosis as a pro-tumorigenic process. (B) Entosis as a tumor-suppressive mechanism.
References
- Overholtzer M, Mailleux AA, Mouneimne G, et al. Cell. 2007. pp. 966–979. [DOI] [PubMed]
- Krajcovic M, Johnson NB, Sun Q, et al. Nat Cell Biol. 2011. pp. 324–330. [DOI] [PMC free article] [PubMed]
- Wen S, Shang Z, Zhu S, et al. Prostate. 2013. pp. 1306–1315. [DOI] [PubMed]
- Cano CE, Sandí MJ, Hamidi T, et al. EMBO Mol Med. 2012. pp. 964–979. [DOI] [PMC free article] [PubMed]
- Sun Q, Luo T, Ren Y, et al. Cell Res. 2014. pp. 1299–1310. [DOI] [PMC free article] [PubMed]
- Sun Q, Cibas ES, Huang H, et al. Cell Res. 2014. pp. 1288–1298. [DOI] [PMC free article] [PubMed]
- Florey O, Kim SE, Sandoval CP, et al. Nat Cell Biol. 2011. pp. 1335–1343. [DOI] [PMC free article] [PubMed]
- Xu W, Mezencev R, Kim B, et al. PLoS One. 2012. p. e46609. [DOI] [PMC free article] [PubMed]
- Krajcovic M, Krishna S, Akkari L, et al. Mol Biol Cell. 2013. pp. 3736–3745. [DOI] [PMC free article] [PubMed]
- Vitale I, Galluzzi L, Senovilla L, et al. Cell Death Differ. 2011. pp. 1403–1413. [DOI] [PMC free article] [PubMed]