Abstract
An ambitious goal in biology is to understand the behaviour of cells during development by imaging—in vivo and with subcellular resolution—changes of the embryonic structure. Important morphogenetic movements occur throughout embryogenesis, but in particular during gastrulation when a series of dramatic, coordinated cell movements drives the reorganization of a simple ball or sheet of cells into a complex multi-layered organism1. In Xenopus laevis, the South African clawed frog and also in zebrafish, cell and tissue movements have been studied in explants2,3, in fixed embryos4, in vivo using fluorescence microscopy5,6 or microscopic magnetic resonance imaging7. None of these methods allows cell behaviours to be observed with micrometre-scale resolution throughout the optically opaque, living embryo over developmental time. Here we use non-invasive in vivo, time-lapse X-ray microtomography, based on single-distance phase contrast and combined with motion analysis, to examine the course of embryonic development. We demonstrate that this powerful four-dimensional imaging technique provides high-resolution views of gastrulation processes in wild-type X. laevis embryos, including vegetal endoderm rotation, archenteron formation, changes in the volumes of cavities within the porous interstitial tissue between archenteron and blastocoel, migration/confrontation of mesendoderm and closure of the blastopore. Differential flow analysis separates collective from relative cell motion to assign propulsion mechanisms. Moreover, digitally determined volume balances confirm that early archenteron inflation occurs through the uptake of external water. A transient ectodermal ridge, formed in association with the confrontation of ventral and head mesendoderm on the blastocoel roof, is identified. When combined with perturbation experiments to investigate molecular and biomechanical underpinnings of morphogenesis, our technique should help to advance our understanding of the fundamentals of development.
Gastrulation is a morphogenetic milestone in which complex cell and tissue rearrangements position the embryonic germ layers. It has been studied in living and fixed gastrulae using time-lapse light microscopy4,8–11, and in fixed, cleaved or dissected embryos3 by electron microscopy. However, these two-dimensional (2D) imaging methods are limited in their capacity to reveal deep tissue dynamics. As a consequence, certain gastrulation movements, such as convergent extension of the mesoderm2 and vegetal rotation of the endoderm12, have been analysed in vitro. Although studies of explants provide valuable insights, they are unlikely to fully recapitulate in vivo morphogenetic behaviours which depend on cell and tissue interactions within the intact embryo1,13,14. Confocal light microscopy15 and digital scanned light-sheet microscopy6,16 have been used to study in vivo developmental processes in four dimensions (4D). However, both techniques are most applicable to optically translucent embryos (for example, zebrafish) and require fluorescence labelling of selected subcellular structures15,17. In vivo microscopic magnetic resonance imaging has been used to investigate opaque embryos7 (for example, mice and X. laevis), but this technique is unable to resolve structure at the cellular level.
Here we use X-ray phase-contrast microtomography to overcome these limitations and to avoid high dose depositions introduced by conventional X-ray absorption imaging18. Exploiting the interference of electromagnetic waves, this imaging technique benefits from their high coherence and intensity when produced by modern synchrotrons. Early developmental stages of vertebrate embryos are composed of light chemical elements. As a consequence, phase shifts in the transmitted wave field dominate attenuation by three orders of magnitude for hard X-rays19–21. This facilitates in vivo X-ray imaging. Due to a high penetration depth, phase-contrast microtomography represents a genuine 3D imaging modality, and it visualizes both cell shape and subcellular structures such as nuclei and yolk platelets (Supplementary Video 1)22–24.
Radiation dose induces heat load and radiolysis of water in living samples. Accordingly, our set-up (Fig. 1 and Methods) was configured to minimise dose deposition while maintaining sufficient image contrast at an optimal X-ray energy E ~30 keV. Under these conditions heat load is predicted to be negligible (Supplementary Fig. 1 and Supplementary Information), which was confirmed by experiment (Supplementary Fig. 2). Radiolysis of water eventually leads to accumulation of lethal radicals resulting in sudden dissolution of tissue structure (Supplementary Video 16). However, before this point, morphogenetic processes and embryonic viability appear unimpaired. For a direct assay of viability, an easily disrupted process—blastopore closure—was examined. Closure rates were found to be comparable in X-rayed embryos (Supplementary Videos 3, 4 and 6) and control gastrulae viewed by light microscopy (Supplementary Fig. 3 and Supplementary Video 13).
Figure 1. Experimental set-up for propagation-based phase-contrast X-ray microtomography.
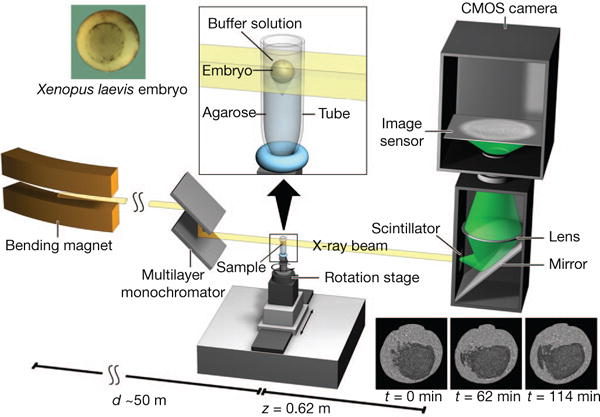
A quasi-parallel photon beam is generated from a synchrotron electron beam traversing the field of a bending magnet. After beam shaping and monochromatization, X-ray wave fronts propagate over a distance d (~50 m) to impinge on the sample (living X. laevis embryo immersed in buffer solution and suspended by agarose) mounted on a rotation stage for tomographic data acquisition. The 2D detector, at a distance z = 62 cm behind the sample, consists of a scintillator, converting X-rays into visible light, followed by a mirror, a lens, and a complementary metal oxide semiconductor (CMOS) camera (effective pixel size Δx = 2.2 μm).
Additional time-lapse in vivo scans, including neurulation, were acquired (Supplementary Videos 5–10 and Supplementary Information). These sequences confirm normal development in X-rayed embryos, even with harsher irradiation conditions than those used to acquire the gastrulation sequence analysed below (Supplementary Video 3).
Figure 2a–c shows mid-sagittally halved tomographic renderings of an embryo at developmental stages 11.5, 12.0 and 12.5 (Supplementary Video 3). Specific embryonic structures are discernible, including the blastopore, Brachet’s cleft, archenteron, ventral and dorsal blastopore lips, blastocoel, blastocoel roof and floor, and the porous, interstitial tissue between archenteron and blastocoel (‘pipe’ system, Supplementary Video 2). Noteworthy processes captured include the progression of epiboly by a thinning of ectoderm, blastopore closure and archenteron formation (Supplementary Videos 3 and 12).
Figure 2. 3D time-lapse series of X. laevis embryo during mid-gastrulation.

a–f, Mid-sagittally (a–c) and mid-horizontally (d–f) halved embryo renderings at stages 11.5 (0 min), 12 (62 min) and 12.5 (114 min). Ectoderm (blue), mesoderm (orange), and endoderm (green). g–i, Velocity fields on a 180-μm-thick 3D slab centred about the cutting planes of (a–c). Colour bar indicates velocity magnitude representation. Animal pole (AP), Archenteron (ARC), Brachet’s cleft (BC), blastocoel (BLC), blastocoel floor (BLCF), blastocoel roof (BLCR), blastopore (BP), dorsal and ventral sides (D, V), dorsal and ventral blastopore lip (DBL, VBL), ‘pipe’ system in-between archenteron and blastocoel (PS), ventral animal pole (VAP) and vegetal pole (VP).
To analyse cell movements, we computed 3D velocity fields from the time-lapse series of reconstructed volumes employing optical flow methods25 adapted to tomographic data. Essentially, the latter determine the displacement of a certain voxel in a given volume towards the associated voxel in the successive volume such that grey-value differences are minimised under certain constraints (Supplementary Information). Morphogenesis is the collective consequence of hierarchical, diverse, and coupled tissue/cell movements, all of which are captured by field . Therefore, we further explored (1) the collective aspects of , (2) its spatially differential behaviour and (3) its implications for individual cell trajectories. Velocity (flow) fields, depicted in Fig. 2g–i, reveal collective cell movements within the gastrula for several morphogenetic processes, including rotation of the vegetal endoderm (white arrowhead in Fig. 2g, Supplementary Video 11)12, involution of the mesendoderm at the dorsal and ventral blastopore lip (white arrows in Fig. 2g), and migration of the mesendoderm on the blastocoel roof towards the ventral animal pole (red arrowhead in Fig. 2g)1. At stage 11.5, involuting cells are fastest (~4 μm min−1) at the dorsal and ventral inner blastopore lips (Fig. 2g). By stage 12, prominent horizontal motion of the endoderm is observed on the ventral side (Fig. 2h and Supplementary Video 11). In addition, rapid migration of presumptive leading-edge head mesendodermal cells on the blastocoel roof, a bulk movement of the mesendoderm towards the animal pole, and localized cell displacements (archenteron formation) are discernible. Once the dorsal and ventral edges of the mesendodermal mantle make contact and enclose the blastocoel, an overall slowing of cell movements is observed on the ventral side (Fig. 2i, stage 12.5). Also, a strong inward displacement of cells in the anterior archenteron wall occurs, probably reflecting the rapid expansion of this cavity taking place towards the end of gastrulation26. Next, differential cell and tissue movements were examined through field (the spatial gradient operator; , , an orthonormal basis). Small values of G indicate collective motion, large values capture gradients in the velocity magnitude and/or variations in the direction of . Unlike the field , G is rotationally non-invariant and thus depends on the choice of Cartesian coordinates. However, G in contrast to S captures directional deviations at constant magnitude , and therefore we favoured it here. Figures 3a, b and 3c, d depict and G, respectively. In computing , optical flow is optimized to feature large, high-contrast mesendodermal or endodermal cells, thereby marginalizing the visualization of posterior involution movements and radial cell intercalation in the ectoderm. Contour C encloses a large, collectively moving cell mass (Fig. 3a), extending from the vegetal region (C1, Fig. 3c) along the posterior-anterior axial mesendoderm and endoderm on the dorsal side (C2, Fig. 3c). The movement of cells within C1 is essentially collective (small values of G), driven mainly by blastopore closure, whereas within C2, a collective-motion component of similar velocity magnitude is due to involution of the dorsal mesendoderm (transition from C1 to C2 in Fig. 3c). In addition, relative cell motion occurs within C2 that is attributable to mediolateral intercalation associated with convergent extension. This explains the acceleration (Fig. 2g, h) of the anterior cells within C (Fig. 3a) towards the animal pole, thus filling the gap (C3, Fig. 3a) in velocity magnitude (compare Fig. 3a and 3b). This gap indicates active migration27 of the leading-edge cells in the small region A (Fig. 3a, b) because their motion cannot be powered by the cell mass within C2, and because A cannot convey sizable convergent extension effects. Individual cells (I1) crawl on the blastocoel floor (Fig. 3c, d, see also Supplementary Video 4), and a single cell (I2) migrates on the endodermal bulk (Fig. 3d). Finally, trajectories of adjacent cells are computed by time integration of 3D flow fields in order to analyse differences in their dynamics (Fig. 3e, Supplementary Information and Supplementary Fig. 4).
Figure 3. Collective versus differential flow and cavity morphogenesis.

a, b, Magnitude of velocity on sagittal slice for two different times. c, d, Field G on same sagittal slice and for the same times. e, Sagittal slice at 52 min, highlighted cell pairs line the archenteron, associated trajectories (period of 30 min) (colour bar: blue, early; red, late). f, g, 3D renderings of cavities within the ‘pipe’ system in between archenteron and blastocoel for times 62 min and 114 min. h, Volume changes of ARC, BLC and gastrula from 52 min to 114 min.
We proceeded with an analysis of morphological changes. Archenteron formation is among the least understood aspects of gastrulation. In the mid-horizontal slice of Fig. 2e, two distinct, hollow regions occur (black arrows) which appear to merge (Fig. 2f). However, rendering of the entire volume (Supplementary Videos 12 and 14) reveals a heart-shaped and contiguous archenteron (Fig. 3f, g), highlighting the importance of 3D imaging. The increased porosity of the ‘pipe’ system may suggest (Fig. 3f, g) that archenteron inflation is driven by fluid transfer from the blastocoel4. To assess this we segmented blastocoel, archenteron, cavities in the ‘pipe’ system, and the entire embryo at different times (Fig. 3f–h and Supplementary Video 14). Strikingly, although the archenteron inflated, the blastocoel volume remained constant (Fig. 3h), whereas the volumes of the ‘pipe’ system and the entire embryo increased. This indicates that early archenteron expansion is not driven by fluid transfer from the blastocoel, but instead by the uptake of external water28. Indeed, this is implied by the conservation of cell–mass volume during gastrulation28, our results concerning volume changes (archenteron, ‘pipe’ system and entire embryo) (Fig. 3f–h), and the absence of additional cavities in the embryonic fluid balance (for example, disappearance of small blastopore cavities well before archenteron inflation, Supplementary Video 17 and Supplementary Information).
An interesting aspect of gastrulation is the active, non-invasive migration of mesendodermal cells on the ectoderm of the blastocoel roof. A model for this was proposed recently in which cycles of Eph/ephrin-dependent attachment and repulsion drive cell migration27. Our data capture, in detail, migration of the leading-edge head and ventral mesendoderm on their ectodermal substrate (Fig. 4f). The sagittal slices in Fig. 4a–e depict regions of contact and non-contact between the mesendoderm and the ectoderm, creating cavities consistent with that model27. Moreover, the ectoderm becomes transiently deformed towards the centre of the blastocoel, forming a cusp where the dorsal and ventral mesendoderm meet and make contact (Fig. 4b, c and Supplementary Video 15). The epiboly or spread of ectodermal cells over the surface of the gastrula runs counter to this. In 3D, the cusp of Fig. 4b, c corresponds to a ridge (Fig. 4g) that may represent a local contraction of the ectoderm itself, or arises from a local, adhesive pull by the dorsal and ventral mesendoderm. When these two eventually overlap, their contact with the ectoderm is lost, relaxing the cusp (Fig. 4c–e and Supplementary Video 18). In turn, new adhesive interactions between mesendodermal cells were observed (Fig. 4c). The formation of this ridge in explants is unlikely, owing to disruption of tissue tension; to our knowledge, it has not been seen before.
Figure 4. Confrontation of head and ventral mesendoderm.

a–e, Sequence of sagittal slices through confrontation zone at 52, 73, 83, 104 and 114 min showing formation and relaxation of a cusp of ectoderm on the blastocoel roof. Dorsal and ventral leading edges (Dors-LE, Vent-LE). Ectoderm (blue), mesoderm (orange), endoderm (green). f, Sequence of shrinking contours (projections of migrating leading edge along posterior–anterior axis onto horizontal plane) starting at 0 min (blue), terminating at 73 min. g, 3D rendering of individual cells and cavities in the confrontation zone at 73 min. Dors-LE cells (shades of green), Vent-LE cells (shades of red).
The present study demonstrates that propagation-based, single-distance phase-contrast X-ray microtomography, combined with optical flow analysis, is a useful tool for 4D in vivo investigation of X. laevis embryonic development. Analysis of differential motion permits active or passive tissue and cell movements to be distinguished, providing insights into how collective motion (convergent extension, collective cell migration on bulk tissue) is powered by individual cell behaviour. Time-lapse series with subcellular resolution can be captured over periods of two hours without adverse effects on development and with an option for longer observation times at lower resolution. When used in high-throughput perturbation experiments, we expect this imaging modality to advance our understanding of the molecular mechanisms and biomechanical processes driving morphogenesis.
METHODS SUMMARY
The time-lapse series analysed in this work relies on phase-contrast X-ray microtomography performed at beamline station 2-BM-B of Advanced Photon Source (monochromatization of ΔE/E=10−2, flux density of ~1012 photons per second per mm2, hutch temperature ~25 °C). The experimental set-up was optimized for low dose deposition (15 ms exposure per projection, N = 1,200 projections, 18 s to record one tomogram with continuous rotation of sample) at sufficient image contrast, avoiding blurring effects due to cell motion and source size (3.4 μm horizontal image blurring at z = 62 cm, horizontal coherence length of ~4.2 μm). Subsequent to phase retrieval22,24 a filtered back-projection algorithm29 was applied to compute the 3D distribution of electron density (Supplementary Video 1 and Supplementary Information). The waiting time between tomographic recordings was 10 min, sufficiently short to perform 3D optical flow analysis (Supplementary Information). In vitro fertilisation, embryo culture and staging were carried out as described30. All experiments were performed in accordance with a protocol approved by Northwestern University’s Committee on Animal Care and Use (Animal assurance number A328301).
METHODS
Sample preparation
In vitro fertilisation, embryo culture and staging were carried out as described30.
Detector system
The scintillator was a LuAG: Ce of 100-μm thickness. The camera system consisted of a pco.dimax CMOS (Cooke) with 2,0163 × 2,016 pixels of size 11 × 11 μm2 and a Mitutoyo long working distance × 5 lens (corresponding to an effective pixel size of Δx = 2.2 μm and Δx = 1.1 μm at 2-BM-B and 32-ID, respectively).
Data acquisition, processing, and analysis
The data was mainly collected at the bending magnet beamline station 2-BM-B of Advanced Photon Source (Argonne National Laboratory) with a monochromatization level of ΔE/E=10–2 and an X-ray energy of E=30 keV, a flux density at the sample of 1012 photons per mm2 per second, a sample–detector distance of z = 62 cm, N = 1,200 projections per tomogram, an exposure per projection of 15 ms, and a waiting time between tomograms of ~10 min when no irradiation of the embryo took place. Time-lapse series were terminated as soon as apoptotic cells or ectoderm ruptures could be observed in the radiographs. Following reconstruction, we concluded from the sequence of volumes, analysed in detail in the present work, that the associated embryo had been imaged in vivo for approximately two hours of development (Supplementary Video 3). To assure that in vivo imaging of gastrulation can be performed with another X-ray source, we also recorded a sequence at beamline 32-ID of Advanced Photon Source (Argonne National Laboratory) with a monochromatization level of ΔE/E = 10−4, an X-ray energy of E = 34 keV, a flux density at the sample of about two times 1012 photons per mm2 per second, a sample-detector distance of z = 70 cm, N = 834 projections per tomogram, an exposure per projection of 20 ms and a waiting time between tomograms of ~8 min (Supplementary Video 4 and Supplementary Fig. 3).
Apart from dose reduction due to an adjustment of X-ray energy, our two-hour time-lapse series, starting at developmental stage 11.5, could be acquired by exploiting the following points: (1) applicability of one exposure per tomographic projection by a single-distance set-up (Fig. 1) owing to a negligible contribution of absorption to the intensity contrast22, (2) use of a large propagation distance, bounded from above by image blurring due to the size of the X-ray source, to provide optimal image contrast24, and (3) tuning down of exposure time per projection to 15 ms, thus scraping the contrast limit for image analysis.
Raw images were flat- and dark-field corrected including removal of hot pixels and monochromator modulation. After phase retrieval22,24, volumes were reconstructed from the in vivo data resulting in a 4D view of mid-gastrulation with micrometre-scale spatial resolution (Supplementary Videos 2 and 3). For both data pre-processing and phase retrieval, MATLAB routines were written. Tomographic reconstruction was performed with a filtered back-projection routine29 inbuilt into PyHST (developed by European Synchrotron Radiation Facility and optimised at Karlsruhe Institute of Technology). We rendered cells, tissues and morphological features manually using Amira/Avizo and ImageJ/Fiji software31,32.
Velocity fields were computed by an automated 3D optical flow algorithm (developed at Karlsruhe Institute of Technology) built to minimise the summed difference of grey values between displaced pixels in successive volumes. This technique relies on the formulation of a variational problem and determines the 3D dense displacement field as its unique solution25,33,34. Cell trajectories were obtained by integrating the flow field, the locations of cellular nuclei serving as initial conditions. We checked the accuracies of the optical flow algorithm and of the integration routine by comparing the trajectories of manually tracked nuclei with the results of flow-field integration subject to the same initial positions of the nuclei. Compared to manually-tracked trajectories, the average error over four successive volumes is about 4 μm at an average distance of 58 μm travelled. Moreover, time-interpolated cell positions of four successive (uncorrupted) volumes between 52 min and 83 min were obtained by determining for each coordinate the coefficients of a cubic in terms of the data. To compute velocities, the according interpolated trajectories were differentiated with respect to time. With future data (longer time-lapse sequences) higher-order time derivatives should be computed of polynomials whose order is significantly smaller than the number of fit points. We estimate the maximum acceleration magnitude throughout mid-gastrulation as 0.2 mμ per min2.
Supplementary Material
Acknowledgments
We would like to acknowledge discussions with J. Wittbrodt, H. Steinbeisser, R. Winklbauer and D. Moss. R. Keller and D. Shook helped us with interpreting the data. D. Wedlich and M. Köhl commented on the manuscript. Discussions with M. Köhl on data analysis are gratefully acknowledged. We also would like to thank T. van de Kamp and D. Karpov for their help visualising the set-up, as well as F. de Carlo and K. Fezza for allocating beamtime at 2-BM-B station and at 32-ID, respectively, of Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract No. DE-AC02-06CH11357. E. Becker, C. Huang, A. Merks and R. Langhe helped analysing blastopore radii. J.K.’s Young Investigator Group received financial support from the ‘Concept for the Future’ programme of Karlsruhe Institute of Technology within the framework of the German Excellence Initiative. This research partially was funded by the German Federal Ministry of Education and Research under grant numbers 05K12CK2 and 05K12VH1.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions J.M., J.K. and R.H. conceived, organised and planned the experiments at beamlines 2-BM-B and 32-ID of Advanced Photon Source including feasibility studies, data management and testing/optimizing the set-up. V.A. established first contact between physicists and developmental biologists at Karlsruhe Institute of Technology to investigate X. laevis embryos. The physics part of the experiment was performed by J.M., X.X. and R.H., the biological parts by J.K. C. LaB. and M.S.P. helped with preparing samples. X.X. conducted heat-load measurements. R.H. investigated heat load theoretically. J.M. and A.E. developed data pre-processing routines. J.M. performed data pre-processing, phase retrieval and tomographic reconstruction. A.E. developed optical flow methods, computed velocity fields and performed image analysis. A. E. and J.M. worked on visualisations. A.E., J.M., V.A. and J.K. performed segmentation of the data. A.E., J.M. and R.H. analysed cell trajectories. J.M., A.E., T.B., X.X., J.K. and R.H. continuously discussed physics and biology problems. R.H. drafted the manuscript, A.E., J.M. and J.K. discussed and contributed text. C.LaB., M.S.P. and T.B. provided critical review. All authors contributed to the final manuscript.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Keller R, Davidson LA, Shook DR. How are we shaped: The biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- 2.Keller R, Danilchik M. Regional expression, pattern and timing of convergence and extension during gastrulation of Xenopus laevis. Development. 1988;103:193–209. doi: 10.1242/dev.103.1.193. [DOI] [PubMed] [Google Scholar]
- 3.Damm EW, Winklbauer R. PDGF-A controls mesoderm cell orientation and radial intercalation during Xenopus gastrulation. Development. 2011;138:565–575. doi: 10.1242/dev.056903. [DOI] [PubMed] [Google Scholar]
- 4.Ewald AJ, Peyrot SM, Tyska JM, Fraser SE, Wallingford JB. Regional requirements for dishevelled signaling during Xenopus gastrulation: separable effects on blastopore closure, mesendoderm internalization, and archenteron formation. Development. 2004;131:6195–6209. doi: 10.1242/dev.01542. [DOI] [PubMed] [Google Scholar]
- 5.Huisken J, Stanier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller PhJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 7.Papan C, Velan SS, Fraser SE, Jacobs RE. 3D time-lapse analysis of Xenopus gastrulation movements using μMRI. Dev Biol. 2001;235:189. [Google Scholar]
- 8.Wallingford JB, Fraser SE, Harland RM. Convergent extension: The molecular control of polarized cell movement during embryonic development. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- 9.Hardin J, Keller R. The behaviour and function of bottle cells during gastrulation of Xenopus laevis. Development. 1988;103:211–230. doi: 10.1242/dev.103.1.211. [DOI] [PubMed] [Google Scholar]
- 10.Weisz PB. The normal stages in the development of the South African clawed toad, Xenopus laevis. Anat Rec. 1945;93:161–169. doi: 10.1002/ar.1090930207. [DOI] [PubMed] [Google Scholar]
- 11.Ewald AJ, McBride H, Reddington M, Fraser SE, Kerschmann R. Surface imaging microscopy, an automated method for visualizing whole embryo samples in three dimensions at high resolution. Dev Dyn. 2002;225:369–375. doi: 10.1002/dvdy.10169. [DOI] [PubMed] [Google Scholar]
- 12.Winklbauer R, Schürfeld M. Vegetal rotation, a new gastrulation movement involved in the internalization of the mesoderm and endoderm in Xenopus. Development. 1999;126:3703–3713. doi: 10.1242/dev.126.16.3703. [DOI] [PubMed] [Google Scholar]
- 13.Keller R, Shook D, Skoglund P. The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys Biol. 2008;5:015007. doi: 10.1088/1478-3975/5/1/015007. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard GB, Adams R. Measuring the multi-scale integration of mechanical forces during morphogenesis. Curr Opin Genet Dev. 2011;21:653–663. doi: 10.1016/j.gde.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Ruffins SW, Russells EJ, Fraser SE. Towards a Tralfamadorian view of the embryo: multidimensional imaging of development. Curr Opin Neurobiol. 2002;12:580–586. doi: 10.1016/s0959-4388(02)00366-5. [DOI] [PubMed] [Google Scholar]
- 16.Greger K, Swoger J, Stelzer EHK. Basic building units and properties of a fluorescence single plane illumination microscope. Rev Sci Instrum. 2007;78:023705. doi: 10.1063/1.2428277. [DOI] [PubMed] [Google Scholar]
- 17.Dubertret B, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 18.Cheng KC, Xin X, Clark DP, La Riviere P. Whole-animal imaging, gene function and the Zebrafish phenome project. Curr Opin Genet Dev. 2011;21:620–629. doi: 10.1016/j.gde.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snigirev A, Snigireva I, Kohn V, Kuznetsov S, Schelokov I. On the possibilities of X-ray phase contrast microimaging by coherent high-energy synchrotron radiation. Rev Sci Instrum. 1995;66:5486–5492. [Google Scholar]
- 20.Wilkins SW, Gureyev TE, Gao D, Pogany A, Stevenson AW. Phase-contrast imaging using polychromatic hard X-rays. Nature. 1996;384:335–338. [Google Scholar]
- 21.Nugent KA, Gureyev TE, Cookson DF, Paganin DM, Barnea Z. Quantitative phase imaging using hard X rays. Phys Rev Lett. 1996;77:2961–2964. doi: 10.1103/PhysRevLett.77.2961. [DOI] [PubMed] [Google Scholar]
- 22.Paganin D, Nugent KA. Noninterferometric phase imaging with partially coherent light. Phys Rev Lett. 1998;80:2586–2589. [Google Scholar]
- 23.Cloetens P, et al. Hard X-ray phase imaging using simple propagation of a coherent synchrotron radiation beam. J Phys D Appl Phys. 1999;32:A145–A151. [Google Scholar]
- 24.Hofmann R, Moosmann J, Baumbach T. Criticality in single-distance phase retrieval. Opt Express. 2011;19:25881–25890. doi: 10.1364/OE.19.025881. [DOI] [PubMed] [Google Scholar]
- 25.Brox T, Bruhn A, Papenberg N, Weickert J. Lecture Notes in Computer Science. Springer; 2004. High accuracy optical flow estimation based on a theory for warping; pp. 25–36. (3024 Part IV). [Google Scholar]
- 26.Keller RE. An experimental analysis of the role of bottle cells and the deep marginal zone in gastrulation of Xenopus laevis. J Exp Zool. 1981;216:81–101. doi: 10.1002/jez.1402160109. [DOI] [PubMed] [Google Scholar]
- 27.Rohani N, Canty L, Luu O, Fagotto F, Winklbauer R. EphrinB/EphB signaling controls embryonic germ layer separation by contact-induced cell detachment. PLoS Biol. 2011;9:e1000597. doi: 10.1371/journal.pbio.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuft PH. The uptake and distribution of water in the embryo of Xenopus laevis (Daudin) J Exp Biol. 1962;39:1–19. doi: 10.1242/jeb.39.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Deans SR. The Radon Transform and Some of Its Applications. Wiley; 1983. (Ch 6). [Google Scholar]
- 30.Kashef J, et al. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 2009;23:1393–1398. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horn BKP, Schunck BG. Determining optical flow. Artif Intell. 1981;17:185–203. [Google Scholar]
- 34.Baker S, et al. A database and evaluation methodology for optical flow. Int J Comput Vis. 2011;92:1–31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


