Abstract
Pigs are ideal organ donors for xenotransplantation and an excellent model for studying human diseases, such as neurodegenerative disease. Transcription activator-like effector nucleases (TALENs) are used widely for gene targeting in various model animals. Here, we developed a strategy using TALENs to target the GGTA1, Parkin and DJ-1 genes in the porcine genome using Large White porcine fibroblast cells without any foreign gene integration. In total, 5% (2/40), 2.5% (2/80), and 22% (11/50) of the obtained colonies of fibroblast cells were mutated for GGTA1, Parkin, and DJ-1, respectively. Among these mutant colonies, over 1/3 were bi-allelic knockouts (KO), and no off-target cleavage was detected. We also successfully used single-strand oligodeoxynucleotides to introduce a short sequence into the DJ-1 locus. Mixed DJ-1 mutant colonies were used as donor cells for somatic cell nuclear transfer (SCNT), and three female piglets were obtained (two were bi-allelically mutated, and one was mono-allelically mutated). Western blot analysis showed that the expression of the DJ-1 protein was disrupted in KO piglets. These results imply that a combination of TALENs technology with SCNT can efficiently generate bi-allelic KO pigs without the integration of exogenous DNA. These DJ-1 KO pigs will provide valuable information for studying Parkinson's disease.
Pigs have been extensively used in biomedical research and are an excellent models for human diseases, especially for cardiovascular disease, atherosclerosis, cutaneous pharmacology, wound repair, cancer, diabetes, ophthalmological studies, toxicology research, lipoprotein metabolism, lung disease, and neurodegenerative diseases, and being considered as a potential source of organs for xenotransplantation1. The similarities of organ size, anatomic and physiologic characteristics as well as genome sequence between pigs and humans have stimulated a wide range of application of pigs in biomedical research. While pigs offer many similarities to human anatomy, metabolism, neurobiology and physiology2,3,4,5, furthermore, pigs have a longer life span, are easily bred, reach puberty at 6–8 months, and generate less ethical concerns when compared with non-human primates.
The ability to make genetic modifications in pigs facilitates the establishment of a human genetic disease model for the exploration and development of new therapies. Because no characterized embryonic stem cells (ESCs) have been generated in pigs that can be used for gene targeting, homologous recombination (HR) in somatic cells followed by somatic cell nuclear transfer (SCNT) is the primary strategy for creating knockout (KO) animals6. However, the very low rate of HR in somatic cells has hindered the establishment of KO animals. Therefore, highly efficient targeting in somatic cells is imperative and will facilitate the ability to genetically modify pigs. The emergence of zinc finger nucleases (ZFNs) has improved the efficiency of gene targeting greatly7, and ZFNs have been used to make various KO animals, such as pigs8,9,10, human cells11, and rats12. Although ZFN-mediated gene targeting has progressed in large animals, some disadvantages of ZFNs, such as off-target cleavage, cytotoxicity, and limited target sites13,14,15, have limited their applications.
Transcription activator-like effector nucleases (TALENs) were first reported in 201016. Similar to ZFNs, TALENs are composed of a TALE DNA binding domain and a FokI endonuclease. TALENs have been shown to possess comparable or higher efficiency for gene targeting, are more straightforward, and can theoretically target any sequence. TALEN-mediated targeting has been used successfully in different organisms, such as zebrafish17,18, rat19, and pig6.
Normally, antibiotic selection was used to screen the targeted cells that were successfully transfected with targeting vectors. The integration of antibiotic genes may cause concerns about food safety and unexpected biological effects. Therefore, highly efficient gene targeting without exogenous DNA integration is worth studying. Additionally, for large animals, making precise genetic modifications in donor cells and then using these donor cells to generate gene KO animals through SCNT technology ensure that all the offspring carry the mutations, thus reducing the cost of maintaining large animals. Few studies related to TALEN-mediated knock-in animals have been reported.
Parkinson's disease (PD) is a common neurodegenerative movement disorder. PD is a progressive disease that is characterized by the degeneration of dopamine neurons in the substantia nigra and the intracytoplasmic inclusion of Lewy bodies. To date, 11 genes from 16 PD loci have been identified in familial PD patients, and 5 of these genes have been studied extensively, namely α-synuclein (also known as SNCA), parkin (PARK2), PINK1 (PARK6), DJ-1 (PARK7), and leucine-rich repeat kinase (also known as LRRK2, PARK8)20. To elucidate the mechanisms of PD, many PD models have been generated by genetic manipulating these 5 genes in different organisms, such as mice, Drosophila melanogaster, and Caenorhabditis elegans. Although many mouse models with different fragment loss of the Parkin and DJ-1 genes have been generated21,22,23,24,25,26,27,28,29, few of these models recapitulate the human disease process well, especially the most important phenotype: dopaminergic loss in the substantia nigra. Considering the high level of similarity between pigs and humans, we hypothesized that pigs are a more suitable model for studying Parkinson's disease.
To overcome the shortage of human organs for organ transplantation, the use of organs from other species is an alternative. Considering the high level of similarity between humans and pigs, pigs are suggested to be most suitable for supplying organs to humans. Gal epitopes encoded by the α-1,3-galactosyltransferase gene exist on the cell surfaces of all porcine tissues; this gene product is the major antigen in pig-to-human transplantation and causes hyperacute rejection. The knockout of this gene may repress hyperacute rejection and GGTA1 disrupted pigs have been made by Lai et al. through homologous recombination method in 200230.
In the current study, we aimed to develop highly efficient gene targeting strategies without the integration of exogenous DNA and site-specific knock-in mediated by TALENs in pigs. Bi-allelic deletions were generated via single transfection of somatic cell with TALENs targeting GGTA1 and DJ-1. Furthermore, TALEN-mediated introduction of a short sequence containing an enzymatic restriction site into the DJ-1 locus was achieved by single-strand oligodeoxynucleotides. Using SCNT and embryo transfer, DJ-1 KO piglets were generated. Western blot analysis verified the disruption of DJ-1 in these KO piglets. These DJ-1 KO piglets can provide valuable information on PD in humans.
Results
TALENs mediated highly efficient gene targeting without the integration of exogenous DNA
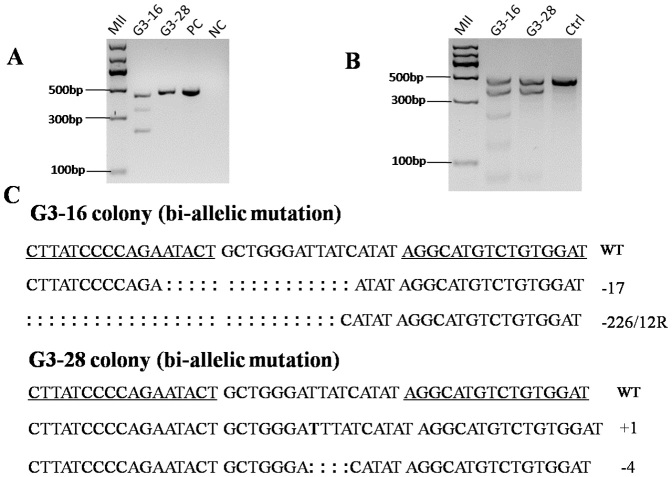
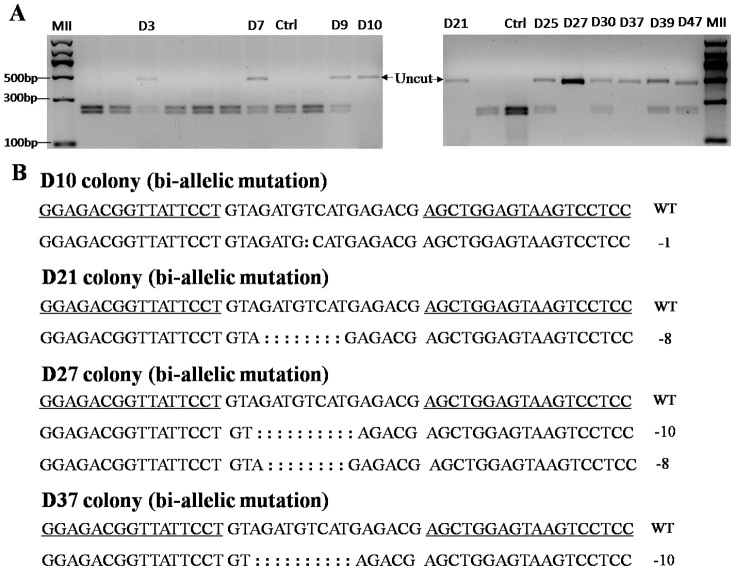
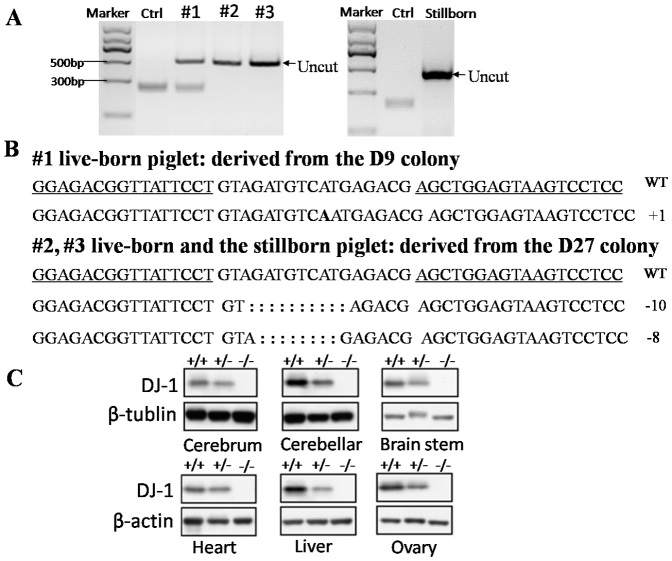
TALEN pair targeting GGTA1 or Parkin was co-transfected with the pCAG-GFP plasmid into male pig fetal fibroblast cells (FFCs), respectively, and single cells were sorted into 96 well plates by FACS. For the GGTA1 TALENs, a total of 40 colonies were obtained, and 2 of them (the G3-16 and G3-28 colony) were shown to be mutated by the T7 endonuclease I (T7EI) assay (Fig. 1A and 1B). Sequencing results showed that both of the colonies were bi-allelically mutated. One allele of G3-16 harbored a 17-bp deletion, and another allele harbored a 226-bp deletion and a 12-bp replacement (Fig. 1C). There was a 1-bp insertion in one allele of the G3-28 colony and a 4-bp deletion in another allele (Fig. 1C). For the Parkin TALENs, 2 of 80 colonies were shown to be modified with mono-allelic mutations (Fig. 2A and 2B). Sequencing results showed that one allele of the P3 colony had a 17-bp deletion, and the P40 colony had a 14-bp deletion (Fig. 2C). The DJ-1 TALENs were transfected into female pig FFCs; 11 of 50 colonies were mutated, and 4 of them were bi-allelically mutated (the D10, D21, D27 and D37 colonies), as shown in Fig. 3. The targeting efficiency is summarized in Table 1. The results showed that the TALEN-mediated targeting efficiency ranged from 2.5% to 22%, and more than 1/3 of the mutant colonies were bi-allelically modified.
Figure 1. GGTA1 TALEN activity in male porcine FFCs.
GGTA1 TALEN pairs were co-transfected with the pCAG-GFP plasmid into male porcine FFCs, and single-cell colonies were screened and genotyped. (A) Agarose gel electrophoresis detection of PCR products from the G3-16 and G3-28 colonies. (B) T7EI digestion of PCR products from the G3-16 and G3-28 colonies. (C) TALEN-mediated indels in the G3-16 colony and the G3-28 colony. The wild-type sequence is shown above with the TALEN binding sites underlined. The deleted bases are indicated by colons, and the inserted base is shown by bold uppercase letters.
Figure 2. Parkin TALEN activity in male porcine FFCs.
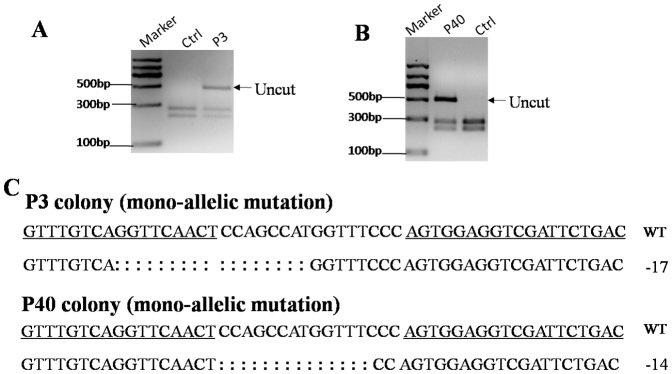
Parkin TALEN pairs were co-transfected with the pCAG-GFP plasmid into male porcine FFCs, and single-cell colonies were screened and genotyped. (A, B) NcoI digestion of PCR products from the P3 colony (A) and P40 colony (B). The arrow indicates the uncut fragment. (C) TALEN-mediated indels in the P3 colony and P40 colony. The wild-type sequence is shown above with TALEN binding sites underlined. The deleted bases are indicated by colons.
Figure 3. DJ-1 TALEN activity in female porcine FFCs.
DJ-1 TALEN pairs were co-transfected with the pCAG-GFP plasmid into female porcine FFCs, and single-cell colonies were screened and genotyped. (A) BspHI digestion of PCR products from female cell colonies. The arrow indicates the uncut fragment. (B) TALEN-mediated indels in bi-allelic mutant colonies. The wild-type sequence is shown above with TALEN binding sites underlined. The deleted bases are indicated by colons.
Table 1. Summary of the targeting efficiency of the 3 TALEN pairs in pig FFCs.
| TALEN pair | Cell type | Mutant colonies (%) | Bi-allelic mutant colonies (%) |
|---|---|---|---|
| GGTA1 | Male | 2/40 (5%) | 2/2 (100%) |
| Parkin | Male | 2/80 (2.5%) | 0 |
| DJ-1 | Female | 11/50 (22%) | 4/11 (36.4%) |
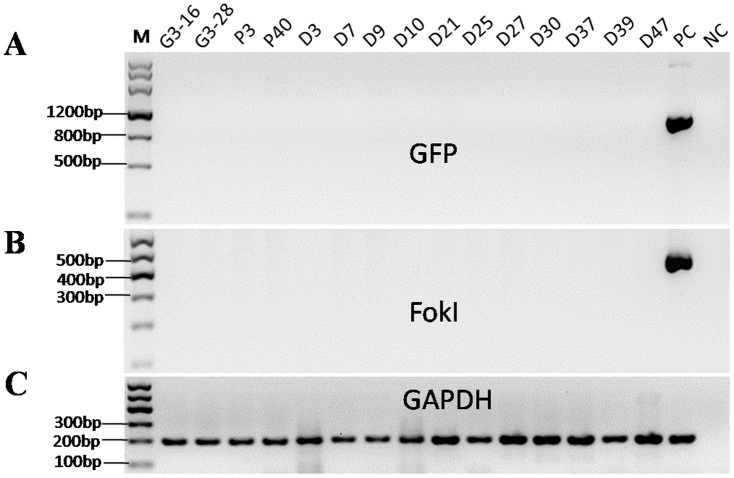
PCR amplification was used to determine the integration of the GFP fragment and TALEN DNA. The mutant cell colonies were negative for GFP fluorescence under a fluorescence microscope. The PCR results confirmed the loss of GFP in all the mutant cell colonies (Fig. 4A). Furthermore, the integration of TALEN plasmids was not detected in any of the mutant cell colonies by PCR amplification of the FokI endonuclease domain (Fig. 4B).
Figure 4. PCR amplification of GFP and FokI confirmed the loss of GFP and no TALEN plasmid integration in mutant colonies.
M represents the DNA marker, PC represents the positive control, and NC represents the negative control. GAPDH was used to confirm the DNA quality of all the samples.
Off-target analysis
To assess the specificity of the 3 pairs of TALEN plasmids used in this study, we used two approaches to scan the porcine genome to predict potential off-target sites. For E-PCR31,32, no off-target sites were predicted for the 3 TALEN pairs. Based on the TALENoffer score33, 5-7 potential off-target sites were verified by PCR amplification and sequencing (Supplementary Tables S1–S3 online). No mutations were observed at these sites in the G3-16 and G3-28 colonies for the GGTA1 TALENs, the P3 and P40 colonies for the Parkin TALENs, and the D9 and D27 colonies for the DJ-1 TALENs. These data suggest that the 3 pairs of TALENs used in the current study have high specificity for their target sequences.
Site-specific insertion via TALENs
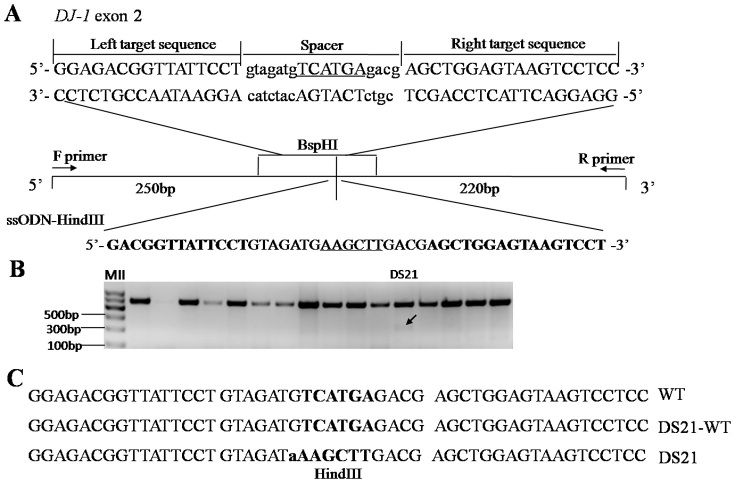
We next investigated the possibility of TALEN-mediated site-specific insertion by co-transfection of TALENs with single-strand oligodeoxynucleotides (ssODN). ssODN was first used in homology-directed repair (HDR) in ZFN-induced gene editing, and at least 20–25 bp homology arms on each side are recommended34. Considering the high targeting efficiency of the DJ-1 TALENs, it was chosen for subsequent investigation in female FFCs. An ssODN was designed to contain a HindIII restriction enzyme site (Fig. 5A) that would facilitate the subsequent detection of an insertion. The ssODN was co-transfected into fibroblasts with the DJ-1 TALENs and the pCAG-GFP plasmid, and single colonies were screened as described above. Ten of 49 colonies were mutated in the DJ-1 gene, and one (the DS21 colony) had the HindIII site insertion (Fig. 5B). Sequencing results confirmed the successful site-specific insertion of ssODN that was mediated by the DJ-1 TALENs (Fig. 5C). However, results of the restriction digest map and sequencing showed that the DS21 colony was not truly mono-colony. It may be mixed with wild type cells, which would be caused by the method we used for cell plating. The insertion rate was 10% among edited.
Figure 5. Site-specific insertion of HindIII site mediated by TALENs.
(A) Schematic of the DJ-1 locus with the ssODN sequence used to introduce the HindIII sequence (underlined) into the genome in vivo. (B) One of the colonies (DS21) could be cut by the HindIII restriction enzyme. The arrow shows the resulting fragments cut by HindIII. (C) DNA sequences of the DS21 colony at the target site. The HindIII site is shown by bold uppercase letters. There is a different base before the HindIII site, as shown in lowercase letters. DS21-WT represents sequence of the mixed wild type cells in DS21 colony.
Production of TALEN-modified piglets by somatic cloning
The DJ-1 mutant female FFCs (a mix of mono-allelic and bi-allelic mutant colonies) were used as donor cells for SCNT and embryo transfer to examine TALEN-mediated KO cells for producing KO animals. A total of 687 embryos were delivered to 5 surrogates, and one pregnancy was established. Three live-born piglets and one stillborn piglet were obtained, and the piglets were genotyped as 1 mono-allelic mutation and 3 bi-allelic mutations (Fig. 6A). The sequencing results showed that the mono-allelic KO piglet was derived from the D9 colony (Fig. 6B), which harbored an insertion-induced frame-shift in one allele, resulting in a prematurely truncated DJ-1 ORF. The two bi-allelic KO live-born piglets and the one stillborn piglet were derived from the D27 colony (Fig. 6B), which harbored an 8-bp deletion in one allele and a 10-bp deletion in another allele, also resulting in a prematurely truncated DJ-1 ORF. Unfortunately, the 2 bi-allelic mutant live-born piglets died within two days after birth, and the mono-allelic mutant piglet died at day 8. No obvious defect in the pig organs was observed by autopsy, and the deaths may have been caused by cloning defects. Tissues were dissected from the dead piglets, and Western blot analysis was performed to confirm the disruption of DJ-1 in these pigs. Compared with wild-type piglets, DJ-1 was partially repressed in the mono-allelic mutant piglet and completely absent in the bi-allelic mutant piglet (Fig. 6C), suggesting the successful generation of DJ-1 KO piglets.
Figure 6. Female piglets obtained from SCNT and embryo transfer using mixed KO colonies.
(A) BspHI digestion of PCR products from piglets #1, #2, #3 and the stillborn piglet. The genotyping result of piglets #1, #2, #3 was cropped from full-length gels indicated in supplementary files by discarding lanes of irrelevant samples. (B) DNA sequence of the DJ-1 gene at the target site in wild-type cells (wt) and KO piglets. The inserted base is shown by bold uppercase letters, and the deleted bases are indicated by colons. (C) Western blot results in six tissues of wild-type and KO piglets. +/− represents mono-allelic KO piglet (8-day-old), −/− represents bi-allelic KO piglet (2-day-old), and +/+ represents wild type pigs (10-day-old, female). β-tubulin and β-actin served as housekeeping gene controls.
Discussion
In the current study, three pairs of TALEN expression plasmids were designed to target the GGTA1, DJ-1 and Parkin genes in pigs. First, the efficiency of TALEN-mediated gene targeting was detected for the GGTA1 gene. Exon 6 was chosen for gene targeting because there is a catalytic domain of the α1, 3-galactosyltransferase enzyme. The targeting efficiency was 5% (2/40), and both colonies were bi-allelically modified, demonstrating the establishment of TALEN-mediated gene targeting in the lab. Then, two more TALEN pairs were designed to target the Parkin and DJ-1 genes, respectively. The efficiency of TALEN pairs designed against the DJ-1 gene was 22% (11/50). Various mutation patterns at the target site were observed, including small deletion (-1 base), large deletions (-226 bases), and small insertions (+1 base), which are characteristic of non-homologous end joining (NHEJ)-induced DNA damage response. The vast majority of mutations were deletions, which were also reported previously in mice and pigs32,35. Among all the targeted cell colonies, more than 1/3 were bi-allelic mutations, and this amount was consistent with previous reports in pigs6,32.
To induce mutations into the pig genome without the integration of exogenous DNA, circular pCAG-GFP plasmid was co-transfected with each TALEN pairs into pig FFCs and then sorted based on GFP expression by FACS to screen the mutant cell colonies instead of the usage of antibiotic markers to screen positive colonies. PCR results confirmed that there was no integration of the GFP or TALEN plasmids in the targeted cell colonies.
Off-target activity is another concern regarding nucleases mediated genome editing. In the current study, no off-target cleavages were identified for all the 3 pairs of TALENs. This lack of off-target cleavage is an advantage of TALENs over ZFNs and CRISPR/Cas9 system, which was reported to cause high off-target mutagenesis13,14,36. Take these data described above, TALENs are a reliable gene targeting tool for high efficient genetic modification in porcine somatic cells.
Site-specific knock-in is another advantage of nuclease-mediated gene targeting in large animals. TALEN-mediated site-specific insertion using ssODN with a HindIII site was successfully introduced to the somatic cell genome, however, a base substitution just before the HindIII site was identified. This substitution may be the result of re-cleavage of TALEN pairs after insertion of HindIII37 because the length of the spacer region between the TALEN pairs was the same as the wild-type sequence, which could be easily re-cleaved in cells cultured for extended periods. ssODN combined with ZFNs34,38,39, TALENs18,40 and Cripsr/Cas941 was used widely for introducing small indels, point mutation, and even deletion of large fragment (up to 100 kb) because of its easy design, rapid synthesis, and comparable efficiency as classical targeting vector34,39. However, since ssODN is very short (tens of nucleotides), it's difficult to detect the random integration of ssODN into genome. Deep sequencing might be necessary to evaluate the random integration of ssDNA. Now, we are trying to obtain longer donor template (>2 kb) to mediate site-specific insertion of long fragments, which will complement the future advances of gene therapy for repairing disease-related genes in humans.
Previous studies have constructed many mouse models with different fragments deleted from the Parkin and DJ-1 genes21,22,23,24,25,26,27,28,29, but few of them recapitulate human PD very well. One of the reasons may be that mice have a short life span, and PD is age-dependent, often occurring in aged people. Considering that pigs have a longer life span than mice and the other similarities between pigs and humans, pigs are ideal for constructing models of PD. Therefore, we used the DJ-1 KO cell colonies combined with SCNT and embryo transfer to generate KO piglets. One DJ-1 +/− and two DJ-1 −/− female piglets were obtained. Western blot analysis showed that the DJ-1 protein was repressed in all the detected tissues of DJ-1 KO piglets, indicating the successful construction of a DJ-1 KO pig model. Unfortunately, the piglets all died due to cloning defect.
In conclusion, TALEN mediated efficiently generation of bi-allelic DJ-1 KO pigs without the integration of exogenous DNA was successfully developed. The highly efficient gene targeting was also verified in the targeting of GGTA1 and Parkin. The results in the current study further confirmed that TALENs can be used conveniently and efficiently for site-specific knock-in for precise gene modification in large animals. The bi-allelic DJ-1 KO pigs could provide valuable information for understanding the mechanisms underlying PD.
Methods
Animals
Pigs were raised at the Beijing Farm Animal Research Center and had ad libitum access to a commercial pig diet (nutrient levels according to the NRC) and water during the entire experimental period. All experiments involving animals were performed according to procedures approved by the Institutional Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences, China.
Generation of TALENs
TALENs expression constructs were assembled by Viewsolid Biotech Company (Beijing, China). Three pairs of TALENs were designed to target exon 6, exon 1, and exon 2 in the porcine GGTA1, Parkin, and DJ-1 genes (ensembl ID: ENSSSCG00000005518, ENSSSCG00000004032 and ENSSSCG00000003385), respectively. The GGTA1 TALENs recognition sequences were as follows: left TALEN 5′- CTTATCCCCAGAATACT-3′ and right TALEN 5′- ATCCACAGACATGCCT -3′. The Parkin TALENs recognition sequences were as follows: left TALEN 5′- GTTTGTCAGGTTCAACT-3′ and right TALEN 5′- GTCAGAATCGACCTCCACT-3′. Between the two binding sites was a 16-bp spacer with a NcoI site (CCAGCCATGGTTTCCC, NcoI underlined). The DJ-1 TALENs recognition sequences were as follows: left TALEN 5′- GGAGACGGTTATTCCT-3′ and right TALEN 5′- GGAGGACTTACTCCAGCT-3′. Between the two binding sites was a 17-bp spacer with a BspHI sequence (GTAGATGTCATGAGACG, BspHI underlined). The TALEN pairs were transfected into porcine fetal fibroblast cells to detect the targeting efficiency.
Cell culture and transfection
Primary fetal fibroblast cells (FFCs) were isolated from 42-day-old Large White porcine fetuses (male and female) and digested by collagenase-DNaseI in cell culture medium (DMEM high glucose (Gibco BRL, Grand Island, NY) supplemented with 15% fetal bovine serum (FBS, Hyclone, Logan, UT), 0.032% collagenase (Sigma, St. Louis, MO), 25 Kunitz units/ml DNaseI (Sigma, St. Louis, MO) and 40 μg/ml gentamicin (Sigma, St. Louis, MO) for 4–6 h at 37°C. Digested cells were washed and plated in 25-cm2 flasks in cell culture medium (DMEM high glucose supplemented with 15% FBS). FFCs were frozen when they became confluent. Two days before transfection, FFCs were thawed and cultured in 75-cm2 flasks. When the cells become 80% confluent, transfection was performed by electroporation as described previously42. To avoid antibiotic gene insertion, the pCAG-GFP plasmid was co-transfected with TALENs to be used as an indicator for FACS sorting and was eliminated after several passages. Forty-eight to seventy-two hours after transfection, cells were subjected to FACS sorting based on the expression of GFP fluorescence. Single cells were plated in each well of 96-well plates and cultured for approximately 10 days in cell culture medium supplemented with 2.5 ng/mL basic fibroblast growth factor42,43,44 (Sigma, St. Louis, MO). The medium was replaced every 4 days. Confluent cell colonies were propagated and genotyped by PCR, restriction enzyme digestion and sequencing. Cell colonies harboring mutations were cryopreserved for SCNT.
For TALEN/ssODN-induced insertion, 0.5 μg of each of the TALEN pairs was co-transfected into female FFCs with 2 nmol ssDNA (a 20-bp 5′ homologous arm and a 20-bp 3′ homologous arm with a HindIII recognition site in the center, synthesized by Invitrogen). The pCAG-GFP plasmid was also included in the transfection system to provide an indicator for FACS sorting. A similar strategy was used to screen single-cell colonies and for genotyping.
Detection of gene mutations
The genomic DNA of each cell colony was extracted using the TIANamp genomic DNA kit (Tiangen, Beijing, China). Genotyping was conducted using PCR followed by restriction enzyme digestion. For GGTA1, the genotyping primers were as follows: 5′-AGCACGAGGTGGACTTC-3′ and 5′-CAGATGTTATTTCTAACCAAAT-3′. The primers for Parkin genotyping were as follows: 5′-AAGGGCAGTCCCACAGGGAAACTC-3′ and 5′- TGGAGTAAAGTTCAAGGAATCCCC-3′. The primers for DJ-1 genotyping were as follows: 5′- GTGCTTCTTCCTATTCATTTGTGC-3′ and 5′- CAGGTCTGTAAGAAATATGGGATG-3′. Mutations in GGTA1 were assessed by the T7 endonuclease I (T7EI) assay. Briefly, the purified PCR products from colonies and wild-type cells (200 ng each) were denatured at 95°C for 5 min and re-annealed at room temperature for 10 min, then digested by T7EI (New England Biolabs) at 37°C for 0.5 h. Different bands were expected for mutant colonies. Mutations in Parkin and DJ-1 were assessed by the loss of restriction enzyme sites harbored in the spacer region. PCR products of colonies were cut by NcoI or BspHI. Uncut bands were expected for mutant colonies. To verify the mutations, the gel-purified PCR products were cloned into the pEASY-T3 vector using the pEASY-T3 cloning kit (Transgen, Beijing, China) and then sequenced. To detect whether there was pCAG-GFP or TALEN plasmid integration in the positive colonies, PCR was conducted using the primers 5′-CAAGCTTCGAATTCTGCAGTCG-3′ and 5′-TGATGAGTTTGGACAAACCACAAC-3′ for GFP detection and the primers 5′-ATTTGGGTGGATCAAGGA-3′ and 5′-GTTTATCTCGCCGTTATT-3′ for FokI detection. One hundred and fifty ng of genomic DNA was used for PCR analysis. Conditions for PCR amplification were one cycle of pre-denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 62.7°C (for GFP primers) or 52°C (for FokI primers) for 30 seconds, and elongation at 72°C for 30 seconds, and one cycle of post-elongation at 72°C for 7 min. To assess the HindIII insertion, the PCR products of each cell colony were digested by the HindIII restriction enzyme, and colonies that could be cut by HindIII were then confirmed by sequencing.
Oocyte maturation
Pig ovaries were collected from a local slaughterhouse and transported to the laboratory within 1 h in 0.9% saline maintained at 37°C. Oocytes were aspirated from antral follicles (3–6 mm in diameter) with an 18-gauge needle fixed to a vacuum pump. Compact cumulus oocyte complexes (COCs) were selected under a stereomicroscope, washed and cultured in maturation medium at 39°C under 5% CO2. The maturation medium was composed of tissue culture medium (TCM) 199 (Hyclone, Logan, UT) supplemented with 0.1% (w/v) polyvinyl alcohol, 3.05 mM D-glucose, 0.91 mM sodium pyruvate, 0.57 mM cysteine, 0.5 μg/ml of luteinizing hormone, 0.5 μg/ml of follicle-stimulating hormone, 10 ng/ml of epidermal growth factor, 10% (v:v) of porcine follicular fluid, and 10 μg/ml of gentamicin. After 42–44 h of in vitro maturation culture, COCs were denuded by vigorous vortexing for 5 min in TL-HEPES supplemented with 0.1% polyvinyl alcohol and 0.1% hyaluronidase. Oocytes with polar bodies were selected under a stereomicroscope and used for somatic cell nuclear transfer (SCNT).
Somatic cell nuclear transfer and embryo transfer
Mature MII oocytes were enucleated by aspirating the first polar body and adjacent cytoplasm with a glass pipette (20 μm in diameter) in manipulation medium supplemented with cytochalasin B (5 mg/ml stock, 4 μl stock per 3 ml manipulation medium). A single DJ-1 KO fibroblast cell (donor cell) was injected into the perivitelline space of an enucleated oocyte. The donor cell membrane should be in contact with the oocyte cytoplasmic membrane. Oocyte cytoplasm-cell complexes were then fused and activated by electric pulse. Reconstructed embryos were cultured in porcine zygote medium (PZM3) in 5% CO2 at 39°C for 14–16 h until embryo transfer. Embryos in good condition (120–200 embryos) were surgically transferred into the oviduct of a surrogate the day after observed estrus. Gestation was detected by ultrasound at day 28 after embryo transfer. If the surrogate was pregnant, the gestation was monitored every 2 weeks. On the day of birth, an ear skin biopsy was isolated from each piglet for genomic DNA extraction and genotyping.
Western blot analysis
Different tissues were dissected from dead piglets, frozen immediately in liquid nitrogen and stored at −80°C until use. Twenty to thirty milligrams of different tissues was lysed in a buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 100 mM NaF, 0.05% SDS, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 2 g/ml aprotinin and leupeptin, 2 mM benzamidine, 1 mM sodium vanadate, 1 mM EGTA and 0.1% deoxycholate for 15 min on ice, and 50 μg of protein was subjected to Western blot analysis. The DJ-1 antibody (DJ-1 (D29E5) XP® Rabbit mAb #5933) was purchased from Cell Signaling Technology (Danvers, MA, USA), and a concentration of 1:1000 was used to detect the expression of DJ-1 in different tissues. β-actin and β-tubulin were used as housekeeping genes to confirm equal sample loading.
Off-target analysis
To examine whether these three pairs of TALENs have off-target effects, we explored two approaches to predict potential off-target sites in the porcine genome, namely E-PCR31,32 and TALENoffer33. For E-PCR (http://www.ncbi.nlm.nih.gov/tools/epcr/), we used the same criteria as Yong Lei and Xiaogang Guo (2012) described. Briefly, up to three mismatches and a 1-bp gap for each binding site was allowed, and the spacer between the two TALEN binding sites was <100 bp because longer spacers could interfere with FokI dimerization45. For TALENoffer, the default parameters were used to perform the predictions. The regions around potential off-target sites were amplified in mutant colonies and sequenced to confirm off-target cleavage.
Author Contributions
Z.Y. and J.Z. conceived and designed the study. J.Y., J.H., T.H., X.W., G.Q., H.Z. and R.W. performed the study. C.C. predicted the off-target sites. J.X. designed the TALEN plasmid pairs. J.Y. and J.Z. analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Full length gel of Fig. 6A
S1-S3
Acknowledgments
The authors thank all the members in Zhao's lab for the help to this project especially of Feng Wang and Xiangnan Miao for somatic nuclear transfer and embryo transfer, thank Fengfeng Zhuang for TALENs plasmid design and synthesis; thank Tong Zhao from Institute of Microbiology, Chinese Academy of Sciences for help on FACS sorting, thank Qi Zhou from Institute of Zoology, Chinese Academy of Sciences for help on conceiving and designing the study. This study was supported by the National High Technology Research and Development Program of China (2012AA020602), the National Basic Research Program of China (2011CBA0100, 2011CB944100) and the National Natural Science Foundation of China (31172281, 31272440).
References
- Prather R. S., Shen M. & Dai Y. Genetically modified pigs for medicine and agriculture. Biotechnol Genet Eng Rev 25, 245–265 (2008). [DOI] [PubMed] [Google Scholar]
- Carter D. B. et al. Phenotyping of transgenic cloned piglets. Cloning Stem Cells 4, 131–145 (2002). [DOI] [PubMed] [Google Scholar]
- Rogers C. S. et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321, 1837–1841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner B. et al. Transgenic pigs as models for translational biomedical research. J Mol Med (Berl) 88, 653–664 (2010). [DOI] [PubMed] [Google Scholar]
- Yang D. et al. Expression of Huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum Mol Genet 19, 3983–3994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson D. F. et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A 109, 17382–17387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J. K. & Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003). [DOI] [PubMed] [Google Scholar]
- Whyte J. J. et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev 78, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. et al. Generation of PPARgamma mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res 21, 979–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild J. et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci U S A 108, 12013–12017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D. et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27, 851–857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts A. M. et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325, 433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu T. I. et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther 16, 352–358 (2008). [DOI] [PubMed] [Google Scholar]
- Radecke S., Radecke F., Cathomen T. & Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol Ther 18, 743–753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson D. F., Fahrenkrug S. C. & Hackett P. B. Targeting DNA With Fingers and TALENs. Mol Ther Nucleic Acids 1, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D. et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29, 697–698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V. M. et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L. et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29, 695–696 (2011). [DOI] [PubMed] [Google Scholar]
- Thomas B. & Beal M. F. Molecular insights into Parkinson's disease. F1000 Med Rep 3, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V. et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259 (2003). [DOI] [PubMed] [Google Scholar]
- Goldberg M. S. et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem 278, 43628–43635 (2003). [DOI] [PubMed] [Google Scholar]
- Itier J. M. et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet 12, 2277–2291 (2003). [DOI] [PubMed] [Google Scholar]
- Von Coelln R. et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A 101, 10744–10749 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. Age-dependent motor deficits and dopaminergic dysfunction in DJ-1 null mice. J Biol Chem 280, 21418–21426 (2005). [DOI] [PubMed] [Google Scholar]
- Perez F. A. & Palmiter R. D. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci U S A 102, 2174–2179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro J. A. et al. Mortality, oxidative stress and tau accumulation during ageing in parkin null mice. J Neurochem 103, 98–114 (2007). [DOI] [PubMed] [Google Scholar]
- Andres-Mateos E. et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A 104, 14807–14812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux M. W. et al. Progressive dopaminergic cell loss with unilateral-to-bilateral progression in a genetic model of Parkinson disease. Proc Natl Acad Sci U S A 109, 15918–15923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L. et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295, 1089–1092 (2002). [DOI] [PubMed] [Google Scholar]
- Lei Y. et al. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci U S A 109, 17484–17489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J. et al. Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS One 8, e84250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau J., Boch J. & Posch S. TALENoffer: genome-wide TALEN off-target prediction. Bioinformatics 29, 2931–2932 (2013). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8, 753–755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. H. et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31, 23–24 (2013). [DOI] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A 110, 16526–16531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F. et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146, 318–331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Ortiz O., Hrabe de Angelis M., Wurst W. & Kuhn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc Natl Acad Sci U S A 109, 9354–9359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers B. et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci U S A 110, 3782–3787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. W. et al. Optimization of square-wave electroporation for transfection of porcine fetal fibroblasts. Transgenic Res 19, 611–620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz D., Ferrara N., Schweigerer L. & Neufeld G. Structural characterization and biological functions of fibroblast growth factor. Endocr Rev 8, 95–114 (1987). [DOI] [PubMed] [Google Scholar]
- Rifkin D. B. & Moscatelli D. Recent developments in the cell biology of basic fibroblast growth factor. J Cell Biol 109, 1–6 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143–148 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full length gel of Fig. 6A
S1-S3