Abstract
The Hedgehog pathway plays important roles in embryonic development, adult stem cell maintenance and tumorigenesis. In mammals these effects are mediated by Sonic, Desert and Indian Hedgehog (Shh, Dhh and Ihh). Shh undergoes autocatalytic cleavage and dual lipidation prior to secretion and forming a response gradient. Post-translational processing and secretion of Dhh and Ihh ligands has not previously been investigated. This study reports on the synthesis, processing, secretion and signaling activities of SHH, IHH and DHH preproteins expressed in cultured cells, providing unexpected evidence that DHH does not undergo substantial autoprocessing or secretion, and does not function in paracrine signaling. Rather, DHH functions as a juxtacrine signaling ligand to activate a cell contact-mediated HH signaling response, consistent with its localised signaling in vivo. Further, the LnCAP prostate cancer cell, when induced to express endogenous DHH and SHH, is active only in juxtacrine signaling. Domain swap studies reveal that the C-terminal domain of HH regulates its processing and secretion. These findings establish a new regulatory role for HHs in cell-mediated juxtacrine signaling in development and cancer.
Keywords: Development, Hedgehog, Signaling, Processing, Secretion
1. Introduction
Hedgehog (Hh) genes encode signaling proteins controlling tissue patterning in the embryo and cell growth and differentiation in regenerating tissues (Ingham et al., 2011; McMahon et al., 2003; Varjosalo and Taipale, 2008). Hedgehog was originally identified in a genetic screen for patterning genes in Drosophila melanogaster (Nusslein-Volhard and Wieschaus, 1980; Varjosalo and Taipale, 2008; Wetmore, 2003). While Drosophila has only a single Hh gene, mammals have three: Desert (DHH), Indian (IHH) and Sonic (SHH). While these isoforms are redundant in some scenarios (Zhang et al., 2001), they largely show differential expression in tissues in the developing embryo and adult, and control induction of different sets of downstream target genes (Hooper and Scott, 2005; Pathi et al., 2001). It is important to note that both Shh and Ihh are expressed in endoderm development (van den Brink, 2007). In the mouse embryo, Shh is expressed and secreted from midline tissues, including the node, notochord, and floor plate, generating a morphogen signaling gradient for patterning of left–right and dorso–ventral axes of the developing embryo (Hooper and Scott, 2005; Varjosalo and Taipale, 2008). By contrast, Ihh is primarily expressed by pre-hypertrophic and early hypotrophic chondrocytes, signaling locally to both proliferating chondrocytes and the overlying perichondrial cells (St-Jacques et al., 1999; Vortkamp et al., 1996). Dhh expression is produced by Sertoli cells to control development of immediately adjacent Leydig cells required for male sexual differentiation (Bitgood et al., 1996; Clark et al., 2000; Kawai et al., 2011), and by Schwann cells to promote the development of the perineural sheath of during neuronal development (Bitgood and McMahon, 1995; Parmantier et al., 1999; Yoshimura and Takeda, 2012).
Hhs are expressed as unprocessed preproproteins that, based on studies of the Drosophila Hh protein, undergo processing and auto-catalytic cleavage (reviewed in Ryan and Chiang (2012) and Ingham et al. (2011)). In brief, human SHH is synthesized as a 462aa protein precursor of approximately 45 kDa designated as ‘preproprotein’. This preproprotein is composed of a 23aa signal peptide ER targeting sequence, a 174 amino acid N-terminal signaling domain, and a 265aa C-terminal autoprocessing domain endowed with autoproteolysis and cholesterol transferase activity. Following cleavage of the signal peptide sequence at the extreme N-terminus, the 19 kDa N-terminal signaling fragment of Hh is autocatalytically cleaved from the C-terminus and a cholesterol moiety is added to the C-terminal end of the signaling fragment. A palmitate group is subsequently added at the N-terminus to produce a dual-lipidated molecule with high signaling capacity (Mann and Beachy, 2004). The processed N-terminal fragment would be retained in the cell by its cholesterol tail, except it is actively secreted by the synergistic actions of Disp and the secreted protein Scube2 (Burke et al., 1999; Tukachinsky et al., 2012). Secreted N-Hh forms a signaling gradient from the producing cells to responsive cells localized near or distant (Mimeault and Batra, 2010). Processed Hh accumulates in producing cells in Disp deficient mice and flies, and is able to activate the pathway in neighboring cells, but is not competent for long range signaling (Burke et al., 1999; Gallet et al., 2006). Studies of Drosophila Hh have identified specific residues in the C-terminal domain required for autoprocessing and lipid modifications (Hall et al., 1997; Lee et al., 1994; Porter et al., 1995), including a conserved cysteine, although prior to this study, cellular autoprocessing and secretion of the remaining mammalian Hh isoforms have not been investigated.
Mammalian Hh signaling, characterized largely in studies of recombinant N-terminal Shh, transduces target gene transcription and/or repression through control of activator and repressor forms of the Gli transcription factors. The Hh signaling response varies with the concentration and duration of the Hh signal. Hh signal transduction begins with the binding of Hh ligand with the transmembrane receptor, Patched (Ptch). Prior to Hh ligand binding, Ptch binds and inhibits the transmembrane protein, Smoothened (Smo), which is the positive transducer of the Hh pathway controlling the function of the Gli family of transcription factors. In the absence of Hh binding and repression of Smo, the Gli3 transcription factor is post-translationally processed to form a potent transcriptional repressor of Hh target genes. Hh ligand binding to Ptch relieves this inhibition of Smo, allowing Smo to activate a signaling cascade through the primary cilium, which results in activation of Gli2 and Gli3 to function as positive regulators of Hh target genes (Singla and Reiter, 2006). Among these primary target genes is Gli1, a most potent transcriptional activator that participates in this strong positive feedback loop for activation of target genes (Stecca and Ruiz, 2010). Other Gli-regulated Hh signaling components are involved in pathway repression, including Ptch1 and Hip (Hedgehog-interacting protein), establishing a negative feedback loop (Chuang and McMahon, 1999). The balance between pathway activation and repression in response to the concentration and duration of Hh signal determines the transcriptional program of the Shh-responding cell.
The N-terminal domains of the three mammalian isoforms are conserved and have similar binding affinities for the receptor Ptch (Pathi et al., 2001), suggesting their function in a common signal transduction pathway. Supporting their conserved functions, N-terminal recombinant Hh ligands activate expression of Gli1 and Ptch1 mRNA transcripts, as direct targets of Gli-mediated pathway activation. However, the N-terminal fragments of three mammalian Hh proteins have different potencies for Gli1 and Ptch1 activation, with N-SHH being most active and N-DHH being least active (Pathi et al., 2001). While recombinant N-terminal SHH, DHH, and IHH proteins are equipotent in their ability to induce Islet-1 expression in chick neural plate explants, the ligands are significantly different in their activities for induction of chondrocyte differentiation and nodal expression in the lateral plate mesoderm in the early chick embryo (Pathi et al., 2001). In support of these results, Echidna Hedghog (shhb) in zebrafish is around 3 times as effective at inducing slow muscle than Shh in zebrafish during development (Norris et al., 2000). These results suggest that the HH N-terminal signaling domains of the different isoforms have specific and individual functional capacities in cell and embryo functional assays.
In this study, we have investigated the processing, secretion and signaling functions of human SHH, DHH and IHH expressed as full length proteins. Current understanding of Hh function derives from genetic studies of Drosophila Hh and cell studies of recombinant N-terminal Hh proteins, primarily N-Shh, which lack the dual lipidation modifications and do not address roles for autoprocessing and secretion in the control of Hh functions. We have investigated the synthesis, processing, secretion and signaling activities of SHH, IHH and DHH preproteins expressed in cultured cells. We report unexpectedly that HH isoforms have remarkably different autoprocessing, secretion and signaling activities. Notably, expressed DHH, unlike SHH and IHH, does not undergo substantial autoprocessing or secretion and does not function in paracrine signaling. Rather, DHH functions in juxtacrine signaling to activate a cell contact-mediated Hh signaling response, consistent with its expression and localized signaling in vivo. We also show that the LnCAP prostate cancer cell, when induced to express endogenous DHH and SHH, is active only in juxtacrine signaling, and not paracrine signaling. Finally, domain swap studies reveal that the C-terminal domain of DHH regulates its processing and secretion, to promote its function as an autocrine signaling regulator. These findings challenge the assumption that HHs function only as secreted morphogens and establish a new regulatory role for HHs in cell-mediated juxtacrine signaling in development and cancer.
2. Results
2.1. Pathway activation activities of expressed full length (FL) Hh isoforms
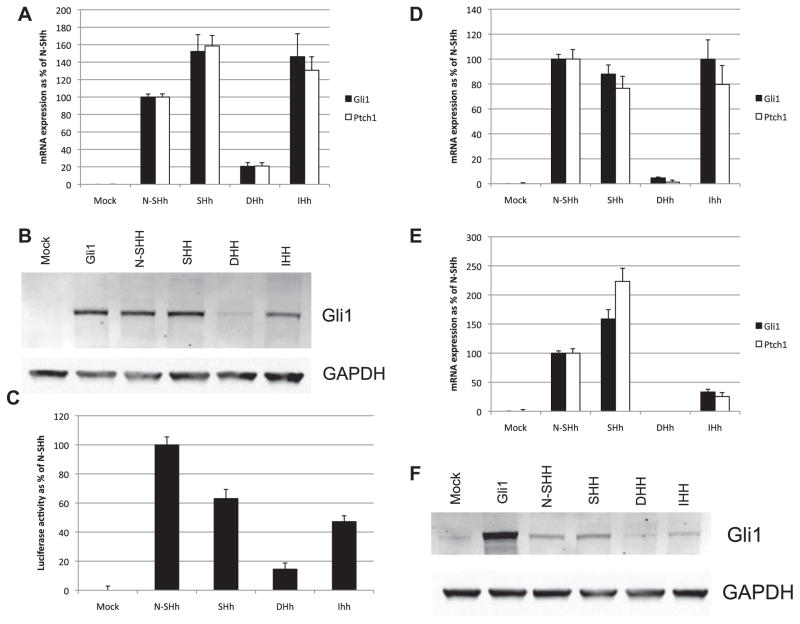
The Hh signaling activities of expressed human Full Length (FL) HH isoforms were assayed in 3T3 cells, an established Hh responsive cell line (Chen et al., 2009; Riobo et al., 2006b; Stanton et al., 2009). FL-cDNAs transfected into 3T3 cells were assayed for Hh pathway activation by induction of Gli1 and Ptch1 mRNA expression (Fig. 1A), by expression of Gli1 protein (Fig. 1B) and by activity of a stably integrated Gli-responsive luciferase reporter gene with an 8X Glibinding site (GBS), when expressed in Light2 cells (Fig. 1C) (Taipale et al., 2000). Mouse N-Shh cDNA transfections served as a positive control, and mock transfected cells as a negative control. Results show that FL-SHH and FL-IHH are strong activators of Hh signaling, with FL-SHH being more active than IHH. FL-DHH has much lower activity for Hh activation than either SHH or IHH in all assays. The activities of FL-SHH and FL-IHH are comparable to or greater that the activities of expressed N-Shh in Ptch1 and Gli1 activation, suggesting that these expressed FL-HH cDNAs are efficiently processed to their active ligand forms. Also, the protein generated from the N-Shh cDNA construct lacks post-translational lipid modifications (Lewis et al., 2001). The signaling activities of recombinant N-IHH and N-DHH are reported to be lower than N-Shh (Pathi et al., 2001), similar to our findings for FL-HH cDNAs. The signaling responses of FL-HH cDNA transfections correspond to the hierarchies of N-Hh signaling activities previously reported for recombinant N-HHs, i.e., SHH > IHH > DHH.
Fig. 1.
Direct transfection of FL-HH cDNA generates a Gli1 response. (A) 3T3 cells were transfected with full-length HH constructs and after 48 h culture in 0.5% FBS medium, mRNA for HH-responsive genes Gli1 and Ptch1 was quantified by qPCR. mRNA expression levels are shown as a % of positive control (N-Shh) expression. Protein levels of Gli1 were confirmed by Western blot (B) of lysates from transfected 3T3 cells. Quantified ratio of Gli1/GAPDH protein is shown below GAPDH for each lane. (C) Light2 cells transfected with full length HH constructs were assayed for luciferase activity after 48 h culture in 0.5% FBS medium. Luciferase activity is shown normalised to 100% of positive control (N-Shh). (D) CH310T1/2 cells and TM3 cells (E) were transfected with full length HH constructs and Gli1 and Ptch1 mRNA assayed by qPCR after 48 h culture in 0.5% FBS medium. (F) Gli1 protein induction in transfected TM3 cells was confirmed by Western blot on cell lysates following HH transfection and 48 h culture in 0.5% FBS medium.
We also tested the signaling activities of FL-HHs in two additional Hh responsive mouse cells lines: in C3H 10T1/2 cells, a multipotential cell line that can be induced to undergo osteogenesis in response to Hh signaling (Nakamura et al., 1997; Spinella-Jaegle et al., 2001), and TM3 cells, a testicular cell line derived from Leydig cells, which in vivo are responsive to Dhh produced by Sertoli cells (Bitgood et al., 1996; Clark et al., 2000; Kawai et al., 2011). FL-SHH induces Gli1 and Ptch1 RNA expression in C3H10T1/2 cells comparable to N-Shh, whereas IHH has significantly lower activity in TM3 cells compared to C3H10T1/2 cells (Fig. 1D and E). Significantly, FL-DHH produced a lowGli1 and Ptch1 induction response in C3H10T1/2 and no induction of Gli1 and Ptch1 mRNA or Gli1 protein in TM3 cells (Fig. 1D and E). The Gli1 mRNA induction results for TM3 cells post-transfection were confirmed at the protein level by Western blot assay (Fig. 1F). These findings show, in three HH responsive cell types, FL-SHH has high signaling activity, IHH has intermediate activity, and DHH has low activity, supporting the conclusion that signaling activity observed in these studies is specific to Hh isoforms.
2.2. Expressed FL-DHH is not processed or secreted from cells for paracrine signaling
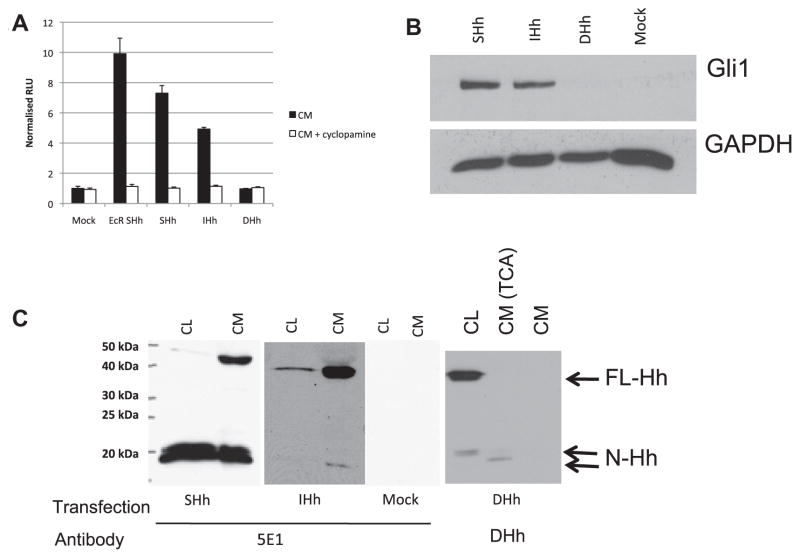
Studies of FL-HH processing and secretion were undertaken to understand the role of these regulatory processes in the control of signaling activities of the HH isoforms. Secretion of FL-HHs was investigated by harvesting conditioned media from 293T cells expressing FL-HHs and assaying cyclopamine-dependent HH signaling activities using Light2 cells with integrated 8X GBS luciferase reporter. All conditioned media results presented here were generated using 1:4 dilutions of harvested media in low serum media. Similar results were obtained with undiluted conditioned media. Cyclopamine inhibits Smoothened-dependent Hh signaling. Conditioned media from 293T cells stably transfected with an inducible N-Shh cDNA expression vector (EcR Shh) (Cooper et al., 1998) served as a positive control, promoting 10-fold induction of luciferase activity, and conditioned medium from mock transfected cells served as a negative control (Fig. 2A). Conditioned media prepared using the FL-SHH cDNA construct induced highest cyclopamine-sensitive reporter activation (7.3-fold), compared to FL-IHH conditioned medium (4.9-fold) and to conditioned media from FL-DHH, which failed to induce any reporter activation. An HH activation response was detected up to and including a 1:25 dilution of SHH conditioned media (data not shown). Gli1 protein induction by FL-SHH and FL-IHH conditioned media were robust (Fig. 2B), whereas DHH conditioned medium was inactive, confirming the results of the 8X GBS luciferase assay that FL-SHH and FL-IHH were secreted and functionally active in HH paracrine signaling, while FL-DHH was not.
Fig. 2.
Conditioned media from cells transfected with full length HH demonstrates isoform-specific autoprocessing and secretion. (A) Light2 cells were assayed for luciferase activity after 48 h treatment with conditioned media (1:4 in medium with 0.5% FBS, with and without cyclopamine as indicated) generated from 293T cells transfected with full length HH cDNA and cultured for 48 h in medium with 2% FBS. EcR Shh was used as a positive control for secreted HH. (B) Gli1 protein levels were determine by Western blot on 3T3 cells after 48 h treatment with conditioned media (1:4 in medium with 0.5% FBS). Quantified ratio of Gli1/GAPDH protein is shown below GAPDH for each lane. (C) HH production and secretion was determined by Western blot of cell lysates (CL) and conditioned media (CM) from 293T transfected cells for HH and cultured in medium with 2% FBS for 48 h, probed with 5E1 anti-HH or anti-DHH primary antibody as indicated. TCA denotes TCA precipitated CM.
HH protein processing was assayed by Western blot assays of expressing cells and conditioned media using HH-specific antibodies (Fig. 2C). Full length and processed N-terminal forms of SHH and IHH were detected on Western blots of cells and media using the polyclonal 5E1 antibody specific to the N-terminal domain of SHH and IHH proteins (DSHB) (Fig. 2C). DHH is not detected by the 5E1 antibody (Beachy et al., 2010) but could be detected with a DHH-specific antibody. 293T cells expressing FL-SHH cDNA were shown to contain trace amounts of ~45 kDa FL-SHH protein, and abundant amounts of a processed 19 kDa N-terminal SHH ligand (Fig. 2C). It is important to note that for all processed HH isoforms multiple bands are likely a result of post-cleavage lipid modifications. Conditioned medium harvested from 293T cells transfected with FL-SHH cDNA also have abundant amounts of processed 19 kDa N-terminal SHH protein, and also contains FL-SHH protein. FL-SHH undergoes efficient intracellular processing and secretion in transfected 293T cells, showing that FL-SHH is actively processed and secreted. By contrast, the FL-IHH protein is less efficiently processed, as abundant levels of FL-IHH are detected in both cell lysates and conditioned medium of expressing cells. Notably, FL-DHH expressing 293T cells express a predominance of unprocessed FL-DHH (Fig. 2C) and trace amounts of processed DHH were detectable in conditioned medium only after concentration of the proteins in conditioned media approximately 16-fold by TCA precipitation. The absence of FL and easily detectable processed DHH in conditioned medium shows that FL-DHH is neither processed nor secreted effectively, thus explaining the absence of DHH signaling activity in conditioned medium from FL-DHH expressing cells. The mechanism of FL-SHH and FL-IHH secretion is currently unclear, although based on these results FL-DHH does not appear to be comparably secreted via this mechanism.
2.3. DHH activates an HH signaling response via cell–cell mediated juxtacrine signaling
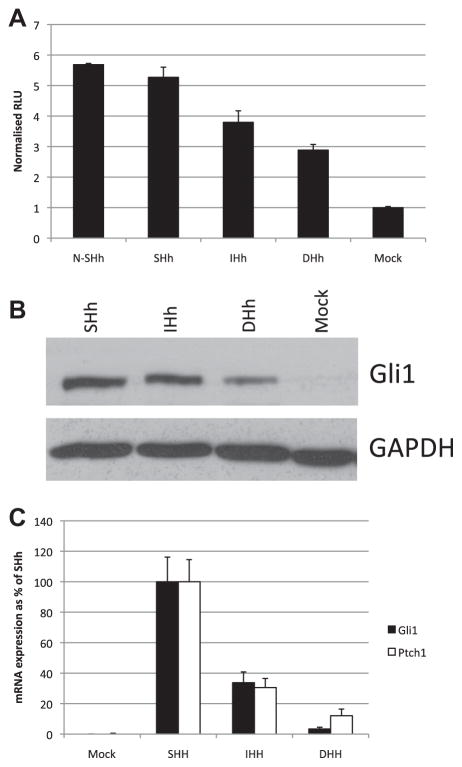
Our finding that DHH is not secreted, but accumulated intracellularly as an unprocessed protein led us to investigate whether cellular FL-DHH functions in cell–cell juxtacrine signaling, suggested by the proximity of Dhh expressing Sertoli and Schwann cells to their responding tissues in vivo. 293T cells expressing FL-HHcDNAs were co-plated at high density with in equal numbers with the HH-responsive Light2 cells to assay juxtacrine induction of 8X GBS luciferase reporter (Fig. 3A). 293T cells were used to generate the HH as they are non-responsive to HH signaling, thus any response in the assay comes fromLight2 cells. The results of these co-plating assays show that DHH expressing 293T cells induce luciferase activity well above control levels, establishing a role for DHH in juxtacrine signaling. FL-SHH and FL-IHH also induce luciferase activity in co-plating assays, which can at least in part be attributed to their processing and secretion for paracrine signaling (Figs. 1 and 2) in addition to any juxtacrine activity. Juxtacrine signaling activity of DHH was confirmed by Western blot assays of Gli1 protein induction in Light2 cells (Fig. 3B). The juxtacrine signaling ability of DHH was also confirmed in co-plating assays with TM3 cells (Fig. 3C), indicating that TM3 cells are responsive to DHH presented by neighbouring cells.
Fig. 3.
Sonic, Indian and Desert HH are able to activate a Gli response in co-plating cultures. (A) Luciferase assays were performed on Light2 cells co-plated with 293T cells transfected with full length HH constructs for 48 h in media containing 0.5% FBS. Luciferase activity is shown relative to mock transfected control. (B) Gli1 protein levels were determined by Western blot of cell lysates from co-plating cultures. (C) qPCR for Gli1 and Ptch1 mRNA was performed on RNA extracted from TM3 cells co-plated for 48 h with 293T cells transfected with full length HH constructs in medium with 0.5% FBS. mRNA expression levels are shown as a % of expression in SHH transfected co-culture.
2.4. The DHH and IHH C-terminal domains control processing for juxtacrine signaling
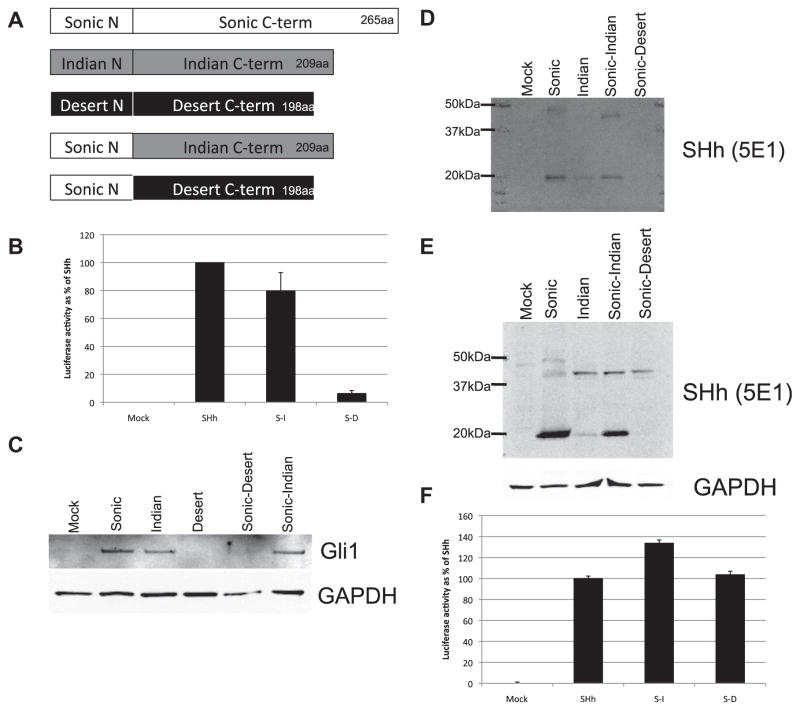
To investigate the mechanisms underlying processing regulation and juxtacrine signaling of DHH, expression constructs of SHH N-terminal domain fused with the C-terminal domains of either DHH or IHH were generated (Fig. 4A). These fusion proteins were expressed in 293T cells to investigate their processing and secretion. Conditioned medium from FL-SHH-DHH fusion protein expressing cells mimicked the inactivity of DHH by not activating HH-Gli signaling as assayed by luciferase assays in Light2 cells above 6.3% of SHH levels, (Fig. 4B) or by Western blot assays of Gli1 protein induction (Fig. 4C). By contrast, conditioned medium produced by FL-SHH-IHH fusion protein was active in HH-Gli signaling in Light 2 cells as well as in the activation of Gli1 expression (Fig. 4B and C). Western blot analyses of conditioned media and cell lysate from transfected 293T cells also revealed that the FL-SHH-DHH fusion protein was not present in conditioned medium (Fig. 4D), but was expressed and remained associated with cells (Fig. 4E), similar to the behavior of FL-DHH protein. By contrast, FL-SHH-IHH fusion protein was partially processed and released into conditioned medium as processed and FL-HH, similar to the behavior of FL-IHH. In co-plating assays, SHH-DHH fusion protein was highly active in juxtacrine signaling (Fig. 4F). Juxtacrine signaling activity of these fusion proteins was significant greater than for the FL-DHH and FL-IHH proteins, likely reflecting the enhanced activity of the N-terminal SHH signaling domain, which has the strongest signaling activity of the three N-terminal HH in expression assays (Pathi et al., 2001). These findings, therefore, establish that C-terminal domains of DHH and IHH control the processing, and in the case of DHH also the juxtacrine signaling functions of these HH proteins.
Fig. 4.
HH fusion constructs demonstrate the HH C-terminus defines auto-processing and secretion events. (A) Schematic representation of fusion cDNA constructs. (B) Luciferase assays were performed on Light2 cells treated for 48 h with conditioned media from 293T cells transfected with full length SHH and HH fusion constructs (1:4 in media with 0.5% FBS). Luciferase activity is shown as a % of activity for full length SHH. (C) Gli1 protein levels were determined by Western blot of cell lysates from 3T3 cells treated with conditioned media from 293T cells transfected with HH and HH fusion constructs (1:4 in media with 0.5% FBS). (D) The presence of secreted HH was determined Western blot of conditioned media from transfected 293T cells cultured for 48 h in media with 2% FBS. (E) HH proteins were detected by Western blot using the 5E1 anti-HH antibody on cell lysates from transfected 293T cells cultured for 48 h in media with 2% FBS. (F) Luciferase assays were performed on Light2 cells co-plated for 48 h in media with 0.5% FBS with transfected 293T cells. Luciferase activity is shown as a % of activity for 293T cells transfected with full length SHH.
2.5. HHs expressed by LnCAP cancer cells engage exclusively in juxtacine signaling
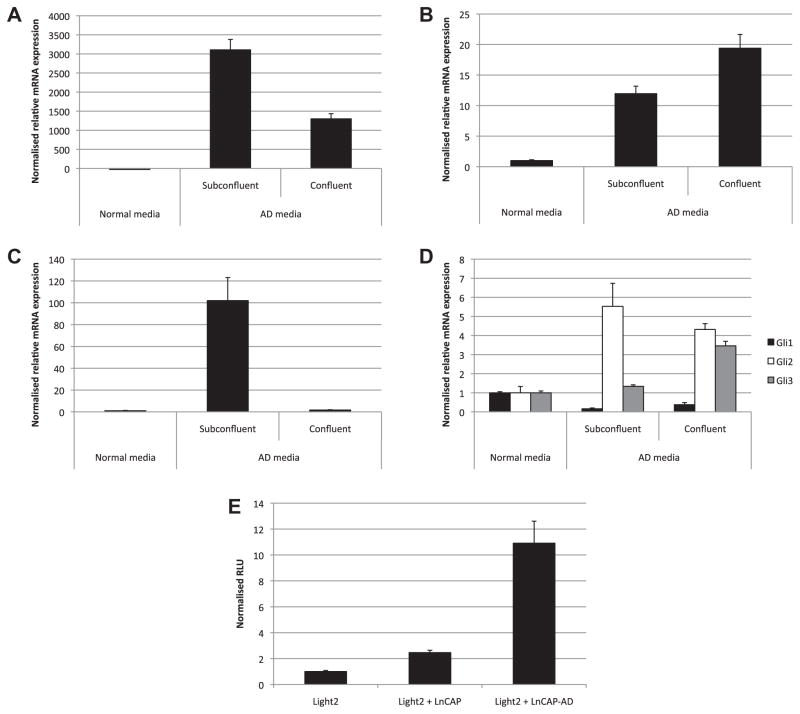
To investigate the juxtacrine and paracrine signaling activities of cells expressing endogenous Hhs we utilized LnCAP human prostate cancer cells, which are reported to express HHs in response to androgen deprivation (Chen et al., 2009). Despite HH production, LnCAP cells are not responsive to HH signaling, as they show no response to cyclopamine or exogenous SHH (Zhang et al., 2007). Expression of the HH isoforms was investigated in LnCAP cells in response to androgen deprivation (AD), under sub-confluent and confluent culture conditions. Expression of mRNA for each of the HH isoforms was activated in LnCAP cells under high and low density conditions in AD medium, with the exception of IHH, which was induced only under low-density culture conditions (Fig. 5A–C). SHH mRNA was very abundantly expressed under all culture conditions. Despite robust activation of HH mRNA expression, Gli1 expression was not induced, showing that cells were not responding to HHs in autocrine or paracrine signaling (Fig. 5D). Interestingly, Gli2 and Gli3 mRNAs were induced by AD conditions, but did not contribute to activation of HH signaling as evidenced by an absence of Gli1 induction (Fig 5D). Significantly, however, coplating assays with Light2 luciferase reporter cells showed that LnCAP cells were active in juxtacrine HH signaling activity, particularly under conditions of AD and high cell density when LnCAP cells express abundant amounts of DHH. High density LnCAP cells cultured in AD conditions activated the 8x GBS luciferase response 10.9-fold over control cells (Fig. 5E). By contrast, conditioned medium from LnCAP cells did not have HH signaling activity, providing evidence that LnCAP cells endogenously expressing HHs function exclusively in the juxtacrine signaling.
Fig. 5.
Prostate cancer cell lines express variable amounts of HH ligand capable of inducing a juxtacrine signaling response in coplating cultures. (A) RNA obtained from LnCAP prostate cancer cells grown in normal growth conditions, and also for 2 weeks in androgen deprived (AD) conditions to confluence or to sub-confluent levels was used in qPCR to determine endogenous SHH, DHH (B) and IHH (C) mRNA levels, as well as Gli1, Gli2 and Gli3 mRNA expression (D). (E) Luciferase assays were performed on Light2 cells co-plated for 48 h in androgen deprived media with LnCAP cells previously grown for 2 weeks in AD media. Luciferase activity is shown relative to Light2 cells plated alone.
3. Discussion
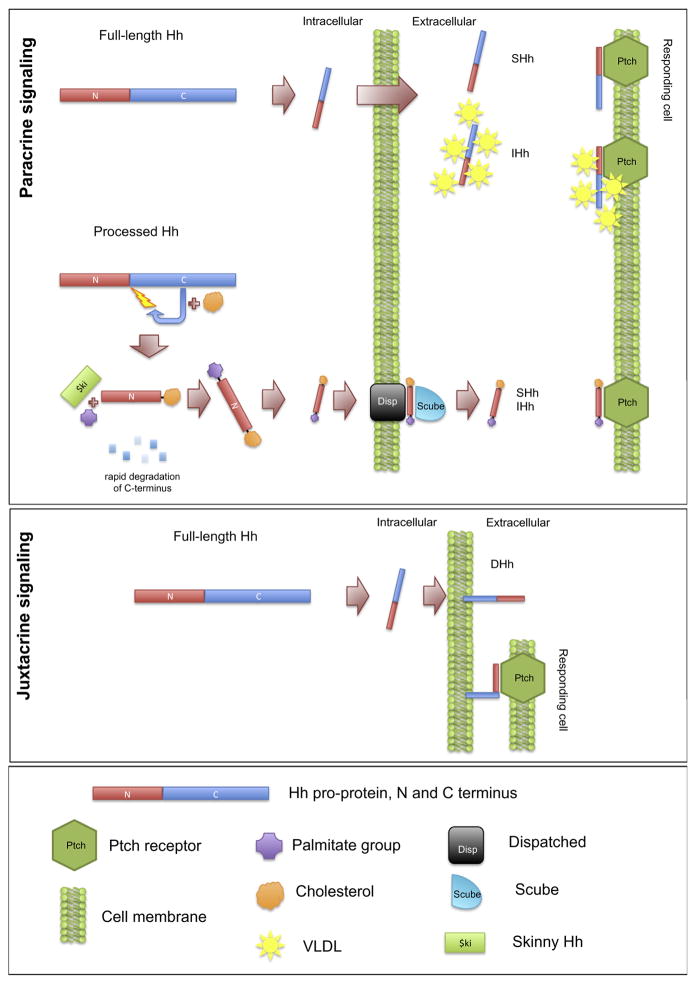
Our findings provide the first evidence that HH isoforms are differentially processed and secreted to enable their dual functions in juxtacrine and paracrine signaling. Shh has well known functions as a secreted ligand for long distance signaling as a developmental morphogen. Our findings (summarised in Fig. 6) show that FL-SHH is efficiently autoprocessed and its N-terminal domain secreted for paracrine signaling (Fig. 2). Additionally, we observed secretion of full-length SHH. By contrast, FL-DHH undergoes little or no autoprocessing, secretion or paracrine signaling, but rather functions in cell contact-mediated juxtacrine signaling (Figs. 2 and 3), consistent with its developmental roles in testis (Bitgood et al., 1996; Clark et al., 2000; Kawai et al., 2011) and neurogenesis (Bitgood and McMahon, 1995; Parmantier et al., 1999; Yoshimura and Takeda, 2012). FL-IHH has intermediate levels of processing and secretion and likely functions in both juxtacrine and paracrine signaling, consistent with its dual functions in localized and long distance signaling for bone repair and chondrogenesis (Joeng and Long, 2009; Karp et al., 2000; Long et al., 2004; St-Jacques et al., 1999; Vortkamp et al., 1996).
Fig. 6.
Schematic models of the three alternative modes of HH processing, secretion and presentation. Paracrine signaling by processed HH is representative of what is known for mammalian SHH. Full length HH secretion has been observed for both IHH (also in association with vLDL) and here for SHH. Juxtacrine full length HH signaling has been observed here for DHH but may also be utilised by SHH and IHH.
The secretion of fully processed and dual-lipid modified Shh has been studied comprehensively (Tukachinsky et al., 2012). This secretion is reliant on both Disp and Scube, and is mediated through both proteins binding the cholesterol tail of N-Shh, thereby overcoming hydrophobic membrane retention of the ligand. Their study raises a number of key points relevant to our study. N-Shh that lacks cholesterol (i.e. generated from truncated cDNA) is secreted in a Disp-independent manner. The membrane retention of processed Hh ligand, which is overcome by Disp and Scube, is mediated by the cholesterol moiety added during the autoprocessing event, and as such does not apply to any full length HH protein secretion or membrane retention observed in our study.
The use of cell lines in our study to overexpress HH ligands may not accurately represent the HH processing and secretion situation in vivo, however the generation of fully processed and secreted SHH ligand suggests all requisite components of the pathway are present in cells used in our experiments. In vivo evidence for secretion and function of FL-IHH has been obtained by mass spectrometry analysis of plasma lipoprotein fractions of human blood. These studies identify IHH, including its C-terminal sequences, in the VLDL fraction (very low density lipoprotein), but not in other plasma lipoprotein fractions (LDL and HDL). Neither SHH nor DHH were detected in any of these three fractions of plasma lipoproteins (Queiroz et al., 2010). IHH associated with the VLDL lipids also generates a HH signaling response, suggesting that the FL-IHH:VLDL complex functions as a circulating morphogen for endothelial cell maintenance and repair. Secreted Drosophila Hh is also associated with a lipid carrier protein required for its extracellular function (Chen et al., 2004; Lin, 2004; Panakova et al., 2005; The et al., 1999). It is notable that primarily full length IHH is secreted from expressing cells (in contrast to DHH and SHH; Fig. 2) suggesting that secretion and lipid interactions of FL-IHH are functionally adaptive for long distance signaling. Processed N-Shh diffusion in tooth development has also been shown to be reliant on proteoglycan/glycosamino-glycan (PG/GAG) complexes (Gritli-Linde et al., 2001). In fact, visualisation of both processed N-Shh and N-Ihh in target tissues was only possible with staining protocols that preserved GAG chains, suggesting that complex formation with GAGs is required for appropriate diffusion and gradient formation of the processed forms of Shh and Ihh.
The cellular mechanisms for trafficking and presentation of FL-DHH to Ptch1 receptors on apposing cells require further investigation. Studies of Shh signaling show that N-Shh, when tethered to the cell membrane by a glycophospholipid (GPI) linkage, is internalized by adjacent Ptch1 expressing cells (Incardona et al., 2000), and unable to form its diffused signaling gradient. Furthermore, mutations introduced into SHH that block autoprocessing enable mutant SHHs to function in cell contact-mediated juxtacrine signaling but not in paracrine signaling, although it is unknown whether such mutant SHHs are secreted in an inactive form (Tokhunts et al., 2010). In Drosophila embryos expressing an autoprocessing deficient mutant form of human SHH (H329A), pathway activation was observed at around 1/3 of the potency of wild type (Lee et al., 1994).
Hh autoprocessing is mechanistically very similar to the first steps of protein splicing, which is reliant on specific protein sequences located in inteins. In Base is an online Blast server (Perler, 2002) that searches for homology to known inteins. Using standard settings SHH yielded 32 hits against known inteins, IHH 36 and Drosophila Hh 36, while DHH was only able to generate 11 hits. This loss of homology to known inteins distinguishes the DHH sequence, despite its high overall homology to other processing competent HHs, including Drosophila Hh (Varjosalo and Taipale, 2008). This suggests that only a small number of key sequence changes are responsible for the loss of autoprocessing in DHH. The C-termini domains of HH isoforms are less conserved than the N-termini, yet little work has been done to show how this affects autoprocessing. Tokhunts et al. analysed a panel of reported holoprosencephaly associated SHH mutations for alterations in HH ligand processing (Tokhunts et al., 2010). Three mutants, D222N, T267I and A373T, displayed significantly different processing efficiency than the wt SHH, clearly indicating a single amino acid change can dramatically alter HH autoprocessing. Two additional reported holoprosencephaly mutations in the SHH C-terminus (S236R and G290D) (Cohen, 2010) were not tested in this experiment. These two residues are of interest as they are not conserved in DHH, and G290D resides in a stretch of 13 amino acids not conserved in DHH or IHH. The functional consequences of non-conserved sequence changes in the Hedgehog C-termini need to be determined through autoprocessing experiments.
Our findings show that unprocessed FL-DHH is not secreted, but is retained in cells by a mechanism mediated by its C-terminal domain (illustrated by our fusion protein studies). One possible mechanism for DHH retention is through a transmembrane domain in the DHH C-terminus. Various algorithms designed to predict transmembrane domains from primary HH protein sequences have yielded divergent results, indicative of the tenuous nature of these predictions. However, DHH does have a higher proportion of hydrophobic amino acids than SHH or IHH in the C-terminus, and therefore an increased likelihood of a transmembrane domain. An alternative retention and presentation mechanism for DHH could involve GPI linkage on the cell membrane. Experiments with a modified Shh N-terminal cDNA have shown that GPI-linkage can retain the ligand on the generating cell surface, where it is able to bind Ptch on neighboring cells and be internalized to initiate signaling (Incardona et al., 2000). Although it must be noted that three independent GPI-anchor prediction programs (PredGPI, (Pierleoni et al., 2008); GPI-SOM (Fankhauser and Maser, 2005); Big-PI (Eisenhaber et al., 1999)) did not find the required signal motifs in the DHH protein sequence.
Studies of HH expression and function in LnCAP prostate cancer cell line further affirm the important role of HH juxtacrine signaling. LnCAP cells express endogenous HH RNAs in response to androgen deprivation, including DHH. Notably, LnCAP cells induced to express HHs do not secrete HHs with paracrine activity, but are active in juxtarine signaling, affirming the importance of juxtacrine signaling controlled by endogenously expressed HHs and highlighting the importance of juxtacrine signaling in HH cancers. A solid tumor mass generating HH protein could stimulate HH pathway activation in adjacent stroma by a juxtacrine signaling mechanism, thereby promoting tumor growth, as has been proposed for HH cancers. These findings have significant implications for development of anti-tumor therapeutics. For instance, anti-tumor drugs targeting the autocatalytic cleavage of FL-HHs are not likely to be successful. In addition, therapies blocking diffusion of HH ligands will not block juxtacrine signaling for pathway activation within tumors. It is also notable that LnCAP cancer cells do not respond to paracrine induction of induction of the HH pathway, as monitored by Gli1 expression, indicating that LnCAP cells do not respond to HH paracrine or autocrine signaling, making them resistant to drugs that disrupt Ptch function in paracrine signal transduction. Development of useful therapeutics for many HH cancers will likely require a deeper understanding of the underlying mechanisms of HH juxtacrine signaling.
The DHH protein and its receptive responding cells may be specifically adapted for juxtacrine signaling. Notably, Dhh functions in vivo are restricted to processes involving highly localized signaling, including expression in Sertoli cells to signal immediately proximal Leydig cells in the testis (Bitgood et al., 1996; Clark et al., 2000; Kawai et al., 2011) and in neural development through its expression in Schwann cells to induce the perinuclear sheath (Bitgood and McMahon, 1995; Parmantier et al., 1999; Yoshimura and Takeda, 2012). It will be of interest in future studies to investigate the possibility of specialized cell surface/receptor mechanisms for DHH signaling as a model for investigating HH juxtacrine signaling mechanisms during development.
4. Materials and methods
4.1. Materials
p8XGBS-luc and full length mGli1 constructs were provided by H. Sasaki (Riken Center for Developmental Biology, Kobe, Japan) (Riobo et al., 2006a,b). pRL-TK was obtained from Promega. Full-length cDNA expression constructs for human SHH, IHH, DHH in pCMV-XL5 were purchased from Origene. pcDNA3. 1-N-Shh was as reported previously (Riobo et al., 2006a,b). Fusion construct cDNA was generated using the above full length cDNA plasmids as templates using a two-step overlap extension high-fidelity PCR protocol, utilising Phusion Taq (Finnzymes). Rabbit anti-Gli1 and anti-rabbit IgG-HRP were obtained from Cell Signaling Technology. Mouse anti-Gapdh was from AbCam. Anti-DHH N-19 and anti-Mouse IgG-HRP were purchased from Santa Cruz Biotechnology. The anti-SHH (5E1) developed by Thomas M. Jessell and Susan Brenner-Morton was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. Anti-goat IgG-HRP was purchased from Jackson Immunolaboratories. Anti-goat IgG IRDye 700DX (Rockland), anti-rabbit IgG Alexa-Fluor 680 (Molecular Probes) and anti-mouse IgG IRDye 800CW (Li-Cor) were also used for imaging blots with the Odyssey imaging system (Li-Cor). Cell lines were obtained from ATCC unless otherwise stated. Chemicals were obtained from Sigma–Aldrich unless stated.
4.2. Cell lines
293T (CRL-11268), 293 EcR Shh (CRL-2782) and NIH3T3 (CRL-1658) cells were maintained in DMEM (Cellgro) with 10% fetal bovine serum (FBS; Thermo Scientific). Light2 cells (CRL-2795; obtained from P. Beachy, Johns Hopkins University, Baltimore, MD) were maintained in the samemedium supplemented with 0.15 mg/mL Zeocin (Invitrogen) and 0.4 mg/mL Geneticin (Gibco). C3H/10T1/2 (CCL-226) cells were maintained in DMEM with 10% fetal bovine serum and 10 mM HEPES (Cellgro). TM3 cells (CRL-1714) were grown in DMEM/F12 (50:50; Cellgro) with 5% equine serum (Thermo Scientific), 2.5% fetal bovine serum, 0.5 mM sodium pyruvate (Gibco), 1.2 g/L sodium bicarbonate (Cellgro) and 2.5 mM L-glutamine (Gibco). LNCaP cells (CRL-1740) were grown in RPMI1640 (Gibco) supplemented with 10% fetal bovine serum. Androgen deprivation media was RPMI1640 supplemented with 2% charcoal stripped FBS (Gibco). All media contained antibiotic/antimycotic (Cellgro), except during transfection protocol as stated, and all cells were grown at 37 °C in a humidified 5% CO2 atmosphere. NIH3T3 and Light2 cells were maintained at less than 70% confluence except for where stated. Cyclopamine (LC Laboratories) treatments were used as a pathway specific inhibitor at 10 μM concentration.
4.3. Transfections, conditioned media and co-plating assays
Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturers instructions. Transfection efficiency and expression of each HH cDNA construct was determined by qPCR on RNA from3T3 and TM3 cell lines. Due to the comparable expression and high degree of similarity between constructs (plasmid backbone, insert size and homology) for all subsequent analysis transfection efficiency and exogenous HH expression levels were deemed to be identical. For luciferase assays, transfection mixes were prepared in each well of a 96 well plate. 2 × 104 cells were added to each well and incubated overnight before media was changed to serum starve medium (DMEM with 0.5% fetal bovine serum with antibiotic/antimycotic) for 48 h. Transfections for RNA and protein extractions were performed in 12 well plates.
For generating HH-conditioned media, 293T cells were plated in a 24 well plate 6 h after transfection the medium was changed to a modified growth medium (2% FBS) and incubated for 48 h. HH-conditioned media was harvested by centrifugation for 20 min at 13,000g at 4 °C, then filtered through 0.2 μm cellulose acetate filter. Conditioned media was diluted 1:4 in 0.5% FBS media for addition to HH responsive cells. For HH-conditioned media derived from 293 EcR Shh cells, cells were grown to 70% confluence whereupon the media was changed to media containing 2% FBS. Shh expression was induced by addition of 1 μM MuristeroneA (Invitrogen). For HH-ligand detection by immunoblotting, conditioned media was centrifuged at 210,000g for 45 min to remove proteins bound to membranous remnants. Co-plating assays were performed using 293T cells transfected as for conditioned media, and were incubated for 24 h before trypinizing and counting, and plating 1:1 with Hh-responsive cells (Light2, 3T3 or TM3 cells). Cells were co-incubated for 48 h before performing harvesting for analysis.
4.4. Luciferase assays
Luciferase assays were performed using the Dual-Glo Luciferase Assay System (Promega) with a modified protocol. Culture media was removed from cells, and the plates frozen at −80 °C for 1 h, then incubated at room temperature for 10 min, 37 °C for 15 min, then room temperature for 10 min. 40 μL of DualGlo Luciferase reagent was added to each well and incubated in the dark for 10 min prior to luminescence reading on a Tecan Safire II plate reader. After the reading was taken, 40 μL of Stop&Glo reagent was added to each well. Plates were then incubated a further 10min prior to a second luminescence reading.
4.5. Quantitative real-time PCR (qPCR)
RNA extractions were performed using TRIzol reagent (Ambion) according to the manufacturer’s instructions. All samples were treated with DNAse1 (Invitrogen) prior to reverse transcription using Superscript III (Invitrogen) primed with random hexamers (Invitrogen) according to the manufacturer’s instructions. qPCRs were performed using Quantitect SYBR Green PCR kit (Qiagen) with primers listed in Table 1. Each RNA sample was analyzed in triplicate, and gene expression relative to control was obtained by the equation 2^(Ct-control)/2^(Ct-Gene). Expression was then normalized to control samples.
Table 1.
qPCR Primer Sequences.
| Genea | Accession number | Sequence | References | |
|---|---|---|---|---|
| m18S rRNA | NR_003278.2 | For | CGGTTCTATTTTGTTGGTTTTCG | De Bortoli et al. (2007) |
| Rev | GCTCTGGTCCGTCTTGCG | |||
| mMyf5 | NM_008656.5 | For | TATGAAGGCTCCTGTATCCC | Han et al. (2011) |
| Rev | ACGTGCTCCTCATCGTCTG | |||
| mPtc1 | NM_008957.2 | For | TCAGTTGACTAAACAGCGTCTGGTA | Guo et al. (2010) |
| Rev | GACCCAAGCGGTCAGGTAGAT | |||
| mGli1 | NM_010296.2 | For | GGCTGTCGGAAGTCCTATTCAC | Guo et al. (2010) |
| Rev | CAACCTTCTTGCTCACACATGTAAG | |||
| hSHH | NM_000193.2 | For | GGTATGCTCGGGACTGGCG | Azoulay et al. (2008) |
| Rev | CAGCCTGTCCGCTCCGGTGT | |||
| hDHH | NM_021044.2 | For | GGAGCCGACCGCCTGATGAC | – |
| Rev | CGTGGACGTGGTTGCGGGAC | |||
| hIHH | NM_002181.3 | For | CCTGAACTCGCTGGCTATCT | Azoulay et al. (2008) |
| Rev | CCACTCTCCAGGCGTACCT | |||
| hGli1 | NM_005269.2 | For | AATGCTGCCATGGATGCTAGA | Zhang et al. (2007) |
| Rev | GAGTATCAGTAGGTGGGAAGTCCATAT | |||
| hGli2 | NM_005270.4 | For | AGCCAGGAGGGCTACCAC | Zhang et al. (2007) |
| Rev | CTAGGCCAAAGCCTGCTGTA | |||
| hGli3 | NM_000168.5 | For | ATCATTCAGAACCTTTCCCATAGC | Zhang et al. (2007) |
| Rev | TAGGGAGGTCAGCAAAGAACTCAT | |||
| hGapdh | NM_002046.3 | For | CCACATCGCTCAGACACCAT | Zhang et al. (2007) |
| Rev | GCAACAATATCCACTTTACCAGAGTTAA |
Prefixes: h denotes human, m denotes mouse.
4.6. Immunoblotting
Cells were lysed in lysis buffer [50 mM Tris HCl pH 7.4, 300 mM NaCl, 2% NP-40, 0.25% sodium deoxycholate, complete protease inhibitors (Roche)] for 30 min on ice. Protein lysate was cleared by centrifugation at 13,000g for 15 min at 4 °C. Protein concentration was determined by BCA (Pierce) or Bradford reagent (BioRad) prior to separation by SDS–PAGE and transfer to nitrocellulose membranes [iBlot Invitrogen]. Membranes were blocked in either 5% milk or BSA in TBS +0.1% Tween20 for 1 h at room temperature. Primary antibodies were incubated in block over night at 4 °C. Secondary HRP- or fluorescent antibodies were incubated 1 h at room temp in block and detected by chemiluminescence (Amersham ECL Western Blotting Detection) or using the Odyssey Imaging System (LiCor). Quantification of Western blotting was done using ImageJ (Schneider et al., 2012).
Acknowledgments
This work is supported by NIH R01 HD007796.
Footnotes
Author contributions
CP and EA designed and performed experiments and analysis. CE contributed to the concept and design of the study. CP wrote the manuscript with input from EA and CE.
Conflict of interest statement
The authors declare they have no conflict of interest.
References
- Azoulay S, Terry S, Chimingqi M, Sirab N, Faucon H, Gil Diez de Medina S, Moutereau S, Maille P, Soyeux P, Abbou C, Salomon L, Vacherot F, de La Taille A, Loric S, Allory Y. Comparative expression of Hedgehog ligands at different stages of prostate carcinoma progression. J Pathol. 2008;216:460–470. doi: 10.1002/path.2427. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24:2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr Biol. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Chen M, Tanner M, Levine AC, Levina E, Ohouo P, Buttyan R. Androgenic regulation of hedgehog signaling pathway components in prostate cancer cells. Cell Cycle. 2009;8:149–157. doi: 10.4161/cc.8.1.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Clark AM, Garland KK, Russell LD. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol Reprod. 2000;63:1825–1838. doi: 10.1095/biolreprod63.6.1825. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Hedgehog signaling update. Am J Med Genet A. 2010;152A:1875–1914. doi: 10.1002/ajmg.a.32909. [DOI] [PubMed] [Google Scholar]
- Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- De Bortoli M, Castellino RC, Skapura DG, Shen JJ, Su JM, Russell HV, Hicks MJ, Man TK, Kim JY. Patched haploinsufficient mouse rhabdomyosarcoma overexpress secreted phosphoprotein 1 and matrix metalloproteinases. Eur J Cancer. 2007;43:1308–1317. doi: 10.1016/j.ejca.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- Fankhauser N, Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- Gallet A, Ruel L, Staccini-Lavenant L, Therond PP. Cholesterol modification is necessary for controlled planar long-range activity of Hedgehog in Drosophila epithelia. Development. 2006;133:407–418. doi: 10.1242/dev.02212. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Lewis P, McMahon AP, Linde A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev Biol. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- Guo S, Zhou J, Gao B, Hu J, Wang H, Meng J, Zhao X, Ma G, Lin C, Xiao Y, Tang W, Zhu X, Cheah KS, Feng G, Chan D, He L. Missense mutations in IHH impair Indian Hedgehog signaling in C3H10T1/2 cells: implications for brachydactyly type A1, and new targets for Hedgehog signaling. Cell Mol Biol Lett. 2010;15:153–176. doi: 10.2478/s11658-009-0040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- Han XH, Jin YR, Seto M, Yoon JK. A WNT/beta-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem. 2011;286:10649–10659. doi: 10.1074/jbc.M110.169391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc Natl Acad Sci USA. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Joeng KS, Long F. The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development. 2009;136:4177–4185. doi: 10.1242/dev.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp SJ, Schipani E, St-Jacques B, Hunzelman J, Kronenberg H, McMahon AP. Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development. 2000;127:543–548. doi: 10.1242/dev.127.3.543. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Noguchi J, Akiyama K, Takeno Y, Fujiwara Y, Kajita S, Tsuji T, Kikuchi K, Kaneko H, Kunieda T. A missense mutation of the Dhh gene is associated with male pseudohermaphroditic rats showing impaired Leydig cell development. Reproduction. 2011;141:217–225. doi: 10.1530/REP-10-0006. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, McMahon AP. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Mimeault M, Batra SK. Frequent deregulations in the hedgehog signaling network and cross-talks with the epidermal growth factor receptor pathway involved in cancer progression and targeted therapies. Pharmacol Rev. 2010;62:497–524. doi: 10.1124/pr.109.002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Aikawa T, Iwamoto-Enomoto M, Iwamoto M, Higuchi Y, Pacifici M, Kinto N, Yamaguchi A, Noji S, Kurisu K, Matsuya T. Induction of osteogenic differentiation by hedgehog proteins. Biochem Biophys Res Commun. 1997;237:465–469. doi: 10.1006/bbrc.1997.7156. [DOI] [PubMed] [Google Scholar]
- Norris W, Neyt C, Ingham PW, Currie PD. Slow muscle induction by Hedgehog signalling in vitro. J Cell Sci. 2000;113 (Pt. 15):2695–2703. doi: 10.1242/jcs.113.15.2695. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, Chakrabarti L, McMahon AP, Jessen KR, Mirsky R. Schwann cell-derived Desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–724. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP. Comparative biological responses to human Sonic, Indian. Mech Dev. 2001;106:107–117. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- Perler FB. InBase: the intein database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- Queiroz KC, Tio RA, Zeebregts CJ, Bijlsma MF, Zijlstra F, Badlou B, de Vries M, Ferreira CV, Spek CA, Peppelenbosch MP, Rezaee F. Human plasma very low density lipoprotein carries Indian hedgehog. J Proteome Res. 2010;9:6052–6059. doi: 10.1021/pr100403q. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Haines GM, Emerson CP., Jr Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006a;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2006b;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. J Biol Chem. 2012;287 (22):17905–17913. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Spinella-Jaegle S, Rawadi G, Kawai S, Gallea S, Faucheu C, Mollat P, Courtois B, Bergaud B, Ramez V, Blanchet AM, Adelmant G, Baron R, Roman-Roman S. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J Cell Sci. 2001;114:2085–2094. doi: 10.1242/jcs.114.11.2085. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, Taveras KM, Hyman JM, Lee SW, Koehler AN, Chen JK, Fox JL, Mandinova A, Schreiber SL. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol. 2009;5:154–156. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- Tokhunts R, Singh S, Chu T, D’Angelo G, Baubet V, Goetz JA, Huang Z, Yuan Z, Ascano M, Zavros Y, Therond PP, Kunes S, Dahmane N, Robbins DJ. The full-length unprocessed hedgehog protein is an active signaling molecule. J Biol Chem. 2010;285:2562–2568. doi: 10.1074/jbc.M109.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012;2:308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87 (4):1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wetmore C. Sonic hedgehog in normal and neoplastic proliferation: insight gained from human tumors and animal models. Curr Opin Genet Dev. 2003;13:34–42. doi: 10.1016/s0959-437x(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Takeda S. Hedgehog signaling regulates myelination in the peripheral nervous system through primary cilia. Differentiation. 2012;83:S78–S85. doi: 10.1016/j.diff.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lipinski R, Shaw A, Gipp J, Bushman W. Lack of demonstrable autocrine hedgehog signaling in human prostate cancer cell lines. J Urol. 2007;177:1179–1185. doi: 10.1016/j.juro.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;105 (6):781–792. [PubMed] [Google Scholar]