Abstract
SAGA is a transcriptional coactivator complex that is conserved across eukaryotes and performs multiple functions during transcriptional activation and elongation. One role is deubiquitination of histone H2B, and this activity resides in a distinct subcomplex called the deubiquitinating module (DUBm), which contains the ubiquitin-specific protease, Ubp8, bound to Sgf11, Sus1 and Sgf73. The deubiquitinating activity depends upon the presence of all four DUBm proteins. We report here the 1.90 Å resolution crystal structure of the DUB module bound to ubiquitin aldehyde, as well as the 2.45 Å resolution structure of the uncomplexed DUB module. The structure reveals an arrangement of protein domains that gives rise to a highly interconnected complex, which is stabilized by eight structural zinc atoms that are critical for enzymatic activity. The structure suggests a model for how interactions with the other DUBm proteins activate Ubp8, and allows us to speculate about how the DUB module binds to monoubiquitinated histone H2B in nucleosomes.
The covalent modification of chromatin is a core feature of eukaryotic transcription (1). The SAGA complex regulates genes transcribed by RNA polymerase II by carrying out multiple functions that include histone acetylation and deubiquitination (2, 3). The 1.8 MDa SAGA complex, which contains 21 proteins conserved from yeast to man (4), plays a role in transcription activation and also couples transcription elongation with export of the nascent RNA through the nuclear pore complex (5). SAGA is recruited to promoter regions by activator proteins, where it catalyzes the cleavage of monoubiquitin from Lys123 of histone H2B, as well as acetylation of histone H3. Both of these activities are thought to be important for evicting nucleosomes from the promoter region, and for facilitating transcription elongation (6). The yeast SAGA complex has been a model for studying SAGA function in the transcription cycle. Histone acetylation by SAGA is mediated by the GCN5 histone acetyltransferase subunit, while histone H2B deubiquitination is mediated by a discrete subcomplex known as the deubiquitinating module (DUBm) (3, 7). The yeast DUBm comprises four proteins: the Ubp8 ubiquitin hydrolase, Sgf11, Sus1 and Sgf73 (8, 9). Sgf73 tethers DUBm to the rest of the SAGA complex through a C-terminal domain, while the N-terminal domain forms an integral part of the DUB module (8). Sgf73, along with Sus1, also facilitates SAGA’s role in nuclear export by binding to components of the nuclear pore complex (10). Although Ubp8 contains a Ubiquitin specific hydrolase (Usp) domain (11), the protein is inactive unless it is in complex with the other three DUBm proteins (8, 12). The structural basis for Ubp8 activation, and its dependence upon the other three DUBm proteins to form a stable complex that can specifically target H2B for deubiquitination, is not understood.
Drosophila and human SAGA include a DUBm that has an analogous function to the yeast DUBm and comprises homologues of the yeast proteins (9, 13, 14). In addition, the human DUBm, consisting of USP22, ATXN7L3, ENY2 and ATXN7 (homologues of Ubp8, Sgf11, Sus1 and Sgf73, respectively), also plays a role in telomere maintenance by regulating the stability of TRF1, a component of the telomeric shelterin complex (14). These additional functions suggest that the human DUBm may have other substrates that remain to be identified. The importance of DUBm function in humans is highlighted by the observations that USP22 is a cancer stem cell marker (15) and that ATXN7 is the affected protein in the polyglutamine expansion disease, spinocerebellar ataxia type 7 (16, 17).
In order to gain insight into the basis for DUBm assembly, activity, and substrate binding, we determined the structure of the heterotetrameric DUBm with and without bound ubiquitin aldehyde (Ubal), an inhibitor that forms a reversible covalent bond with the active site cysteine (18). Soluble, enzymatically active complex was obtained by co-expressing the intact Ubp8 (474 a.a.), Sgf11 (99 a.a.), and Sus1 (96 a.a.) proteins along with an N-terminal fragment of Sgf73 (residues 1-96) that is sufficient for enzymatic activity (8). The co-expressed DUBm used in the structural study cleaves ubiquitin-AMC with a kcat of 0.17 s−1 and a KM of 1.5 μM, giving a kcat/KM value of 1.1 × 105 M−1 s−1 (Figure S1, supplementary data). The activity is inhibited by Ubal with an apparent Ki of 0.2 μM (Figure S2). The coexpressed DUBm can also cleave ubiquitin from the free histone substrates, H2B and H2A (Figure S3), demonstrating that the complex retains the reported deubiquitinating activity of the complete SAGA complex. The crystal structure of the DUBm bound to Ubal was determined by selenomethionine multiwavelength anomalous dispersion (MAD) phasing and refined at 1.90 Å resolution to an R/Rfree of 15.5/20.3 %. The 2.45 Å resolution structure of free DUBm was determined by molecular replacement using the high-resolution DUB module structure as a search model, and refined to an R/Rfree of 20.4/28.1 %. Data collection and refinement statistics are shown in Table S1. Sequence alignments for all four DUBm proteins, annotated with the positions of secondary structural elements, are shown in Figure S4. The description of the structure that follows focuses on the higher resolution model of the Ubal-bound complex, except where noted.
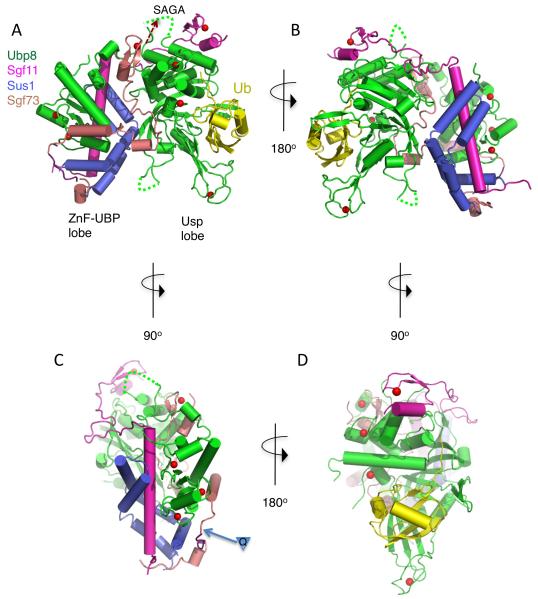
The structure of the DUB module shows Ubp8, Sgf11, Sus1 and Sgf73 to be remarkably intertwined (Figs. 1A and 1B). Each polypeptide contacts the other three proteins in the complex, thereby explaining why active complex formation requires the presence of all four DUBm subunits (8, 12). With the exception of Ubp8, which contains two globular domains separated by a short linker, the conformations of the other three proteins are largely dependent upon their interactions with other DUBm proteins. The complex is organized into two lobes, each containing one of the two domains of Ubp8: the N-terminal Zinc Finger-Ubiquitin Binding Protein (ZnF-UBP) domain and the C-terminal enzymatic Usp domain (Figs. 1A, 1B). The ZnF-UBP lobe (Fig. 1C) is organized around the long N-terminal helix of Sgf11, which binds the Ubp8 ZnF-UBP domain on one face and Sus1 on the opposing face. The 40 N-terminal residues of Sgf73 comprise the remainder of this lobe, primarily contacting the Ubp8 ZnF-UBP domain and Sus1 with short helices (Fig. 1A, C). The second lobe of DUBm contains the ~350 amino acid Usp domain of Ubp8 (Fig. 1A, 1D). Ubiquitin aldehyde binds as expected to the “fingers” region of the Usp domain (19), which consists of an antiparallel β sheet and a zinc-binding domain (Fig. 1D), and forms a covalent bond between the modified C-terminal glycine and the active site cysteine. To the opposite side of the active site lies the N-terminal zinc finger domain of Sgf11, which binds to the face of the Usp domain (Fig. 1D). An extended well-ordered linker region joins the Sgf11 zinc finger with its N-terminal helix (Fig. 1B), thereby bridging the two lobes of the DUBm while forming additional contacts with Ubp8. The interactions at the interface between the two lobes are mediated primarily by 50 C-terminal residues of the Sgf73 fragment (45-95), which are buried between the two lobes. This portion of Sgf73 contains a short α helix and a zinc finger domain that are connected by 20 residues that lack secondary structure, yet are well ordered and have low temperature factors. Sus1 helices α2 and α3 mediate additional interface contacts with Ubp8 on the back face of the Usp domain. Together, Sgf11, Sus1 and Sgf73 bury a total of 7,910 Å2 of the Ubp8 surface. The overall features of the DUBm complex are preserved in the structure of apo DUBm, which crystallizes with different crystal packing, indicating that the unusual arrangement of proteins in the DUBm complex is not an artifact of crystal contacts.
Figure 1. Overall structure of the SAGA DUB module.
(A) View of DUB module bound to ubiquitin aldehyde (Ubal). The ZnF-UBP lobe and Usp lobe are labeled. Coloring scheme is Ubp8, green, Sgf11, magenta, Sus1, blue, Sgf73, salmon, Ubal, yellow, with zinc atoms shown as red spheres. Disordered loop residues are indicated with dotted lines. Residues in Sgf73 that are ordered in the structure of the apo DUBm are shown in gray.
(B) View of DUB module rotated 180° about the vertical as compared to panel B.
(C) View of the ZnF-UBP lobe. The Ubp8 ZnF-UBP domain and Sus1 bind on opposite faces of the Sgf11 N-terminal helix. The Sgf73 residues shown in gray are disordered in the Ubal-bound complex but are ordered in the apo complex. The insertion point for polyglutamine expansion in the human homolog, ATXN7, is indicated by an inverted triangle containing the letter, Q.
(D) View of the Usp lobe. The bound ubiquitin is shown in yellow.
The binding of Sgf73 between the two lobes of the DUBm is consistent with its role in anchoring the DUB module to the rest of the SAGA complex (8, 9) and in helping to activate DUB activity. As compared with Sgf11 and Sus1, which are also non-globular, Sgf731-96 appears to be the most dependent on other DUBm proteins for its observed conformation, since it contains the fewest intrachain contacts. In the intact protein, the remaining C-terminal residues of Sgf73 link the DUBm to the SAGA complex (Fig. 1A), with residues 227-402 mediating interactions with the other SAGA proteins (8, 9). There is a low complexity acid region (residues 130-162) between the SAGA-binding and DUBm-binding regions of Sgf73, suggesting that the DUBm might be flexibly tethered to the remainder of the SAGA complex. Sgf731-96 contains a zinc finger that is coordinated by two cysteines (C78, C81), and a histidine (H93). The fourth ligand in the DUBm-Ubal complex is an aspartic acid (D95) that is conserved in the human homologue, ATXN7 (Fig. S4); however, this residue does not coordinate the zinc in the apo DUBm structure and instead is part of an extended C-terminal helix. It is likely that, in the full-length protein, the histidine (H97) or cysteine (C98) residues that are missing from the crystallized fragment serve as the fourth zinc-coordinating ligand.
The incorporation of Sgf73 into the DUBm depends upon the integrity of the zinc finger, since a slightly shorter fragment containing residues 1-92 disrupts binding to the other DUBm subunits (8). We therefore tested the effect of zinc chelation on the incorporation of Sgf73 and the other DUBm subunits into the complex. In the absence of EDTA, the DUBm sediments with an S20,w value of 4.9 (Figure 2), as expected for a single globular 90 kDa species with a somewhat asymmetric axial ratio. The addition of 2 mM EDTA causes the complex to dissociate into two principal species corresponding to Sgf73 and the Ubp8/Sgf11/Sus1 heterotrimer (Fig. 2). We note that Sgf11/Sus1 sediments as multiple species (Fig. S5) that do not account for any of the observed peaks. Based on the DUBm structure, the stable association between Ubp8, Sgf11 and Sus1 is most likely maintained by contacts between the long N-terminal helix of Sgf11, Sus1 and the ZnF-UBP domain. The loss of complex integrity was accompanied by a loss of activity, which was observed upon addition of increasing concentrations of EDTA as well as increasing incubation time (Fig. S6). In addition to dissociating Sgf73, which is known to compromise DUBm activity (9), it is also possible that the EDTA further affects enzymatic activity by chelating the zinc atoms in the enzymatic domain of Ubp8 (Fig. 3A and 3B). The importance of wild-type Sgf73 function is highlighted by the effect of polyglutamine expansions in the N-terminus of the human homologue, ATXN7 (17). These insertions of up to 300 glutamine residues compromise SAGA activity and give rise to defects in transcription (20). The polyglutamine expansion occurs near the N-terminus of Sgf73 (17), in the coil region between the first two helices (Fig. 1C). It is possible that large insertions in this region disrupt Sgf73 association with the other DUB proteins, compromising enzymatic activity or causing them to dissociate from the SAGA complex. Alternatively, polyglutamine tracts may cause Sgf73 to aggregate and therefore fail to bind to the other DUBm proteins.
Figure 2. Dissociation of the Sgf73 fragment from the DUBm complex.
Velocity sedimentation of the DUBm in the presence (blue) and absence (black) of 2 mM EDTA, superimposed with the sedimentation of Sgf73 (1-106) alone.
Figure 3. Structural details of the DUB module.
(A) Structural zinc coordination in the Ubp8 Usp domain located between α9 and α10.
(B) Structural zinc coordination in the Ubp8 Usp domain located between α12 and α13.
(C) Superposition of the structure of Sus1/Sgf111-33 (gold) (PDB ID 3KIK) with the corresponding residues of Sus1 (blue) and Sgf11 (magenta/pink) in the DUBm. The arrow indicates the additional helical residues (pink) in Sgf11 seen in the DUBm structure.
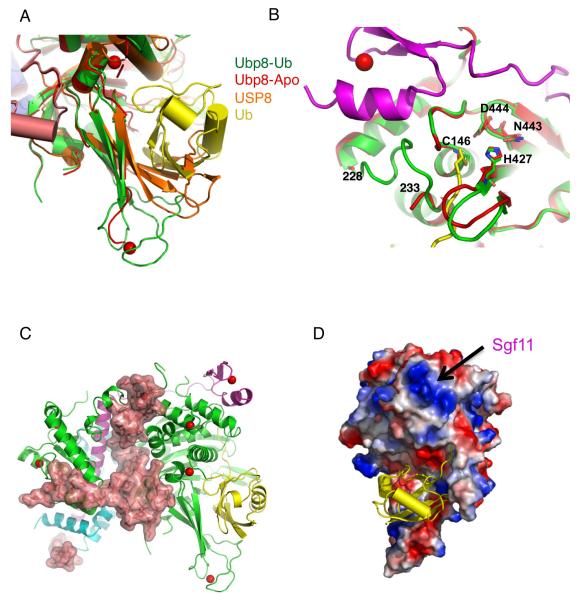
The structures of the individual DUBm subunits contain some unexpected features. The overall fold of the Ubp8 catalytic domain is very similar to that of other Usp proteins (11). However, the catalytic domain of Ubp8 contains two bound structural zinc atoms (Fig. 3A and 3B) that have not been previously observed (11). One of the bound zinc atoms (Fig. 3A) is located between helices α9 and α10 (see Fig. S4 for numbering) and is coordinated by H170, C174, C182 and C185, while the second bound zinc (Fig. 3B) is located in a non-conserved loop region between helices α12 and α13 and is coordinated by H250, C271, C273 and H276. Although these bound zinc atoms are not conserved among the vast majority of USP enzymes, sequence comparisons indicate that both of these sites are conserved in the Ubp8 homologues, human USP22 (Fig. S4) and Drosophila Nonstop, which are SAGA DUBm subunits (13, 14). Another interesting feature of Ubp8 is that the ZnF-UBP domain, which in other USP enzymes such as Usp5 binds ubiquitin (21), has its ubiquitin-binding face occluded in the DUBm structure. That region of the domain, which is centered around beta strands 2 – 5 (Fig. S4), is largely buried at the interface with Sgf11. While a role for ubiquitin binding to this domain cannot be ruled out when Ubp8 is not part of the observed complex, the absence of conserved ubiquitin-binding residues in Ubp8, as well as in the human homologue, USP22 (22), makes this unlikely. Other ZnF-UBP domains that similarly lack conservation in the ubiquitin-binding residues and do not bind ubiquitin, such as USP33 (23), may also play structural role in those enzymes.
A structure of Sus1 that was previously determined in complex with residues 1-33 of Sgf11 (24) is virtually superimposable with the corresponding residues of Sgf11 and Sus1 in the DUBm complex (Fig. 3C). In the DUBm structure, however, which contains the intact 96-amino acid Sgf11 protein, the N-terminal Sgf11 helix is extended by an additional 10 amino acids at its C-terminus; this presumably is due to the additional contacts with the Ubp8 ZnF-UBP domain (Fig. 1C and 3C). The ZnF-UBP domain also contacts helix α1 of Sus1 and likely helps to stabilize its interaction with Sgf11. In the structure of Sus1 bound to Sgf111-33, helix α1 of Sus1 adopts widely variable orientations in the four complexes found in the asymmetric unit (24), only one of which contacts Sgf11 and matches the closed conformation observed in the DUBm. The closed conformation of Sus1 is also observed when it binds to Sac3 as part of the TREX-2 complex, which functions in mRNA export (25). Thus, the N-terminal helix of Sgf11 plays a scaffolding role similar to Sac3, which also forms a long α-helix to which multiple proteins bind.
A comparison of the ubiquitin aldehyde (Ubal) DUBm complex with the apo DUBm complex reveals that a number of structural changes occur in Ubp8 when ubiquitin binds to the DUBm. The overall features of ubiquitin binding, including the arrangement of the catalytic residues, C146, H427, N443 and D444 (see Fig. S7 for view of active site residues in electron density), are similar to those seen in structures of other Usp enzymes bound to Ubal (19, 26, 27). Ubiquitin binds to the fingers domain of Ubp8, with its hydrophobic face (including I44) interacting with the beta sheet, while the C-terminal tail of ubiquitin binds in a conserved groove that leads to the active site (Fig. 1A and 1D). In the absence of ubiquitin, a number of structural changes occur in Ubp8. The fingers region that binds ubiquitin in the complex remains in an open conformation in apo DUBm (Fig. 4A), but the zinc-binding module and a several beta strand residues become less well-ordered, resulting in weak electron density that could not be completely traced (Fig. S8A). Importantly, the fingers domain does not appear to collapse and occlude the ubiquitin-binding pocket, as is seen in the structure of the human USP8 apoenzyme (a Usp family member that is not an ortholog of yeast Ubp8) (28) (Fig. 4A). Most striking, however, are the changes that occur near the enzyme active site (Fig. 4B; electron density shown in Fig. S8). Seven Ubp8 residues located adjacent to the ubiquitin C-terminus, 228-233, are disordered in the absence of ubiquitin binding (Fig. 4B). In addition, loop residues 421-426 move inward by up to 2.0 Å towards the tail-binding groove (Fig. 4B). Since the loop and the disordered region contain residues that contact ubiquitin directly, this suggests that binding of the ubiquitin C-terminal tail triggers the observed conformational change. Despite these conformational differences, the active site residues (C146, H427, N443 and D444) are in their catalytically competent orientation in both the presence and absence of ubiquitin (Fig. 4B and S8B). This is in contrast to deubiquitinating enzymes such as HAUSP/USP7 (19), in which the active site residues are not in a catalytically competent arrangement in the apoenzyme.
Figure 4. Interaction between the DUB module and substrate.
(A) Alignment of apo Ubp8 (red), Ubp8 (green)-Ubal (yellow), and USP8 (orange). In the apo USP8 structure, the fingers region is collapsed and cannot accommodate ubiquitin without a conformational change.
(B) Structural changes in the region of the Ubp8 active site. Residues 228-233 of the Ubp8 apoenzyme (red) are disordered in the absence of ubiquitin and the loop, 421-426, shifts into the groove where the C-terminal tail of ubiquitin (yellow) would bind. The loop, which contains the active site cysteine, C146, maintains the same conformation in the presence and absence of ubiquitin and is stabilized by contacts with the Sgf11 zinc finger (magenta). The other active site residues, H427, N443, and D444, also remain in the same orientation in both structures.
(C) Structure of the DUBm showing a surface representation of Sgf73. The color scheme used is the same is in Figure 1.
(D) Electrostatic surface potential of the ubiquitin-binding face of the DUBm. The region of strong electropositive surface potential (blue) is due to the Sgf11 zinc finger.
How does complex assembly activate Ubp8? In the absence of the other DUBm proteins, we find that Ubp8 is only about 1/200,000th as active as the intact DUBm (Fig. S9) and shows significant aggregation (Fig. S10). A comparison of the DUBm structures with and without Ubal suggests that interactions among the DUBm proteins may stabilize a conformation of Ubp8 that is catalytically competent and able to bind ubiquitin. In the active site region, the Sgf11 zinc finger directly contacts the loop in Ubp8 that contains the active site cysteine, C146 (Fig. 4B). These contacts with Sgf11 may help to maintain the active site residues in a catalytically competent conformation even when the neighboring residues (229-235) become disordered. Another way in which the other DUBm proteins may activate Ubp8 is by stabilizing a conformation of the fingers domain that favors binding of the globular portion of ubiquitin. The human USP8 apoenzyme (not an ortholog of yeast Ubp8), for example, has a partially occluded ubiquitin-binding pocket that would require a conformational change to accommodate ubiquitin (Fig. 4A). Since the β sheet of the Ubp8 fingers region is in an open conformation even in the absence of ubiquitin (Fig. 4A), it is possible that interactions between Ubp8 with the other DUBm subunits help to maintain a conformation that favors ubiquitin binding. Interactions of the upper portion of the fingers domain with Sgf73 helix α2 and with Sus1(Fig. 1A and 4C) form extensive contacts that may favor the open conformation of Ubp8. Sgf73 may also play a general role in stabilizing the conformation of the Usp domain. Ubp8 forms the most extensive interface with Sgf73, with a total area of 3,075 Å2, as compared with the other pairwise domain interactions in the DUBm. Importantly, Sgf73 is the “mortar” that holds together the two lobes of the DUBm complex (Fig. 4C), promoting interactions between the two lobes as well as aligning Sgf11 and Sus1, which also contact the Usp domain at the interface between the two lobes. Although the structure of the Ubp8 Usp domain on its own is not known, it is possible that these extensive interactions may also help to stabilize the overall USP fold or affect protein dynamics in a way that favors the catalytically competent structure.
The structure of the SAGA DUBm suggests how this module interacts with its natural, in vivo substrate, monoubiquitinated histone H2B within a nucleosome. The electrostatic surface potential of the DUBm (Fig. 4D) reveals a basic region that could favor interactions with the negatively charged DNA when the nucleosome is positioned with ubiquitinated K123 of H2B in the active site of Ubp8. This positively charged region of the DUBm is due to the zinc finger module of Sgf11, which contains four basic residues (R78, R84, R91, R95; Fig. S11) that are conserved in ATXN7L3, the human homologue of Sgf11. Two additional C-terminal arginine residues, R98 and R99, which are disordered in the structure but are also conserved in the human homologue, would further contribute to the strong positive charge in this region. Figure S12 shows a model for how a yeast nucleosome (29) monoubiquitinated at K123 of H2B can dock on the DUBm, with ubiquitin in the active site as seen in the Ubal-bound structure (Fig. 1D). This arrangement brings the basic patch on the DUBm in close approach with the DNA, thereby favoring interactions with the sugar-phosphate backbone. The interdependent structural and functional roles of the four SAGA DUBm proteins in mediating biochemical activity, substrate binding and incorporation into the larger SAGA complex is likely an example of how other subcomplexes of SAGA, and other coactivator and corepressor complexes, cooperate to regulate transcription.
Supplementary Material
Acknowledgements
We thank Anthony DiBello for helping with the ubiquitin aldehyde synthesis and Ed Hurt for providing the Sus1 clone. We thank Stephen Corcoran at the GM/CA-CAT beamline at the Advanced Photon Source for assistance with data collection and processing, and Mario Bianchet for advice and discussions. C.E.B. is supported by a Ruth Kirchstein Fellowship from the National Institute of General Medical Science (F32GM089037) and T.Y. is a Special Fellow of the Leukemia and Lymphoma Society. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3MHS (DUBm bound to Ubal) and 3MHH (apo DUBm).
REFERENCES
- 1.Campos EI, Reinberg D. Annu Rev Genet. 2009;43:559. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 2.Koutelou E, Hirsch CL, Dent SY. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.03.005. epub DOI:10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Navarro S. EMBO Rep. 2009;10:843. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel JA, Grant PA. Mutat Res. 2007;618:135. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luthra R, et al. J Biol Chem. 2007;282:3042. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 6.Pascual-Garcia P, et al. Genes Dev. 2008;22:2811. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry KW, et al. Genes Dev. 2003;17:2648. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Nat Cell Biol. 2008;10:707. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 9.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler A, et al. Mol Biol Cell. 2006;17:4228. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Y, Scheel H, Hofmann K, Komander D. Mol Biosyst. 2009;5:1797. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- 12.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. Mol Cell Biol. 2005;25:1173. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weake VM, et al. EMBO J. 2008;27:394. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atanassov BS, et al. Mol Cell. 2009;35:352. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HJ, et al. Gene Expr Patterns. 2006;6:277. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Lindblad K, et al. Genome Res. 1996;6:965. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]
- 17.Latouche M, et al. Mol Cell Neurosci. 2006;31:438. doi: 10.1016/j.mcn.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 18.Pickart CM, Rose IA. J Biol Chem. 1986;261:10210. [PubMed] [Google Scholar]
- 19.Hu M, et al. Cell. 2002;111:1041. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 20.Helmlinger D, Tora L, Devys D. Trends Genet. 2006;22:562. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Reyes-Turcu FE, et al. Cell. 2006;124:1197. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 22.Bonnet J, Romier C, Tora L, Devys D. Trends Biochem Sci. 2008;33:369. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Allen MD, Bycroft M. Protein Sci. 2007;16:2072. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellisdon AM, Jani D, Koehler A, Hurt E, Stewart M. J Biol Chem. 2009 doi: 10.1074/jbc.M109.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jani D, et al. Mol Cell. 2009;33:727. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renatus M, et al. Structure. 2006;14:1293. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu M, et al. EMBO J. 2005;24:3747. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avvakumov GV, et al. J Biol Chem. 2006;281:38061. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 29.White CL, Suto RK, Luger K. EMBO J. 2001;20:5207. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.