Abstract
The final cleavage event that terminates cell division, abscission of the small, dense intercellular bridge, has been particularly challenging to resolve. Here, we describe imaging innovations that helped answer long-standing questions about the mechanism of abscission. We further explain how computational modeling of high-resolution data was employed to test hypotheses and generate additional insights. We present the model that emerges from application of these complimentary approaches. Similar experimental strategies will undoubtedly reveal exciting details about other underresolved cellular structures.
Introduction
Cytokinesis, the last step of cell division, involves large-scale constriction of the plasma membrane, which leads to partitioning of cellular content into two daughter cells. An actomyosin contractile ring causes ingression of the plasma membrane at the cell equator (Eggert et al., 2006; Green et al., 2012). This divides the organelles and most of the cytoplasm equally between the two daughter cells, but the microtubules that formed the spindle linger inside a narrow membrane bridge that connects the cells, called the intercellular bridge (see Figure 1A). To separate the daughter cells and complete the division process, the microtubules must be severed and the plasma membrane must be sealed. While much of mitosis proceeds rapidly (less than 30 min from metaphase to telophase), the intercellular bridge usually persists for over an hour prior to the final cleavage event, termed abscission (Dambournet et al., 2011; Elia et al., 2011; Gromley et al., 2005; Guizetti et al., 2011).
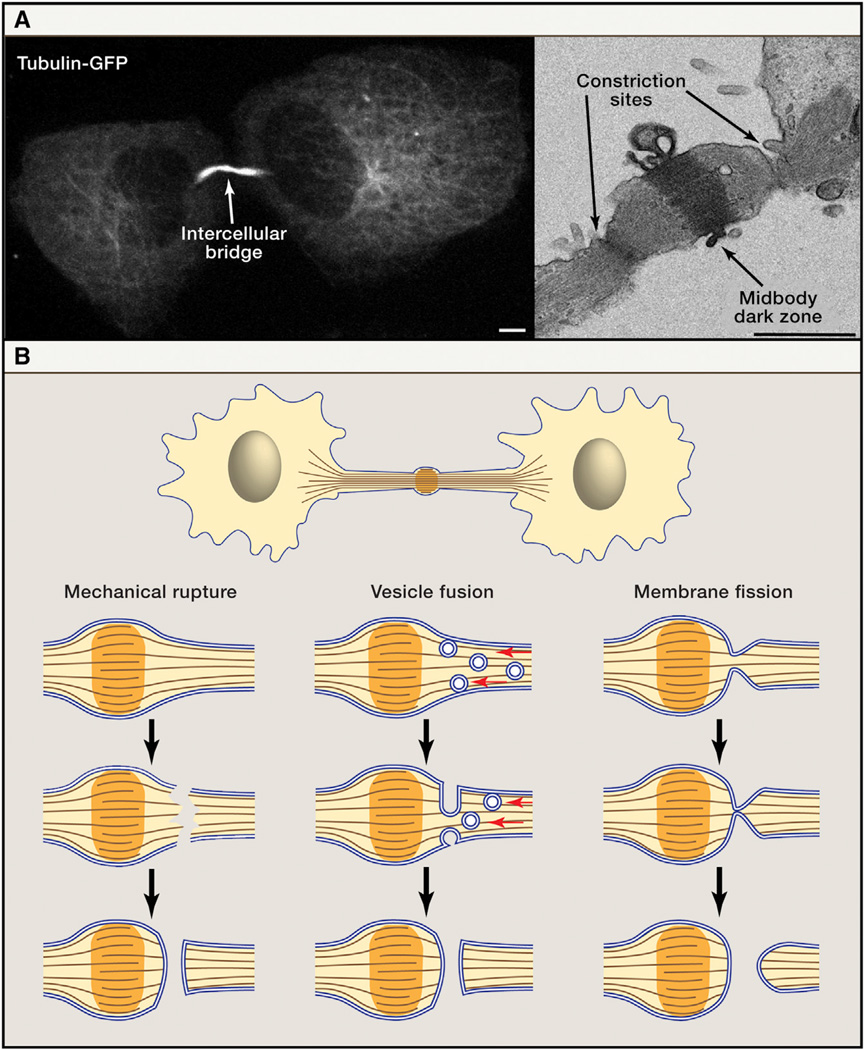
Figure 1. Abscission of the Intercellular Bridge.
(A) The intercellular bridge connecting two daughter cells must be resolved to complete cytokinesis. The microtubules in the intercellular bridge are visible using both confocal microscopy and TEM. Shown here are live MDCK cells expressing tubulin-GFP (left) and fixed MDCK cells imaged by TEM (right) (Elia et al., 2011). The bridge can be severed on either or both sides of the dark zone. Scale bars: (left) 5 µm, (right) 1 µm.
(B) Schematic representation of the three classic models for abscission of the intercellular bridge. The mechanical rupture model (left) posited that as daughter cells move away from one another, the intercellular bridge is torn. The opening is then sealed by cellular wound healing mechanisms. The vesicle fusion model (center) proposed that vesicles delivered to the midbody fuse with each other and with the plasma membrane to separate the daughter cells. Themembrane fission model (right) suggested that acute reduction in the diameter of the bridge membrane brings the lipid bilayers close enough to fuse. Prior to the technical innovations discussed in this review, poor spatial and temporal resolution made it difficult to differentiate between these models experimentally.
Conventional light microscopy methods have been employed over the years to investigate the mechanism of cytokinetic abscission. Key pathway components were identified using assays such as protein localization, bridge persistence, and cytokinetic failure (reviewed in Barr and Gruneberg, 2007; Schiel and Prekeris, 2010). Based on these results, researchers formulated models in an effort to understand how different proteins contribute to abscission of the intercellular bridge. However, because the bridge is only about 1 µm in diameter, and the densely packed microtubules fill much of that space (Elia et al., 2011; Guizetti et al., 2011; Mullins and Biesele, 1977), many molecular details have been difficult to visualize and resolve. This made testing model-based predictions problematic.
To understand how and when two daughter cells fully separate, several key questions need to be answered. The vintage models of abscission (described below) each attempted to answer some of these questions using data from conventional microscopy experiments. We do not yet have every answer; however, like a child gazing through his first pair of glasses, the increased spatial and temporal resolution provided by recent advances in cryo-electron microscopy (cryo-EM) tomography, structured illumination microscopy (SIM), and high-speed quantitative fluorescent microscopy have enabled researchers to look anew at the process of cytokinetic abscission (Elad et al., 2011; Elia et al., 2011; Guizetti et al., 2011; Schiel et al., 2011). Consequently, the critical protein complexes for driving abscission have been identified and a revised model has emerged. In this review, we describe each technological advance and explain how it shed new light on these long-standing questions. We also describe how computational modeling using the new imaging data resulted in additional insights. Together, the answers provided through employing these imaging innovations have led to creation of the modern model of cytokinetic abscission presented here. Undoubtedly, application of additional innovations will be needed to fully understand the regulated scission of the intercellular bridge, but the advances described here represent considerable refinement of the old views of abscission. Applying the same strategy to other persistent cell biological questions will likely lead to unexpected insights and novel revisions of current models.
Five Key Questions
The physical separation of two daughter cells requires significant, highly coordinated rearrangements of both the cytoskeleton and the membrane that comprise the intercellular bridge. To facilitate a mechanistic understanding of this process, five fundamental questions need to be addressed.
Where Is the Site of Separation?
Microtubules are visible by transmission electron microscopy (TEM) throughout the intercellular bridge, but the density of microtubules (and potentially other proteins) increases at the center. This region, termed the midbody, is highly enriched in proteins (Skop et al., 2004) and takes up stains that are visible as an electron dense dark zone (see Figure 1A). Identifying whether cleavage occurs inside or outside the dark zone is a prerequisite for characterizing the mechanism of abscission.
When Does Cytokinetic Abscission Occur?
As mentioned earlier, the intercellular bridge remains intact for over an hour before final separation occurs (Dambournet et al., 2011; Elia et al., 2011; Gromley et al., 2005; Guizetti et al., 2011). The decrease in bridge diameter that will ultimately lead to abscission could therefore occur gradually, in steps, or acutely.
How Are the Necessary Proteins Organized in Space and Time during Abscission?
Once characterized, the spatial and temporal changes during abscission can be correlated with changes in localization and dynamics of microtubules and other intercellular bridge components to dissect the role of potential cleavage mediators. This information can provide key evidence to build a detailed mechanistic description of the abscission machinery.
What Changes Does the Cytoskeleton Undergo?
Although there is some evidence that a population of the tightly packed microtubules in the midbody is dynamic (Hu et al., 2012), most are acetylated (Piperno et al., 1987), which is indicative of stable microtubules that do not turn over quickly. In order to separate the daughter cells, the microtubules must either disassemble or they must be cut. Actin filaments from the actomyosin contractile ring that drive the early phase of cytokinesis may also need to be cleared from the intercellular bridge in order for abscission to take place.
What Changes Does the Membrane Bilayer Undergo?
The final cell separation requires dramatic changes in the morphology of the intercellular bridge membrane. It is critical to understand how lengthening, narrowing, and, eventually, constriction and scission of the membrane bridge are accomplished. Proteins impact these processes; however, changes in membrane composition, membrane curvatures, and membrane tension are also likely to directly influence the topology and timing of abscission.
A true vision of abscission will require knowledge of how these five aspects of abscission are coordinated and regulated. Changes in membranes and microtubules have to be coupled with the assembly and activity of the scission machinery. The answers to individual questions all contribute to creating a comprehensive model of the mechanism of cytokinetic bridge abscission.
Classic Models of Abscission
Many studies have sought to answer the above questions using conventional microscopy and biochemical methods. Three main models emerged from evaluation of the experimental data (see Figure 1B). These historic models provide a framework for understanding the more recent developments.
The Mechanical Rupture Model
The first abscission model suggested was the mechanical rupture model (Figure 1B, left). This model was based on early studies that used transmitted light microscopy and transmission electron microscopy (TEM) to characterize the midbody, and examine abscission timing (Mullins and Biesele, 1973, 1977). In these studies lengthening and constriction of the intercellular bridge on either side of the dark zone was described. Based on these observations, it was suggested that the connection between the two daughter cells is stretched and eventually broken by mechanical rupture as the daughter cells pull in opposite directions. Mullins proposed that a wound healing mechanism seals the membrane opening. Evidence that cell adhesion molecules (such as integrins) and cellular wound healing components are required for successful cytokinesis in some cells supports the mechanical rupture model (Kanada et al., 2005; LaFlamme et al., 2008; Reverte et al., 2006). However, because abscission can occur in nonadherent cells (e.g., leukocytes and lymphocytes) and in nonmotile cells that do not migrate away from one another under physiological conditions, researchers began to look for alternate mechanisms (Schiel and Prekeris, 2010).
The Vesicle Fusion Model
Inspired by plant cell division, a second model for abscission emerged: the vesicle fusion model (Figure 1B, center). In plants, cell separation is induced by the directed delivery of recycling endosomes and secretory vesicles to the midline between daughter cells. There, the vesicles fuse to form a new cell wall (Barr and Gruneberg, 2007). Endocytic and secretory vesicles also accumulate at the center of the intercellular bridge in mammalian cells (Gromley et al., 2005). Membrane trafficking components, such as Rab proteins (Rab 11, Rab 8, Rab 35) (Dambournet et al., 2011; Wilson et al., 2005), and SNARE proteins (Vamp8, syntaxin2) (Gromley et al., 2005) were identified in a midbody proteomic study (Skop et al., 2004) and subsequently shown to be involved in abscission (reviewed in Neto et al., 2011). Thus, it was proposed that a common mechanism mediates cytokinesis in plant and mammalian cells (Gromley et al., 2005; Neto et al., 2011). According to this model, vesicles transported to the center of the intercellular bridge fuse with one another and with the plasma membrane to induce final separation between the cells. This model of abscission predominated for many years; however, no direct evidence for vesicle fusion events in the intercellular bridge at the time of abscission could be found.
The Membrane Fission Model
The third model—the membrane fission model—proposed that the midbody membrane is acutely constricted, and then sealed and severed (Figure 1B, right). This mechanism seemed plausible after fluorescent protein localization, mRNA depletion, yeast two-hybrid, and biochemical purification experiments indicated the involvement of the ESCRT (endosomal sorting complex required for transport) fission machinery in abscission (Carlton and Martin-Serrano, 2007; Morita et al., 2007). The ESCRT machinery mediates topologically equivalent constriction and fission events during both multivesicular body (MVB) formation (i.e., the inward budding of intraluminal vesicles) and retrovirus budding from the plasma membrane (reviewed in (McCullough et al., 2013; McDonald and Martin-Serrano, 2009; Wollert et al., 2009). While bacterial and some archael cell divisions are mediated by the FtsZ contractile machine, several archael lineages that lack FtsZ utilize ESCRT homologs to complete cell division, suggesting an evolutionary conserved role for ESCRT complex proteins in cell division (Lindås et al., 2008; Samson et al., 2008). The membrane fission model was further enriched by the finding that spastin, a microtubule-severing enzyme, directly binds to an ESCRT complex protein in the intercellular bridge (Yang et al., 2008). It was proposed that the ESCRT machinery couples microtubule severing to the physical cleavage of the membrane.
Critics of the membrane fission model had several concerns (Schiel and Prekeris, 2010). First, although the archael ESCRT homologs mediate constriction and scission of membrane tubes with similar diameter to that of the mammalian intercellular bridge (~one micron) (Elia et al., 2011; Guizetti et al., 2011; Lindås et al., 2008; Samson et al., 2008), the established role of the ESCRT complex was to mediate fission of small vesicles (~50–100 nm diameter) (Lindås et al., 2008; McDonald and Martin-Serrano, 2009; Samson et al., 2008). Second, it seemed plausible that ESCRT depletion could indirectly cause failure of vesicle delivery and fusion by obstructing late endosome formation.
Challenges for Abscission Researchers
Each classic model relied upon a subset of experimental observations and differentiating between the models remained a challenge. The unique features of the midbody made some experimental approaches ineffectual. Some of the challenges included the following: (1) immunolocalization studies have been difficult because many antibodies cannot penetrate the densely packed midbody. (2) Because the midbody is small, it has not been possible to resolve protein localization in detail. (3) It has been difficult to discriminate which of the plethora of midbody-localized proteins directly mediate abscission and which have an indirect effect. In addition to proteins mentioned above, kinesins (Barr and Gruneberg, 2007; Glotzer, 2009), kinases (Glotzer, 2009), septins (Estey et al., 2010; Spiliotis et al., 2005), exocyst complex proteins (Gromley et al., 2005; Neto et al., 2011), autophagy proteins (Pohl and Jentsch, 2009), and ubiquitin-conjugating proteins (Pohl and Jentsch, 2008) are all found at the midbody and have been shown to be essential for completion of cytokinesis. In order to overcome these challenges and discriminate the precise role of individual components improved temporal and spatial resolution was needed (Schiel and Prekeris, 2010; Steigemann and Gerlich, 2009).
New Imaging Tools and New Information
Using three innovative technologies, cryo-EM tomography, SIM, and high-speed quantitative fluorescent imaging, researchers have been able to begin addressing the five key questions described above in different ways. Below, we describe each technology and explain how experimental data from each method address aspects of the key questions and reveal new details about the mechanism of cytokinetic abscission. (Table 1 summarizes the advantages of each technology and how it has contributed to the key questions.) No single method can answer every question, but combining the information obtained through the various imaging techniques provides a new, more detailed view of the process of cytokinetic abscission. In later sections, we explain how computational methods utilized the new data to test predictions. Finally, we synthesize the findings and describe the new working model that emerged as a result of creative application of these technologies.
Table 1.
Imaging Method Advantages and Contributions
| Cryo-EM-Tomography | Structured Illumination Microscopy |
Spinning Disk Confocal Microscopy |
|
|---|---|---|---|
| Spatial resolution (x and y) | 1 nm | 100 nm | 200 nm |
| Spatial resolution (z) | 2–8 nm | 350 nm | 500 nm |
| Temporal resolution | none | seconds | milliseconds |
| Main application in cell biology | Subnanometer 3D spatial architecture of the entire cellular content | 3D structure analysis of selected macromolecular complexes and organelles in cells | 3D protein and membrane dynamics in live cells |
| General advantages for applications in cell biology |
|
|
|
| Insights into cytokinetic abscission |
|
|
|
Cryo-EM Tomography
Capable of 1–2 nm lateral resolution, TEM is probably the most widely used technique for mapping cellular ultrastructure. To obtain three-dimensional information, serial sectioning has traditionally been employed, but the axial (Z) resolution is limited by the thickness of each slice (usually >20 nm). Further, chemical fixation methods have the potential to introduce artifacts, which limits the physiological relevance of information obtained. Two major breakthroughs overcame the limitations of poor sample preservation and limited axial resolution. By tilting the specimen 1°–2° through a range up to 70° and back projecting the series of tilted images, EM tomography can be used to generate a 3D reconstruction of a cell at 2–8 nm resolution in all dimensions. Combining tomography with cryo-fixation, a rapid freezing method that preserves cellular structures, and imaging samples in a frozen state using a cryo-EM provides near-to-native information about cellular architecture at nanometer-scale resolution. In addition, it is now possible to correlate images of live cells fixed at different time points to cryo-EM-tomograms to gain temporal insights (for more detailed description of cryo-EM tomography, please refer to Fridman et al., 2012).
Cryo-EM-tomography probably represents today’s most powerful, high-resolution, technique for resolving the ultrastructure of a cell. However, the information obtained by this technique presents the overall structure and does not provide detailed information about the organization of specific proteins in a defined cellular structure. In spite of this limitation, cryo-EM tomography has been used to obtain answers to some of the key questions about cytokinetic abscission.
Insights Gained from Cryo-EM Tomography
To address the first key question (where cytokinetic abscission occurs), researchers combined cryo-EM tomography and conventional fluorescence imaging. They collected fluorescence images of midbodies at different stages in cytokinesis, fixed the cells and then imaged the same locations using cryo-EM tomography. Through this correlative approach researchers observed that constriction sites appear peripheral to the midbody dark zone late in cytokinesis (Guizetti et al., 2011). Measurements from an alternate technique, atomic force microscopy, were consistent: a 2-µm-long area was identified at the center of the intercellular bridge that is bordered by two areas of lower height—the constriction zones (Elia et al., 2011). The new data provided evidence that the actual event of cell separation does not take place at the midbody dark zone as initially thought, but rather at two peripheral sites located about 1 µm away from the midbody center, as postulated by Mullins many years ago (Mullins and Biesele, 1977).
Correlative time-lapse imaging and cryo-EM tomography experiments also led to insights related to the question of the spatial organization of proteins and membranes during abscission (Elad et al., 2011; Guizetti et al., 2011; Schiel et al., 2011). Although specific proteins could not be identified, helical cortical filaments were documented at the constriction sites (Figure 2A, left) (Guizetti et al., 2011). It was proposed that contractile forces generated by filamentous cellular machinery help facilitate abscission. More recently, cryo-EM tomography revealed cortical filaments at the constriction furrow in ESCRT-dependent dividing archea. Comparison of these structures to in vitro ESCRT assemblies is suggestive of a helical filament configuration (Dobro et al., 2013).
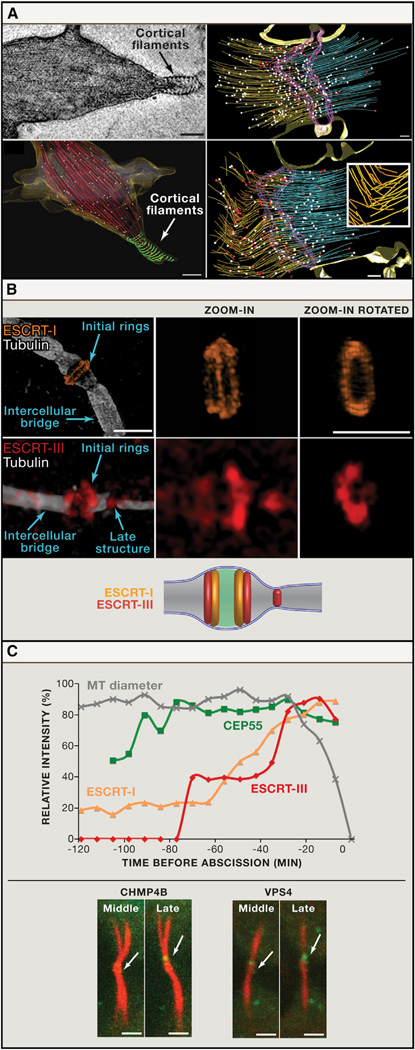
Figure 2. Visualization of Cytokinetic Abscission by High-Resolution Imaging.
(A) Cryo-EM tomography of the intercellular bridge. Left: the identification of cortical filaments at the membrane of the intercellular bridge in cells at late cytokinesis. The original cryo-EM image (top) and a 3D reconstruction of the entire tomogram series (bottom) are shown. Red: microtubules, green: cortical filaments. Scale bar, 200 nm. Images are from Guizetti et al. (2011) with permission. Right: Reconstructions of EM tomogram from cells at early (top) and late telophase (bottom). The number of microtubules with flared ends indicative of rapidly depolymerizing microtubules (red dots), increases in late telophase (relative to microtubules with nonflared ends; white dots). Also note the appearance of buckled microtubules in cells at late telophase. (Purple: midbody dark zone; yellow and blue: microtubules). Scale bar, 5 µm. Inset: enlarged view from the buckled microtubules zone. Images are Schiel et al. (2011) with permission.
(B) Structural organization of ESCRT complex components at the intercellular bridge as revealed by SIM. ESCRT-I component TSG101 (top) localizes to ring structures on either side of the dark zone. The ESCRT-III protein CHMP4B (bottom) is visible both in rings adjacent to the dark zone and at a late structure closer to the cell body. An integrated model of ESCRT localization based on measurements from SIM images is shown below. Scale bar, 2 µm.
(C) Kinetics of cytokinetic abscission assessed using high-speed quantitative fluorescence imaging. ESCRT components are recruited to the intercellular bridge at different times prior to abscission (time 0). As shown in the graph, the relative intensity values CEP55 increases early. ESCRT-I and ESCRT-III levels increase later in abscission. An acute increase in ESCRT-III levels correlates with a decrease in the diameter of the intercellular bridge. Lower panel: representative middle and late-phase images from time series of cells expressing either the ESCRT-III protein CHMP4B or the AAA-ATPase VPS4. Note that both CHMP4B and VPS4 have similar localization patterns (arrows). Scale bar, 2 µm.
Because individual microtubules can be resolved by cryo-EM tomography this method was additionally used to investigate the question of how midbody microtubules change as abscission progresses. Using 3D reconstruction of individual fibers, researchers classified the microtubules into several subpopulations (Elad et al., 2011). The microtubules in the midbody form early in cell division as part of the mitotic spindle, so many of the microtubules from each daughter cell terminate at the midbody. However, other microtubules appear to cross through the midbody structure and extend to the other side of the intercellular bridge. While the continuous microtubules appear to remain intact throughout cytokinesis, the truncated microtubules reorganize during the late stages of cytokinesis. Prekeris and colleagues used cryo-EM tomography to document both microtubule buckling (which might precede or promote microtubule severing), and an increase in the number of depolymerizing microtubules just outside the dark zone (as evidenced by visible flared ends; see Figure 2A right) (Schiel et al., 2011). Interestingly, microtubule buckling was also observed in cells depleted of the microtubule- severing protein, spastin, suggesting that alternate molecules may be required to promote buckling (Schiel et al., 2011).
Cryo-EM tomography has been useful to address questions related to changes in the intercellular bridge membrane. It was known that during cytokinesis vesicles accumulate near the midbody dark zone (Schiel et al., 2011). Careful quantification of cryo-EM tomograms revealed that the calculated sum volume of organelles within the intercellular bridge can account for only ~25% of the membrane that would be needed to fully seal off the abscission site (Schiel et al., 2011). This suggested that vesicle fusion would have to be supported by other mechanisms to achieve membrane separation. Perhaps vesicles deliver lipids and proteins that promote the necessary membrane remodeling.
Cryo-EM tomography experiments provided crucial insights that address several of the dominant questions in the field. However, protein-specific information was also needed, so alternate methods were employed.
Structured Illumination Microscopy
SIM is a fluorescence imaging method that enables researchers to gain protein-specific information with spatial resolution around 100 nm in the×and y dimensions, and 350 nm in z—twice the resolution of conventional fluorescent microscopy methods such as wide-field and confocal microscopy (Gustafsson, 2000; Heintzmann and Cremer, 1999). The spatial resolution of conventional fluorescence microscopes is limited to approximately half the wavelength of light used to excite fluorophores, due to the physical properties of the light and optics. In practical terms this means that only structures that are more than 200–350 nm apart (depending on the wavelength used) can be resolved. In conventional wide-field imaging, the image created by the camera is the result of illuminating the entire field. But this image is not an accurate representation of the location of each fluorophore. In fact, the detected image results from a pattern of the constructive and destructive interference of light emitted from individual fluorophores across the entire field of view. Although structures are visible, the interference pattern contains additional information about fine sample details that cannot be extracted. In SIM, the image detected by the camera results from masking the unknown sample pattern with a known, structured illumination pattern. A series of images that together contain interference information from all fluorophores is created by illuminating the sample 15–25 times using a grated (structured) pattern of light that is rotated and phase shifted between each image acquisition. To transform the data from the interference information into improved resolution, the raw data are converted by a Fourier transform into frequency space. (Every image is a visual representation of numeric pixel values at specific locations; therefore, mathematical functions can be applied to images.) In frequency space, details about the structural information in the unknown sample pattern can be calculated using the known structured illumination pattern and the observed pattern in the image (which results from the combination of the known and unknown patterns). By combining the data from all rotations and phase shifts, a single data set is created that contains detailed information about each location in the field. The previously unusable information from the sample interference pattern is translated into improved resolution by reversing the Fourier transformation and generating a new, high-resolution image. SIM is versatile because it can be used with any fluorescent dye or protein. Combining SIM images captured at consecutive focal planes can create three-dimensional data sets (for more detailed explanation on SIM please refer to Gustafsson, 2000; Gustafsson et al., 2008).
The improved spatial resolution in SIM is achieved at the expense of temporal resolution. It takes more time to acquire 15–25 images with interference patterns than it does to acquire a single image by conventional methods. In addition, structures that would be faint using conventional fluorescent methods may be hard to visualize using SIM because high-frequency information can be lost in a subset of the images due to photobleaching during multiple exposures of the sample. However, new innovations are already being developed to mitigate these limitations (Fiolka et al., 2012; York et al., 2012).
Insights Gained from SIM
SIM is an ideal method for addressing the question of how proteins are organized during abscission because it provides increased protein-specific spatial resolution. The ESCRT machinery (composed of ESCRT-0, -I, -II, -III, and VPS4) was among the first abscission candidates examined. Both ESCRT-I and ESCRT-III proteins were found to organize in well-defined cortical rings located on either side of the dark-zone center (Figure 2B) (Elia et al., 2011; Guizetti et al., 2011). ESCRT-I rings appear to overlap partially with ESCRT-III rings located closer to the cell body. These cortical ESCRT ring complexes obtain a diameter of over one micron indicating that the ESCRT complex is indeed capable of assembling into large diameter structures and is not restricted to small diameters complexes (50–100 nm) such as MVBs and HIV (human immunodeficiency virus) buds. It is also notable that the ESCRT complex has not been found associated with internal structures within the intercellular bridge, such as vesicles. Using SIM microscopy, ESCRT-III, VPS4, and spastin were found to be associated with the constriction/abscission sites in late intercellular bridges suggesting that both the membrane fission and microtubule severing machinery can be recruited to the same site (Guizetti et al., 2011).
Only a small fraction of the known midbody proteins have been analyzed using SIM. Visualization of additional proteins will surely lead to many more insights into the mechanism of abscission. Much of the published SIM data were collected on systems designed for fixed-cell imaging. The recent emergence of technologies that enable SIM in living samples will enable researchers to add temporal information to the structures they observe (Fiolka et al., 2012; York et al., 2012). However, the speed and sensitivity of confocal imaging methods (described below) have already provided critical insights into the organization and coordination of events during abscission.
High-Speed Quantitative Fluorescence Microscopy
Cameras provide one of the fastest methods for capturing an entire field of view and technological advances have led to increased capture speeds and signal sensitivity. Electron multiplying charged couple device (EM-CCD) cameras capture a 150–300 µm2 field of view in 2 ms. Combining spinning disk confocal microscopy (which illuminates the entire field simultaneously using an array of rapidly rotating pinholes) with EM-CCD detection enables fast 3D imaging of protein and organelle dynamics in live cells. The dynamic range of EM-CCD cameras has improved due to decreases in camera noise and increases in pixel bit depth. Intensity values at each pixel in a 16-bit image are assigned a value between 0 and 65,000. In practical terms this means that very dim signals can be detected and quantified alongside intense signals. Analysis of these images is more quantitative because both mild and dramatic changes in protein dynamics can be measured in a single experiment. (For more information on spinning disk microscopy please refer to Maddox et al., 2003; Stehbens et al., 2012.)
Although spinning disk confocal microscopy is currently the most powerful technique for 4D imaging, the spatial resolution of the image is often suboptimal due to the use of a fixed pinhole size, the relatively small distance between pinholes, and the large pixel size of the camera that is needed for signal sensitivity. The resolution in z is even more limited than in x and y. In spite of these drawbacks, this high-speed fluorescent imaging method has been used extensively to assess cytokinetic abscission.
Insights from High-Speed Quantitative Fluorescence Microscopy
High-speed quantitative live-cell imaging experiments have provided insights relevant to all of the key questions in cytokinetic abscission. Measurements of the distance between the site of microtubule disassembly and the midbody center revealed that microtubule disassembly occurs about 1 µm away from the center on each side of the intercellular bridge (Elia et al., 2011). This finding, which is consistent with the cryo-EM tomography and atomic force microscopy data mentioned earlier, addresses the key question of where cell separation occurs. By analyzing the change in bridge diameter during cytokinesis progression the question of when abscission occurs was assessed. Although the intercellular bridge remains intact for over an hour, constriction of the bridge occurs acutely 20 min prior to final cleavage (Elia et al., 2011; Guizetti et al., 2011) (Figure 2C). This means that the machinery that mediates the abscission process per se probably comes into action only about 20 min prior to the final separation events.
While SIM provided critical information about the spatial organization of proteins in the intercellular bridge, high-speed confocal imaging helped address questions of temporal changes in the location of ESCRT complex assembly (Elia et al., 2011). 4D time sequence analysis established that fluorescent-tagged ESCRT proteins assemble sequentially in the intercellular bridge (Figure 2C). Arrival of CEP55, which directly recruits ESCRT components to the midbody (Carlton and Martin-Serrano, 2007; Lee et al., 2008; Morita et al., 2007), appears early in cytokinesis. The ESCRT-I component TSG101 arrives at midphase of cytokinesis and is followed by the arrival of ESCRT-III components (Elia et al., 2011). Fluorescence intensity quantification demonstrated that the levels of ESCRT-III and VPS4 proteins at the intercellular bridge increase acutely approximately 20 min prior to cell separation; coincident with the timing of microtubule bridge thinning (Elia et al., 2011; Guizetti et al., 2011). Importantly, the acute recruitment of ESCRT-III was restricted to the constriction/abscission site. Detailed analysis of time-lapse images revealed that a portion of the ESCRT-III proteins at the dark zone migrate to the abscission site as the diameter of the microtubule bundle shrinks. Activity of the ATPase, VPS4, may facilitate ESCRT-III redistribution because upon ATP depletion the ESCRT-III proteins, CHMP2A, CHMP4B, and CHMP5, extend toward the constriction sites but do not break off into two pools and the cells remain connected (Elia et al., 2012).
The question of cytoskeleton dynamics was also addressed using quantitative live-cell imaging. The end-binding protein, EB3, which binds to the plus end of growing microtubules, was visualized to assess microtubule dynamics within the bridge (Hu et al., 2012). EB3 is present along most of the intracellular bridge but is excluded from the midbody dark zone. These data indicate that in addition to stable, acetylated microtubules, a population of dynamic microtubules is present within the intercellular bridge. Actin can also be detected within the intercellular bridge during early stages; however, levels of actin decrease as cytokinesis progresses (Dambournet et al., 2011; Guizetti et al., 2011).
Quantitative, high-speed imaging has also been used to assess the spatial and temporal changes of membranes as the intercellular bridge matures. High-speed imaging of cells containing fluorescently labeled transferrin demonstrated that endocytic vesicles redistribute toward the constriction site late in cytokinesis (Hu et al., 2012). Levels of the endosomal marker Rab 8 decrease at the intercellular bridge as cytokinesis progresses (Guizetti et al., 2011), while Rab 11 containing endosomes accumulate at the bridge close to abscission (Schiel et al., 2012).
Recently, the effects of changes in lipid bilayer tension on abscission timing were investigated using high-speed confocal imaging and other microscopy tools. These studies revealed that tension release promotes abscission progression. In addition, release of membrane tension by ablation on one side of the dark zone prompted accumulation of the ESCRT-III complex at the opposite constriction site (Lafaurie-Janvore et al., 2013).
The ability to see where and when proteins are moving as abscission progresses has led to several critical insights. A limitation of this and other fluorescent-based methods is that although many proteins contribute to abscission, only a few can be monitored simultaneously. Perhaps future insights will come from application of emerging imaging methods such as spectral unmixing (Zimmermann et al., 2003).
Together, cryo-EM tomography, SIM, and high-speed confocal imaging have each contributed to answering the key questions about the location, timing, mechanism, and regulation of cytokinetic abscission. To help synthesize the results obtained through these different methods, researchers turned to advanced computational modeling that is capable of integrating measured experimental values and known physical parameters to test the feasibility of models.
Computational Modeling to Generate and Test Predictions
With improved spatial and temporal resolution from the methods just described, researchers found that although physical tension and vesicle delivery (two of the classic models described above) may contribute to daughter cell separation, ESCRT-mediated membrane fission was likely fundamental to the mechanism of abscission. Therefore, computational modeling was used to investigate specifically how ESCRT proteins contribute to cytokinetic abscission. Data on the size of the midbody, the location of the constriction sites, and the temporal and spatial organization of specific ESCRT proteins, was coupled with established knowledge, such as the energetics of membrane bending, to create and test a detailed mechanistic model of ESCRT complex function in abscission.
SIM imaging identified ESCRT components in large diameter cortical rings at the center of the intercellular bridge (Elia et al., 2011; Guizetti et al., 2011) and highly ordered helical filamentous structures were observed at the abscission site using EMtomography (Guizetti et al., 2011). These helical spirals resemble the morphology of ESCRT-III spirals observed in vitro by EM (Guizetti and Gerlich, 2012), and their appearance is dependent on expression of the ESCRT-III component CHMP2A (Guizetti et al., 2011). Ring formation on the inner surface of the membrane tubes could potentially induce constriction of the membrane and facilitate abscission. Yet the exact factors that determined the site for abscission, and how the ESCRT machinery could cause dramatic changes in the membrane diameter remained unclear.
The spatiotemporal information about ESCRT proteins in cytokinesis was integrated with previous in vitro characterization of ESCRT proteins to generate a plausible mechanistic model for cytokinetic abscission (Elia et al., 2012). Spinning disk confocal imaging had established that a portion of the ESCRT-III proteins move away from the dark zone and collect at the abscission site, indicating that the directionality of the abscission process is from the midbody center toward the constriction sites (Elia et al., 2012; Elia et al., 2011). The constriction sites are likely to form at a distance from the midbody center where the energy required to bend the membrane is minimal. When the bridge diameter is 1.2 µm, in silico calculations of the energetically preferred distance between the dark-zone bulge and the constriction site—0.71 µm, computed based on tube shape and membrane bending energy—matched the observed distance between the two pools of CHMP proteins and the distance between the end of the dark zone and site of microtubule constriction in MDCK cells (Figure 3A) (Elia et al., 2012). A new, experimentally testable prediction based on these findings is that the distance between where ESCRT-III filaments first assemble on a membrane tubule and where they constrict and cleave is a direct function of the tube diameter and membrane bending energy.
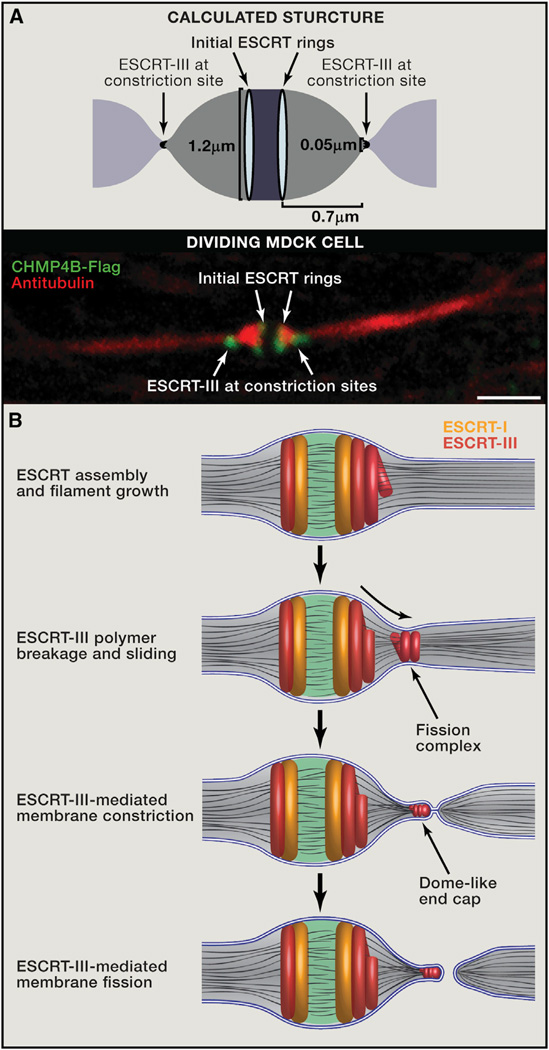
Figure 3. Mechanistic Model for ESCRT-Mediated Abscission.
(A) The calculated shape of the intercellular bridge from computational modeling (top) and the morphology of the bridge as observed by tubulin and ESCRT-III staining using spinning disk confocal microscopy in dividing MDCK cells (bottom) are very similar. The model predicts that when the bridge diameter is 1.2 µm, the optimal distance between the initial and late ESCRT-III pools will be 0.7 µm, which is similar to the distance measured experimentally. Scale bar, 2 µm.
(B) Suggested mechanistic model for ESCRT-driven cytokinetic abscission based on high-resolution microscopy data and computational modeling. As detailed in the text, cytokinetic abscission begins with the assembly of early and late ESCRT proteins into a series of partially overlapping cortical rings located at the center of the intercellular bridge. Ring formation is followed by ESCRT-III polymerization and remodeling into 3D helical spirals. Breakage and sliding of the membrane-associated ESCRT-III spiral away from the dark zone, results in acute constriction of the cytokinetic tube. This continues until the ESCRT-III spiral reaches an equilibrium distance where it relaxes to a spontaneous diameter of 50 nm. At this point fission of the 50 nmdiameter constricted membrane tube occurs, mediated by a dome-like end-cap structure, finalizing cell separation. Similar events occur on the other side of the bridge (not illustrated).
Another open question was could ESCRT-mediated membrane constriction actually bring the opposing membranes close enough to fuse? In vitro experiments have revealed that ESCRT-III proteins in membrane tubes form helical polymers with terminal dome structures that associate tightly with lipid head groups (Lata et al., 2008). It seemed possible that the helical structures observed by cryo-EM tomography could terminate in a similar dome cap. The ability of ESCRT-III proteins to bring the opposing lipid bilayers together would be dependent on how tightly the membrane associates with the ESCRT proteins as a dome-like cap forms. Calculations indicate that even if the affinity of CHMP proteins for the membrane was ~10 times less than the in vitro observed value, the association of the membrane with the helical CHMP protein structure would still be sufficient to bring the membranes close enough to undergo spontaneous fission (Elia et al., 2012; Fabrikant et al., 2009).
The information obtained from computational analysis supports the role of ESCRT proteins in mediating abscission and provides novel insights into how the biophysical properties of ESCRT proteins could influence key steps in abscission: scission site selection, membrane constriction, and membrane fission. Application of computational methods could also be used to test other mechanistic scenarios for cytokinetic abscission, for example, determining the biophysical properties of spiraled ESCRT-III filaments that enable them to bring the membranes close enough for abscission. Perhaps in the future computational analysis can be employed to investigate how changes in membrane tension and lipid composition influence the membrane tube bending energy. Additional studies could also address how removal of cytoskeletal elements from the bridge facilitates abscission. Experimental and computational methods are complimentary—additional data from cryo-EM-tomography, SIM, quantitative high-speed imaging, and other methods will facilitate the modeling of these and other questions. Future computational modeling will undoubtedly generate new predictions that can be experimentally tested.
A New Mechanistic Model
Data from cryo-EM-tomography, high-speed quantitative fluorescence imaging, structured illumination, and computational modeling experiments have revealed answers to many questions and, together, suggested a new, mechanistic, working model of cytokinetic abscission driven by ESCRT-mediated fission (Figure 3B). Early in cytokinesis, CEP55 localization to the dark zone appears to facilitate sequential recruitment of ESCRT-I and ESCRT-III proteins. Abscission could then be driven by polymerization of the central ESCRT-III ring near the dark zone into helical filaments, which polymerize away from the dark zone toward the cell body. To reach the energetically preferred diameter of approximately 50 nm, the filaments constrict the bridge membrane. Changes in membrane composition facilitated by regulation of phoshoinositides levels and endosome delivery could also contribute to membrane constriction at this step. At the same time microtubules are severed in the constricted area and actin is removed from the bridge. The AAATPase, VPS4, could mediate ESCRT-III helix remodeling during this process, which includes breaking the helix into two rings—one that remains associated with the dark zone, and a second that may slide outward, constricting the membrane further and forming an ESCRT-III fission complex. Positioning of the fission complex at the abscission site would be determined by elastic forces in the membrane, which dictate the distance from the dark zone that minimizes the energy required for membrane bending. Here, the tight association of the membrane with the ESCRT-III helix dome-like end cap could further constrict the membrane and the two bilayers would be brought into such close proximity that membrane fusion occurs in the absence of SNAREs or other conventional membrane fusion machinery. In other biological contexts, ESCRT-III proteins have been proposed to form helical polymers that constrict and facilitate cleavage of membrane buds (for example, in MVB formation and HIV particle budding) (McCullough et al., 2013). During cytokinetic abscission, the ESCRT-III helical complex acts on a membrane tube instead of a membrane bud to facilitate membrane scission.
Several aspects of this model have yet to be fully demonstrated in cells. Alternate mechanistic explanations, supported by some of the new imaging data, have been proposed to explain how the ESCRT-III complex induces membrane constriction and fission during abscission. For example, it was suggested that continuous outgrowth of ESCRT-III filaments, which decrease their diameter as they grow from the dark zone toward the constriction sites, drive constriction of the bridge (Guizetti and Gerlich, 2012). Another possibility is that nucleation of ESCRT-III filaments at the constriction sites drive membrane fission independent of ESCRT-III midzone rings (Guizetti and Gerlich, 2012). However, the agreement of the computational modeling and the experimental data supports the ESCRT-III filaments constriction and remodeling scenario described above. Resolving the dynamics of ESCRT-III filaments in the intercellular bridge with increased spatiotemporal resolution will help clarifying this point.
Irrespectively of the exact mechanistic details of cytokinetic abscission, the emergence of a revised ESCRT-mediated membrane fission model facilitated a shift in the cytokinetic field, which has already led to new discoveries. The observation that during cytokinesis ESCRT-III proteins redistribute from the midbody dark zone toward the abscission sites initiated a search for additional previously described components of the midbody dark zone at the constriction site. In a systematic characterization of the spatial dynamics of multiple midbody proteins at the intercellular bridge during different stages of cytokinesis Mitchison and colleagues discovered that several proteins relocate within the intercellular bridge during midbody maturation (Hu et al., 2012). As ingression begins in the early stages of cytokinesis, anillin, RhoA, and Septin7 localize to the midzone between the two daughter cells but by late cytokinesis, the proteins redistribute to both the dark zone and the constriction site. AuroraB kinase, a master regulator of cytokinesis, is located on both rims of the dark zone in early stages and spreads from the edge of the dark zone to the constriction sites in late bridges (and possibly further toward the cell bodies Elia et al., 2011).
The realization that the midbody protein CEP55 directly recruits ESCRT components to the membrane of the midbody dark zone further highlights the intimate association between the midbody structure and the plasma membrane surrounding it. Consistent with this, a novel membrane-binding domain was recently discovered in the conserved midbody protein complex centralspindlin (Lekomtsev et al., 2012). Mutations in this domain disrupt midbody-membrane interactions and are associated with cytokinetic failure. In addition, the ESCRT-III-binding protein MITD1 was found to tightly associate with the membrane and depletion of MITD1 causes premature abscission (Hadders et al., 2012).
The evidence that the ESCRT machinery is involved in driving abscission has further pressed the field to reconsider data that support the classic abscission models. Pulling forces generated as daughter cells move apart, which were thought to facilitate abscission in the classic mechanical rupture model, have now been shown to inhibit abscission. Release of membrane tension in late intercellular bridges leads to rapid ESCRT-III recruitment at the constriction site, followed by abscission (Lafaurie-Janvore et al., 2013). Efforts are now underway to reconcile data that contributed to the membrane fusion model. Prekeris and colleagues have recently shown that endosomes containing the Rab11 effector protein, FIP3, are essential for actin depolymerization, for secondary ingression of the plasma membrane before abscission, and for ESCRT-III recruitment to the constriction site (Schiel et al., 2012). The SNARE protein Syntaxin 16 was shown to be required for efficient delivery of endosome-associated Exocyst complexes to the intercellular bridge and for proper localization of the early ESCRT component Alix at the cytokinetic bridge (Neto et al., 2013). Together, these studies provide the first steps toward a unified inclusive model for abscission.
The newly revealed details described above indicate that temporal regulation of cytokinetic abscission is tightly coordinated. Experimental investigations have, therefore, begun to test hypotheses about how abscission timing is integrated with other aspects of cell cycle regulation. It was recently found that reorganization of the ESCRT-III component CHMP4C at the intercellular bridge is involved in abscission timing. These studies further show that CHMP4C redistribution at the intercellular bridge is AuroraB dependent, linking for the first time the ESCRT complex to the cytokinesis regulatory machinery (Capalbo et al., 2012; Carlton et al., 2012). The actin/tubulin crosslinking protein Gas2l3 was identified as a novel, constriction site protein (Pe’er et al., 2013). Because modulations of Gas2l3 cellular levels are controlled by the major cell cycle regulator, the anaphase-promoting complex/cyclosome (APC/C), it is tempting to speculate that Gas2l3 activity coordinates cell cycle regulation and abscission timing.
Future Frontiers
The results of many of the cutting-edge experiments described here have begun to answer the five key questions, and have led to significant mechanistic insights; however, many details remain unresolved. The current model suggests a possible mechanism for the selection of the abscission site, and new data indicate that abscission timing is dependent on AuroraB, but the question of why there is such a lag between the initial cytokinetic constriction and abscission initiation is still unclear. It is unknown how structural changes, like membrane tension, coordinate with changes in lipid composition to help stimulate abscission initiation. Questions also remain regarding the coordination of the cleavage on both sides of the dark zone. In many cell types, cleavage of one side is followed by cleavage on the other (Dubreuil et al., 2007; Elia et al., 2011; Guizetti et al., 2011). However, in stem cells and various cancer cells, cleavage only occurs on one side of the dark zone and the attached cell takes up the midbody remnant (Kuo et al., 2011). Additional studies are needed to understand how abscission at the second site is regulated.
Additional experiments are also needed to further define the mechanism of ESCRT-mediated membrane scission. Are the cortical spirals identified at the abscission site ESCRT-III based, as predicted? What triggers the remodeling of the ESCRT-III spiral and causes the complex to move away from the dark zone? Is VPS4 involved in remodeling individual polymers, and if so how? Are there changes in the biophysical properties of the intercellular bridge membrane that affect ESCRT recruitment or function? Our new understanding of the spatio-temporal organization of ESCRT proteins in abscission provides a framework for additional investigations. What are the roles of proteins like anillin, Rho A, and septins that also localize to the abscission site? What additional proteins function at the abscission site? Employment of the methods described here and other emerging techniques will likely reveal how these proteins coordinate with ESCRT proteins to facilitate membrane fission, cleave microtubules, and modify the lipid composition of the midbody plasma membrane.
The methods described here have provided little insight into the fifth key question regarding changes occurring within the lipid bilayer. It has been shown that reduced levels of PI3P lead to cytokinetic failure and abnormal accumulation PI(4,5)P2 leads to inhibition of cytokinetic abscission (Dambournet et al., 2011; Sagona et al., 2010) (detailed overview of the involvements of phosphatidyl inositols in cytokinesis can be found at Echard, 2012). High-speed confocal imaging did reveal that vesicles redistribute late in cytokinesis (Hu et al., 2012). It is therefore possible that the vesicles are delivering lipids (and possibly proteins) that transform the bilayer at the constriction sites, to facilitate membrane abscission. A detailed characterization of the changes in membrane composition and curvature will likely provide insights into how the opposing membranes come together in the final steps.
Improved resolution, more direct labeling approaches, and development of new tools to acutely manipulate ESCRT function in cells will lead to additional insights. New technologies are available that can be used to address some of these questions. In addition to SIM, other fluorescent superresolution methods have been developed that can resolve structures that are closer than 50 nm. These include STED (stimulated emission depletion) microscopy and point localization methods such as PALM (photoactivation localization microscopy), and STORM (stochastic optical reconstruction microscopy; for a comprehensive description of these methods, see Patterson, 2009; Schermelleh et al., 2010; Sengupta et al., 2012). Correlative fluorescence based super resolution imaging and cryo-EM tomography will undoubtedly provide unique insights, especially into the morphological changes that accompany ESCRT localization and function in abscission.
Concluding Remarks
The employment of new, advanced imaging technologies has revitalized the field of cytokinesis and led to a new understanding of the molecular components that promote the complete physical separation between two dividing cells. Together, the results from quantitative light microscopy, cryo-EM-tomography, super resolution fluorescence imaging, and computational modeling not only helped clarify the mechanism for cytokinetic abscission but also opened new research avenues to be addressed in the future. Recently, these methodologies were successfully applied to resolve another compact, dynamic cellular structure: the centrosome (Lawo et al., 2012; Lüders, 2012; Mennella et al., 2012). Application of technologic imaging innovations to other pressing cell biological questions will likely reconcile old controversies, reveal surprising insights, and generate new questions for future investigation as it has in the field of cytokinetic abscission.
ACKNOWLEDGMENTS
The authors thank Erin Fincher and Nicole Jonas from the NICHD Unit on Computer Support Services for creating the graphics used in the figures. We also thank Christian Hellriegel (Zeiss), and members of the Lippincott-Schwartz laboratory, Schuyler VanEngelenberg, Prabuddha Sengupta, Angelika Rambold, and Bennet Waxse for helpful comments on the text. N.E. is supported by the Marie Curie Intergration grant (CIG), EU and by the Israel Science foundadtion (ISF).
REFERENCES
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Capalbo L, Montembault E, Takeda T, Bassi ZI, Glover DM, D’Avino PP. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2012;2:120070. doi: 10.1098/rsob.120070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, Formstecher E, Salomon R, Goud B, Echard A. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 2011;13:981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL, Bell SD, Jensen GJ. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol. Biol. Cell. 2013;24:2319–2327. doi: 10.1091/mbc.E12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco AM, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J. Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A. Phosphoinositides and cytokinesis: the “PIP” of the iceberg. Cytoskeleton (Hoboken) 2012;69:893–912. doi: 10.1002/cm.21067. [DOI] [PubMed] [Google Scholar]
- Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- Elad N, Abramovitch S, Sabanay H, Medalia O. Microtubule organization in the final stages of cytokinesis as revealed by cryo-electron tomography. J. Cell Sci. 2011;124:207–215. doi: 10.1242/jcs.073486. [DOI] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J. 2012;102:2309–2320. doi: 10.1016/j.bpj.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, Trimble WS. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 2010;191:741–749. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG. Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc. Natl. Acad. Sci. USA. 2012;109:5311–5315. doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman K, Mader A, Zwerger M, Elia N, Medalia O. Advances in tomography: probing the molecular architecture of cells. Nat. Rev. Mol. Cell Biol. 2012;13:736–742. doi: 10.1038/nrm3453. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly:microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadders MA, Agromayor M, Obita T, Perisic O, Caballe A, Kloc M, Lamers MH, Williams RL, Martin-Serrano J. ESCRT-III binding protein MITD1 is involved in cytokinesis and has an unanticipated PLD fold that binds membranes. Proc. Natl. Acad. Sci. USA. 2012;109:17424–17429. doi: 10.1073/pnas.1206839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzmann R, Cremer CG. Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. Proc. SPIE. 1999;3568:185–196. [Google Scholar]
- Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell. 2012;23:1024–1034. doi: 10.1091/mbc.E11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada M, Nagasaki A, Uyeda TQ. Adhesion-dependent and contractile ring-independent equatorial furrowing during cytokinesis in mammalian cells. Mol. Biol. Cell. 2005;16:3865–3872. doi: 10.1091/mbc.E05-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, Weismann CM, Xu P, Houghton JM, Gao FB, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaurie-Janvore J, Maiuri P, Wang I, Pinot M, Manneville JB, Betz T, Balland M, Piel M. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 2013;339:1625–1629. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- LaFlamme SE, Nieves B, Colello D, Reverte CG. Integrins as regulators of the mitotic machinery. Curr. Opin. Cell Biol. 2008;20:576–582. doi: 10.1016/j.ceb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, Cherepanov P, Divecha N, Petronczki M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 2012;492:276–279. doi: 10.1038/nature11773. [DOI] [PubMed] [Google Scholar]
- Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J. The amorphous pericentriolar cloud takes shape. Nat. Cell Biol. 2012;14:1126–1128. doi: 10.1038/ncb2617. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Moree B, Canman JC, Salmon ED. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 2003;360:597–617. doi: 10.1016/s0076-6879(03)60130-8. [DOI] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Cytokinetic activities in a human cell line: the midbody and intercellular bridge. Tissue Cell. 1973;5:47–61. doi: 10.1016/s0040-8166(73)80005-9. [DOI] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto H, Collins LL, Gould GW. Vesicle trafficking and membrane remodelling in cytokinesis. Biochem. J. 2011;437:13–24. doi: 10.1042/BJ20110153. [DOI] [PubMed] [Google Scholar]
- Neto H, Kaupisch A, Collins LL, Gould GW. Syntaxin 16 is a master recruitment factor for cytokinesis. [Published online October 9, 2013];Mol Biol Cell. 2013 doi: 10.1091/mbc.E13-06-0302. http://dx.doi.org/10.1091/mbc.E13-06-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH. Fluorescence microscopy below the diffraction limit. Semin. Cell Dev. Biol. 2009;20:886–893. doi: 10.1016/j.semcdb.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er T, Lahmi R, Sharaby Y, Chorni E, Noach M, Vecsler M, Zlotorynski E, Steen H, Steen JA, Tzur A. Gas2l3, a novel constriction site-associated protein whose regulation is mediated by the APC/C Cdh1 complex. PLoS ONE. 2013;8:e57532. doi: 10.1371/journal.pone.0057532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- Reverte CG, Benware A, Jones CW, LaFlamme SE. Perturbing integrin function inhibits microtubule growth from centrosomes, spindle assembly, and cytokinesis. J. Cell Biol. 2006;174:491–497. doi: 10.1083/jcb.200603069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Prekeris R. Making the final cut - mechanisms mediating the abscission step of cytokinesis. ScientificWorldJournal. 2010;10:1424–1434. doi: 10.1100/tsw.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Park K, Morphew MK, Reid E, Hoenger A, Prekeris R. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. J. Cell Sci. 2011;124:1411–1424. doi: 10.1242/jcs.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol. 2012;14:1068–1078. doi: 10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Van Engelenburg S, Lippincott-Schwartz J. Visualizing cell structure and function with point-localization superresolution imaging. Dev. Cell. 2012;23:1092–1102. doi: 10.1016/j.devcel.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Pemble H, Murrow L, Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–616. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley JH. The ESCRT machinery at a glance. J. Cell Sci. 2009;122:2163–2166. doi: 10.1242/jcs.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York AG, Parekh SH, Dalle Nogare D, Fischer RS, Temprine K, Mione M, Chitnis AB, Combs CA, Shroff H. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat. Methods. 2012;9:749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T, Rietdorf J, Pepperkok R. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 2003;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]