Abstract
Purpose
Previous studies suggest that solar UV exposure in early life is predictive of cutaneous melanoma risk in adulthood, whereas the relation of BRAF mutation with sun exposure and disease prognosis has been less certain. We investigated the associations between BRAFV600E and NRASQ61R mutations and known risk factors, clinicopathologic characteristics and clinical outcomes of melanoma in a case series of primary invasive cutaneous melanoma from the Nurses' Health Study (NHS).
Methods
Somatic BRAFV600E and NRASQ61R mutations of 127 primary invasive melanomas from the NHS cohort were determined by pyrosequencing using formalin-fixed, paraffin-embedded block tissues. Logistic regression analyses were performed to detect the associations of mutations with melanoma risk factors, and Kaplan-Meier method was used to examine associations between mutations and survival.
Results
The odds ratios for harboring BRAFV600E mutations was 5.54 (95% CI, 1.19-25.8, Ptrend=0.02) for women residing in states with UV index ≥7 versus those residing in states with UV index ≤5 at 30 years of age. Patients with BRAFV600E mutations tended to have shorter melanoma-specific survival when compared to patients wild-type at both loci (median survival time 110 months vs. 159 months) (P=0.03).
Conclusions
BRAF V600E mutations in primary cutaneous melanomas were associated with residence in locations with medium and high UV indices in mid life. BRAFV600E mutation may be associated with an unfavorable prognosis among melanoma patients.
Keywords: BRAF, melanoma, NRAS, sun exposure, survival
Introduction
Cutaneous melanoma is a common form of cutanous malignancy arising from the pigment cell of the skin, and its incidence is increasing in the US as well as in other parts of the Western world (1-3). An individual's risk of developing melanoma depends on both constitutional and environmental factors. The constitutional characteristics found to be associated with increased melanoma risk include fair skin color, red hair color, increased number of atypical nevi, sun exposure sensitivity, tanning ability, etc. (4-7), whereas solar ultraviolet (UV) radiation is an established environmental risk factor (8). However, melanoma is a heterogeneous disease involved complex risk factors including genetic alterations, and previous studies support the concept that cutaneous melanoma may develop through several divergent pathways (9,10). Findings during recent years suggest that the genetic profile of cutaneous melanoma, with a particular emphasis on oncogenes BRAF and NRAS, is actively involved in these causal pathways (11-13).
BRAF mutations have been reported to occur in more than 60% of cultured melanoma cell lines and 50% of primary human melanomas, with a majority in or around codon 600 in exon 15 (V600E) (14,15). NRAS mutations have been reported to occur in 15% of human melanomas, mainly in codon 61 in exon 2 (Q61R) (15,16). Although BRAF and NRAS mutations were rarely found to occur in the same melanomas (17), both mutations have been shown to activate the RAS-RAF-MEK-ERK mitogen-activated protein kinase (MAPK) pathway and thus may play important roles in cancer initiation and progression (18,19). Furthermore, since the discovery of activating BRAF mutation in cutaneous melanoma, it has become a favored target for drug design (20), and several previous studies have tried to correlate BRAF as well as NRAS mutations with constitutional and clinical features of melanoma (6,7,17,21). Specifically, evidence from previous epidemiological studies suggest that solar UV exposure in early life is predictive of cutaneous melanoma risk in adulthood (22-24), whereas the relation of BRAF mutation with sun exposure has been less certain (12,25,26).
In the present study, we aimed to study the relations of the most common somatic BRAF (V600E) and NRAS (Q61R) mutations with a number of constitutional and environmental risk factors, including UV exposures associated with geographic variation in early to mid life, and clinical features of melanomas based on a case series from the Nurses' Health Study (NHS), a female cohort which has been prospectively followed for over 30 years since 1976.
Materials and Methods
Study participants
The participants in the study were confirmed melanoma patients with primary invasive cutaneous melanoma from the Nurses' Health Study (NHS) cohort. The NHS was established in 1976 when 121,701 married, registered, female nurses aged 30-55 years and residing in the United States at the time of enrollment responded to a baseline questionnaire that included questions about their medical history and lifestyle risk factors. Eligible participants for the present study were diagnosed with a first incident cutaneous invasive melanoma between June 1, 1978 and August 30, 2004. Hospitals across the United States where these participants were diagnosed with melanoma were asked for the participants' pathological reports and histopathology specimens (diagnostic slides and/or melanoma recuts) being sent. The study was approved by the Institutional Review Board of Brigham and Women's Hospital, and each participant's consent was obtained before her histopathology specimen was sent to our institution.
We received both pathological reports and formalin-fixed, paraffin-embedded (FFPE) block tissues from a total of 210 melanoma patients. Each FFPE block was first cut for one 4-micron hematoxylin and eosin stained section and reviewed by a pathologist for ascertainment of primary invasive melanoma. Of these samples, 59 were excluded after histopathologic review based on ineligible diagnosis or absence/insufficient of melanoma tissue for laboratory analysis. Eligible tumors (n=151) were sent to laboratory for BRAFV600E and NRASQ61R genotyping, and 127 tumors were successfully genotyped for both mutations.
Phenotypic characteristics and sun exposure history in early to mid life
Information on phenotypic characteristics and sun exposure history of the eligible melanoma patients was abstracted from the NHS cohort database. Phenotypic characteristics include family history of melanoma in first-degree relatives, natural hair color at age 21, the number of moles with a diameter ≥3mm on left arm from shoulder to wrist, propensity to tan as a child/adolescent, skin reaction to sun exposure for 2 hours or more as a child/adolescent, and the number of severe or blistering sunburns. We also asked about locations of residence (US state) at birth and at 15 and 30 years of age. The UV index is a method used to estimate UV radiation reaching the earth's surface and is important for effects on human skin on a non-cloudy day (23). Based on the mean UV index in August in North America provided by the National Oceanic and Atmospheric Administration, states (and the District of Columbia) were divided into the following 3 UV index groups: 5 or less (low UV index group: Alaska, Maine, Michigan, Minnesota, New Hampshire, Oregon, Pennsylvania, Vermont, Washington, and Wisconsin); 6 (medium UV index group: Connecticut, Delaware, Illinois, Indiana, Iowa, Maryland, Massachusetts, Missouri, Nebraska, New Jersey, New York, North Dakota, Ohio, Rhode Island, South Dakota, and West Virginia); and 7 or more (high UV index group: Alabama, Arizona, Arkansas, California, Colorado, Florida, Georgia, Hawaii, Idaho, Kansas, Kentucky, Louisiana, Montana, Mississippi, Nevada, NewMexico, North Carolina, Oklahoma, South Carolina, Tennessee, Texas, Utah, Virginia, Washington, DC, and Wyoming) (23). This grouping for northern, middle, and southern states remains the same for other months throughout the year.
BRAFV600E and NRASQ61R genotyping
Each FFPE block with confirmed diagnosis of primary invasive melanoma was recut for four 10-µm sections for DNA extraction. These sections were deparaffinized using xylene and purified with ethanol (100%). Qiagen QIAamp DNA FFPE Tissue Kit (Qiagen, Strasse, Germany) was used for DNA extraction and quantified using NanoDrop Spectrophotometer (ThermoScientific, Wilmington, DE). PCR was performed using Promega GoTaq Flexi DNA Polymerase (Promega, Madison, WI) and 10x Buffer A from Fisher Scientific Taq DNA Polymerase (Fisher Scientific, Fair Lawn, NJ). Amplified PCR products were checked for quality with 1% agarose gels (Fisher Scientific, Fair Lawn, NJ). Pyrosequencing of the BRAF and NRAS mutations was performed using the Qiagen PyroMark Q24 platform (Qiagen, Strauss, Germany). Based on the pyrosequencing results, each of the 127 tumors was assigned one of the following genotype: BRAFV600E mutation, 31 patients (24.4%); NRASQ61R mutation, 31 patients (24.4%); and wild-type at both loci, 72 patients (56.7%). Among tumors with mutations, 7 tumors (5.5%) had mutations at both loci. For analytic purposes, the 7 patients with both mutations were examined separately, and therefore the final analysis included 120 subjects.
Statistical analysis
Associations of melanoma clinicopathlogic characteristics, risk factors and clinical outcome with mutations were evaluated using the 3 major genotypes (BRAFV600E mutation, NRASQ61R mutation, and wild-type at both loci). Fisher's exact test was used to examine associations between mutation status and the following factors: histological subtype, Breslow thickness <1 mm, Clark level, and body site. The associations between genotype and ordinal variables (patient age and Breslow thickness in mm) were determined using the Kruskal–Wallis test.
Odd ratios (ORs) and 95% confidence intervals (CIs) for harboring BRAFV600E mutation or NRASQ61R mutation associated with melanoma risk factors were calculated in separate logistic regression models in SAS software (version 9.2, SAS Institute, Cary, NC) with adjustment for age as a continuous variable. Trend tests were performed by using medians of different categories (number of severe or blistering sunburns and number of moles with a diameter ≥3mm on left arm) or by modeling each variable as a single categorical variable (natural hair color, propensity to tan as a child/adolescent, skin reaction to sun exposure as a child/adolescent, and UV indices in early to mid life).
In addition, overall survival was computed from the date of diagnosis until the date of death, and women alive at the end of the study were censored at the date of last follow-up (January, 2013). Melanoma-specific survival (MSS) was identified when the diagnosis of melanoma was determined to be the primary cause of death. Survival curves were drafted using the Kaplan-Meier method and the log-rank test was used to evaluate the statistical difference between groups. Significant level was set at P<0.05 (two sided) for all statistical analyses.
Results
The eligible melanoma patients (n=120) had an overall median age at diagnosis of 61.0 years (range, 37-82 years). Table 1 shows the clinicopathologic characteristics of the melanoma patients. Patients with only NRASQ61R mutation appeared to have a higher median age at diagnosis (66 years) compared to patients with BRAFV600E mutations alone (64 years) and wild-type at both loci (61 years). The percentages of histological subtypes were 77.5% superficial spreading melanoma (SSM, n=93), 12.5% nodular melanoma (NM, n=15), 4.2% lentigo maligna melanoma (LMM, n=5), 2.5% acral lentiginous melanoma (ALM, n=3), and 3.3% unclassified melanoma (n=4). Among the tumors, 59.2% had Breslow thickness <1.00 mm (n=71), 33.3% had thickness ≥1.00 mm (n=40), and 7.5% unspecified (n=9). Tumors harboring BRAFV600E mutation were more likely to occur on the trunk whereas tumors harboring NRASQ61R mutation were more likely to occur on the lower extremity (Figure 1). However, none of the above differences between melanoma patients in different genotype subgroups reached statistical significance.
Table 1. Clinicopathologic characteristics of the melanoma patients (n=120).
| BRAFV600E (n=24) | NRASQ61R (n=24) | Wild-type (n=72) | P | |
|---|---|---|---|---|
| Median age at diagnosis, years | 64 | 66 | 61 | |
| Range, years | 37-82 | 46-81 | 37-81 | 0.80 |
| Histological subtypea, n (%)b | ||||
| SSM | 17 (71) | 19 (79) | 57 (79) | 0.74 |
| NM | 4 (17) | 3 (13) | 8 (11) | |
| LMM | 2 (8) | 0 | 3 (4) | |
| ALM | 0 | 0 | 3 (4) | |
| Missing | 1 (4) | 2 (8) | 1 (1) | |
| Breslow thickness, n (%)b | ||||
| <1.00 mm | 12 (50) | 14 (58) | 45 (63) | 0.79 |
| ≥1.00 mm | 8 (33) | 9 (38) | 23 (32) | |
| Missing | 4 (17) | 1 (4) | 4 (6) | |
| Breslow thickness | ||||
| Median (range), mm | 0.77 (0.23-9.50) | 0.83 (0.15-4.80) | 0.72 (0.24-6.00) | 0.90 |
| Clark level, n (%)b | ||||
| 2 | 7 (29) | 7 (29) | 13 (18) | 0.40 |
| 3 | 4 (17) | 5 (21) | 24 (33) | |
| 4 | 9 (38) | 9 (38) | 28 (39) | |
| 5 | 0 | 2 (8) | 2 (3) | |
| Missing | 4 (17) | 1 (4) | 5 (7) |
SMM, superficial spreading melanoma; NM, nodular melanoma; LMM, lentigo maligna melanoma; ALM, acral lentiginous melanoma.
Counts (percentage) may not sum to the total number of study subjects (100%) due to rounding.
Figure 1.
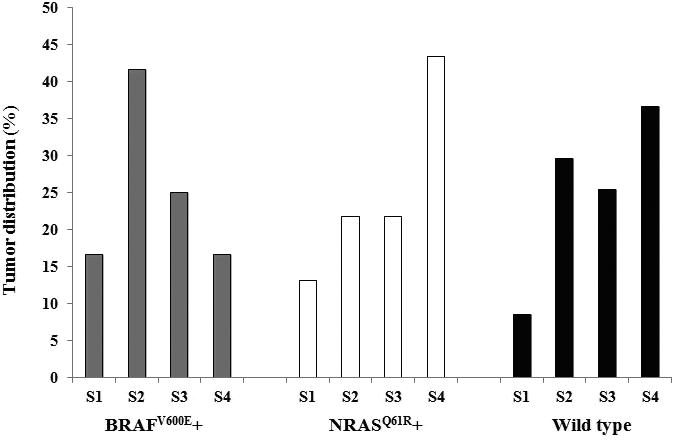
Influence of genotypes on body site distribution of the melanomas (N=118). “S1” indicates head and neck; “S2” indicates trunk (back, shoulder, chest, abdomen, and buttock); “S3” indicates upper extremity (arm, elbow, and wrist); and “S4” indicates lower extremity (thigh, knee, leg, ankle, and foot).
Table 2 shows the associations between BRAFV600E and NRASQ61R mutations and phenotypic characteristics and geographic residence variation in early to mid life. Generally, there were no clear associations between mutations and phenotypic characteristics. Compared to melanoma patients residing in states with UV index ≤5 in mid life (30 years of age), the ORs for harboring BRAFV600E mutation were 1.82 (95% CI, 0.43-7.59) and 5.54 (95% CI, 1.19-25.8) for patients residing in states with UV index of 6 and ≥7 (Ptrend=0.02), respectively. Melanoma patients residing in states with higher UV index at 15 years of age also appeared to have higher ORs for harboring BRAFV600E mutation, though the estimates were not statistically significant. In contrast, there is no significant association between NRASQ61R mutation and residence at any time point.
Table 2. Association of BRAFV600E and NRASQ61R mutations with melanoma risk factors.
| BRAFV600E (N=24) | NRASQ61R (N=24) | Wild-type (N=72) | BRAFV600E vs wild-type ORa (95% CI) | NRASQ61R vs wild-type ORa (95% CI) | |
|---|---|---|---|---|---|
|
| |||||
| n | |||||
| Family history of melanoma | |||||
| No | 22 | 21 | 63 | 1.00 | 1.00 |
| Yes | 2 | 3 | 9 | 0.65 (0.13, 3.23) | 0.98 (0.24, 3.99) |
| Natural hair color | |||||
| Dark brown/black | 9 | 8 | 23 | 1.00 | 1.00 |
| Light brown | 8 | 4 | 21 | 1.01 (0.33, 3.12) | 0.55 (0.14, 2.10) |
| Blonde/red | 3 | 9 | 18 | 0.43 (0.10, 1.84) | 1.50 (0.48, 4.72) |
| Ptrend | 0.30 | 0.51 | |||
| Number of moles with a diameter ≥3mm on left arm | |||||
| None | 6 | 14 | 32 | 1.00 | 1.00 |
| 1-2 | 3 | 3 | 11 | 1.45 (0.31, 6.81) | 0.60 (0.14, 2.53) |
| ≥3 | 5 | 2 | 12 | 2.26 (0.56, 9.17) | 0.40 (0.08, 2.07) |
| Ptrend | 0.25 | 0.22 | |||
| Propensity to tan as a child/adolescent | |||||
| No tan or light tan | 7 | 10 | 33 | 1.00 | 1.00 |
| Average tan | 8 | 9 | 21 | 1.86 (0.58, 5.92) | 1.44 (0.50, 4.13) |
| Deep tan | 5 | 2 | 8 | 3.08 (0.76, 12.4) | 0.84 (0.15, 4.64) |
| Ptrend | 0.10 | 0.88 | |||
| Skin reaction to sun exposure as a child/adolescent | |||||
| None or some redness | 10 | 10 | 35 | 1.00 | 1.00 |
| Burn | 6 | 6 | 12 | 1.65 (0.48, 5.68) | 1.72 (0.51, 5.78) |
| Painful burn or blisters | 4 | 5 | 15 | 0.93 (0.25, 3.43) | 1.17 (0.34, 4.02) |
| Ptrend | 0.95 | ||||
| Number of severe/blistering sunburns | |||||
| None | 7 | 9 | 29 | 1.00 | 1.00 |
| 1-2 times | 7 | 5 | 14 | 2.37 (0.67, 8.38) | 1.29 (0.33, 5.00) |
| ≥3 times | 4 | 4 | 13 | 1.38 (0.34, 5.63) | 1.09 (0.27, 4.44) |
| Ptrend | 0.51 | 0.87 | |||
| UV index at birth | |||||
| ≤5 | 5 | 4 | 17 | 1.00 | 1.00 |
| 6 | 14 | 11 | 27 | 1.72 (0.52, 5.67) | 1.62 (0.43, 6.05) |
| ≥7 | 3 | 2 | 12 | 0.86 (0.17, 4.29) | 0.70 (0.11, 4.47) |
| Ptrend | 0.99 | 0.85 | |||
| UV index at age 15 | |||||
| ≤5 | 3 | 4 | 19 | 1.00 | 1.00 |
| 6 | 15 | 11 | 27 | 3.43 (0.86, 13.6) | 1.84 (0.50, 6.72) |
| ≥7 | 4 | 2 | 10 | 2.45 (0.45, 13.3) | 0.88 (0.14, 5.75) |
| Ptrend | 0.25 | 0.90 | |||
| UV index at age 30 | |||||
| ≤5 | 3 | 5 | 17 | 1.00 | 1.00 |
| 6 | 10 | 10 | 30 | 1.82 (0.43, 7.59) | 1.04 (0.30, 3.64) |
| ≥7 | 9 | 2 | 9 | 5.54 (1.19, 25.8) | 0.69 (0.11, 4.37) |
| Ptrend | 0.02 | 0.76 | |||
OR values are adjusted for age at diagnosis as a continuous variable.
Forty three melanoma patients died during the follow-up, and 19 died of melanoma. The median survival time was 110 months (range, 4-366 months) for melanoma patients with BRAFV600E mutation, 128 months (range, 13-365 months) for patients with NRASQ61R mutation, and 159 months (range, 11-379 months) for patients with wild-type at both loci. The MSS showed an increasing trend over the above three genotypes (P=0.039, log-rank test for trend, Figure 2). In particular, the difference between MSS of melanoma patients with BRAFV600E mutation and patients with wild-type also reached statistical significance (P=0.03, log-rank test).
Figure 2.
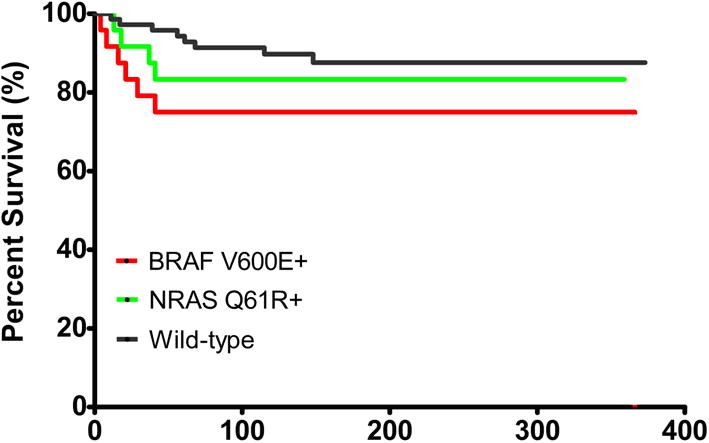
Kaplan–Meier curve for melanoma-specific survival among melanoma patients, according to mutation status of BRAFV600E and NRASQ61R. Red line: survival for patients with BRAFV600E mutation; green line: survival for patients with NRASQ61R mutation; and black line: survival for patients with wild-type at both loci.
Seven tumors were found to harbor both BRAFV600E and NRASQ61R mutations (Supplemental Table S1). The median age at diagnosis was 61 years for melanoma patients with both mutations. Other features of the both mutated tumors were not distinct from those of the other 3 genotypes. Six patients remain alive at the end of follow-up, with a median survival time of 145 months since melanoma diagnosis.
Discussion
In the present study, we provide a comprehensive analysis on the association between BRAFV600E and NRASQ61R mutations and clinicopathologic characteristics, conventional risk factors, and clinical outcome of primary cutaneous melanomas in a case series from the NHS. Our results showed that melanoma patients with BRAFV600E mutation were characterized by residence in locations with medium and high UV indices in early to mid life when compared with melanoma patients wild-type at both loci, suggesting that the risk to develop BRAF V600E mutation in participants with primary invasive melanoma is affected by UV radiation exposure received in early to mid life. Furthermore, patients with BRAFV600E mutated melanomas appeared to experience shorter survival as compared to patient with wild-type at both loci, suggesting an unfavorable prognosis associated with the mutation.
Although evidence from previous studies have identified solar UV exposure as the main environmental risk factor of melanoma (8,22-24), evidence on the association between BRAF mutation and sun exposure has been less certain. A previous study found a high mutation frequency (59%) in cutaneous melanomas without chronic sun-induced damage and much lower mutation frequencies (11-23%) in melanomas on mucosal surfaces, soles, sub-ungual sites and palms, suggesting that sun exposure may be necessary for the development of BRAF mutation (11). Another study also found the fraction of alleles carrying BRAFV600E substitution was variable but strongly associated with sun exposure (27). However, older subjects after accumulating sun exposure sufficient to produce chronic sun-induced damage in the surrounding skin also exhibit lower BRAF mutation frequencies, arguing against a simple positive link between UV exposure and BRAF mutation (11,12). Alternatively, there is evidence showing that BRAF mutations are more likely to develop in tumors located on skin subject to intermittent sun exposure (13), whereas NRAS mutations are more commonly found in tumors on skin with continuous sun exposure (17,21). In line with the previous evidence, our data also suggest that melanomas harboring BRAFV600E mutations tended to distribute more commonly on skin subject to intermittent sun exposure (trunk), and melanomas harboring NRASQ61R mutations tended to distribute more commonly on skin with continuous sun exposure (lower extremity). In addition, we found that BRAFV600E mutation is associated with residence in locations with medium and high UV indices in early to mid life (15 and 30 years of age). Thomas et al. (2007) found a high BRAFV600E mutation rate associated with UV exposure (based on residential history) before age 20 but not UV exposure after age 30 (26), and Liu et al. (2007) also reported a higher BRAFV600E mutation rate among melanoma patients who had high sun exposure as compared to those who had low sun exposure in childhood (28). It has been suggested that error-prone replication of UV-induced DNA damage could underlie the acquisition of BRAF mutations in melanocytic tumors (29). Therefore, our results together with previous findings suggest that BRAFV600E mutations may emerge in melanocytes at younger ages due to early-to-mid-life UV exposure, and these mutated cells may have a high potential to develop melanoma in subsequent adulthood.
We found a similar overall prevalence rate of somatic NRAS mutation (24.4%, 31/127) and a lower rate of overall BRAF mutation (24.4%, 31/127) as compared with previous studies (6,17,26). Interestingly, we also found that a small portion of melanoma patients had both mutations (n=7) in our samples. Previous studies generally found little overlap between BRAF and NRAS mutations (15,17,26). For example, a previous study found 3 patients with both BRAF and NRAS mutations among 233 primary melanomas (17). Several reasons may help explain the lower prevalence of BRAF mutation and relatively high prevalence of both mutations in the present study. First, we focused on the most common BRAF mutation in or around codon 600 in exon 15 (V600E), and may miss a few other BRAF mutations (e.g., V600K). However, BRAFV600E mutation accounts for more than 80% of all BRAF mutations (14,30). Second, we used data on a case series of melanoma patients from a longitudinal cohort study, which is different from sporadic patients used in other studies. Third, the different technologies used in the present study (pyrosequencing) and previous studies (e.g., PCR/single-strand conformation polymorphism analysis) to detect the mutations may also account for the difference rates in part. The NRAS mutation is generally found to be less common than BRAF mutation in cutaneous melanoma (15,31), and individuals with NRAS mutations are characterized by older age at diagnosis (21,26). Similarly, our melanoma patients with NRASQ61R mutations also had an older median age at diagnosis, though the difference between groups was not statistically significant. Due to a limited number of patients with both mutations, we were unable to identify a clear characteristic profile for these patients. Nevertheless, more advanced techniques including single cell digital PCR, next generation sequencing and immune-staining with mutation specific antibodies could assist in determining whether co-existent mutations are present in individual melanomas and single cells with a higher accuracy in future studies.
Previous reports on the associations between BRAF and NRAS mutations and survival of melanoma patients have been inconsistent. A previous study reported shorter survival associated with BRAF mutation among patients with metastatic melanomas (32), and another study also reported poor prognosis associated with NRAS mutation among patients with primary melanomas (33). However, other studies also reported a null relationship (17,21,34). In the present study, patients with BRAFV600E mutated melanomas appeared to experience shorter melanoma-specific survival when compared to melanoma wild-type at both loci (P=0.03). BRAF mutation has become a favored target for drug design since its discovery in cutaneous melanoma (20), and recent clinical studies have identified several drugs which may improve survival among melanoma patients with BRAF mutated tumors (35,36). The unfavorable prognosis of patients with BRAF mutated melanomas suggests that drug development targeting the mutated BRAF locus would be necessary to improve the survival of melanoma patients among this subgroup.
Our study has several limitations. The present analysis used data on a case series of primary melanomas from a cohort of women, and thus may limit the generalizability of the results to men. Our sample size is relatively small due to difficulties in obtaining the melanoma tissues in a prospective study setting over a long duration as well as the strict criteria in tissue processing. In addition, we did not measure sun exposure directly and used state of residence as a surrogate for sun exposure. Participants' real sun exposure may vary due to differences in personal activities (e.g., vacationing and traveling between areas with different intensities of sun exposure) which may not be captured by the measure of UV index. However, our previous study has demonstrated an increasing risk of squamous cell carcinoma, a form of skin cancer which is more likely to be sun-exposure dependent than melanoma, across the gradient of UV index (23). Therefore, case misclassification is possible but would not likely to be substantial and bias the observed high odds ratios. Finally, a few less common mutations (e.g., BRAFV600K) were not examined in the study, and we did not validate the results of pyrosequencing by other technologies and therefore may lead to potential mutation misclassification. However, previous studies have demonstrated that pyrosequencing is an efficient method for detecting point mutations in BRAF and NRAS with excellent concordance to PCR/single-stand conformation polymorphism analysis and conventional DNA sequencing of identical samples (21,37).
In conclusion, our data provide evidence that the risk of developing BRAFV600E mutation in primary cutaneous melanoma is associated with residence in locations with high UV index in mid life, patient with BRAFV600E mutated melanomas tended to have shorter survival time after diagnosis when compared to patients wild-type at both loci (BRAFV600E and NRASQ61R). These findings are supportive of sun exposure prevention in early to mid life as well as drug development targeting the mutated BRAF locus. However, due to the relatively small sample size in our study, future studies are warranted to replicate these findings.
Supplementary Material
Acknowledgments
We are deeply indebted to the staff of the Nurses' Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. This work was supported by the Department of Dermatology, Brigham and Women's Hospital, Boston, Massachusetts, and two grants from NCI (P01 CA87969 and R01 CA137365).
Footnotes
Disclosure of Potential Conflicts of Interest: AAQ serves as a consultant for Abbott, Centocor, Novartis and the Centres for Disease Control and Prevention. The other authors state no conflict of interest.
References
- 1.Geller AC, Miller DR, Annas GD, et al. Melanoma incidence and mortality among US whites, 1969-1999. JAMA. 2002;288:1719–20. doi: 10.1001/jama.288.14.1719. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Devesa SS, Hartge P, et al. Recent trends in cutaneous melanoma incidence among Whites in the United States. J Natl Cancer Inst. 2001;93:678–83. doi: 10.1093/jnci/93.9.678. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: I Common and atypical naevi. Eur J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: III Family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41:2040–59. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 6.Hacker E, Hayward NK, Dumenil T, et al. The association between MC1R genotype and BRAF mutation status in cutaneous melanoma: findings from an Australian population. J Invest Dermatol. 2010;130(1):241–8. doi: 10.1038/jid.2009.182. [DOI] [PubMed] [Google Scholar]
- 7.Qureshi AA, Zhang M, Han J. Heterogeneity in host risk factors for incident melanoma and non-melanoma skin cancer in a cohort of US women. J Epidemiol. 2011;21(3):197–203. doi: 10.2188/jea.JE20100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: II Sun exposure. Eur J Cancer. 2005;41:45–60. doi: 10.1016/j.ejca.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Whiteman DC, Watt P, Purdie DM, et al. Melanocytic nevi, solar keratoses, and divergent pathways to cutaneous melanoma. J Natl Cancer Inst. 2003;95:806–12. doi: 10.1093/jnci/95.11.806. [DOI] [PubMed] [Google Scholar]
- 10.Whiteman DC, Stickley M, Watt P, et al. Anatomic site, sun exposure, and risk of cutaneous melanoma. J Clin Oncol. 2006;24:3172–7. doi: 10.1200/JCO.2006.06.1325. [DOI] [PubMed] [Google Scholar]
- 11.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 12.Landi MT, Bauer J, Pfeiffer RM, et al. MC1R germline variants confer risk for BRAF -mutant melanoma. Science. 2006;313:521–2. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 13.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–90. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 14.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Goydos JS, MannB, Kim HJ, et al. Detection of B-RAF and N-RAS mutations in human melanoma. J Am Coll Surg. 2005;200:362–70. doi: 10.1016/j.jamcollsurg.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 16.van Elsas A, Zerp SF, van der Flier S, et al. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am J Pathol. 1996;149:883–93. [PMC free article] [PubMed] [Google Scholar]
- 17.Ellerhorst JA, Greene VR, Ekmekcioglu S, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011;17(2):229–235. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell PM, Der CJ. Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol. 2004;14:105–14. doi: 10.1016/j.semcancer.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Meier F, Schittek B, Busch S, et al. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front Biosci. 2005;10:2986–3001. doi: 10.2741/1755. [DOI] [PubMed] [Google Scholar]
- 20.Kudchadkar RR, Smalley KS, Glass LF, et al. Targeted therapy in melanoma. Clin Dermatol. 2013;31(2):200–8. doi: 10.1016/j.clindermatol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Edlundh-Rose E, Egyhazi S, Omholt K, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16:471–8. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 22.Autier P, Doré JF. Influence of sun exposures during childhood and during adulthood on melanoma risk. EPIMEL and EORTC Melanoma Cooperative Group European Organisation for Research and Treatment of Cancer. Int J Cancer. 1998;77(4):533–7. doi: 10.1002/(sici)1097-0215(19980812)77:4<533::aid-ijc10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Qureshi AA, Laden F, Colditz GA, et al. Geographic variation and risk of skin cancer in US women. Differences between melanoma, squamous cell carcinoma, and basal cell carcinoma. Arch Intern Med. 2008;168(5):501–7. doi: 10.1001/archinte.168.5.501. [DOI] [PubMed] [Google Scholar]
- 24.Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001;12:69–82. doi: 10.1023/a:1008980919928. [DOI] [PubMed] [Google Scholar]
- 25.Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164(4):776–84. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- 26.Thomas NE, Edmiston SN, Alexander A, et al. Number of nevi and early-life ambient UV exposure are associated with BRAF-mutant melanoma. Cancer Epidemiol Biomarkers Prev. 2007;16:991–7. doi: 10.1158/1055-9965.EPI-06-1038. [DOI] [PubMed] [Google Scholar]
- 27.Venesio T, Chiorino G, Balsamo A, et al. In melanocytic lesions the fraction of BRAF V600E alleles is associated with sun exposure but unrelated to ERK phosphorylation. Modern Pathology. 2008;21:716–26. doi: 10.1038/modpathol.2008.41. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, Kelly JW, Trivett M, Murray WK, Dowling JP, Wolfe R, et al. Distinct clinical and pathological features are associated with the BRAF (T1799A(V600E)) mutation in primarymelanoma. J Invest Dermatol. 2007;127(4):900–5. doi: 10.1038/sj.jid.5700632. [DOI] [PubMed] [Google Scholar]
- 29.Thomas NE, Berwick M, Cordeiro-Stone M. Could BRAF mutations in melanocytic lesions arise from DNA damage induced by ultraviolet radiation? J Invest Dermatol. 2006;126:1693–6. doi: 10.1038/sj.jid.5700458. [DOI] [PubMed] [Google Scholar]
- 30.Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36(5):486–93. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Poynter JN, Elder JT, Fullen DR, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16(4):267–73. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- 32.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29(10):1239–46. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 33.Devitt B, Liu W, Salemi R, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. 2011;24(4):666–72. doi: 10.1111/j.1755-148X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 34.Akslen LA, Angelini S, Straume O, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Invest Dermatol. 2005;125:312–7. doi: 10.1111/j.0022-202X.2005.23788.x. [DOI] [PubMed] [Google Scholar]
- 35.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 37.Sivertsson A, Platz A, Hansson J, Lundeberg J. Pyrosequencing as an alternative to single-strand conformation polymorphism analysis for detection of N-ras mutations in human melanoma metastases. Clin Chem. 2002;48:2164–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


