Abstract
We recently demonstrated that ex vivo activation of SMAD-independent BMP4 signaling in hematopoietic stem/progenitor cells (HSPCs) influences their homing into the bone marrow (BM). We here assessed if alterations in BMP signaling in vivo affects adult hematopoiesis by affecting the BM niche. We demonstrate that systemic inhibition of SMAD-dependent BMP signaling by infusion of the BMP antagonist Noggin (NGN) significantly increased CXCL12 levels in BM plasma leading to enhanced homing and engraftment of transplanted HSPCs. Conversely, the infusion of BMP7 but not BMP4, resulted in decreased HSPC homing. Using ST2 cells as an in vitro model of BM niche, we found that incubation with neutralizing anti-BMP4 antibodies, NGN or dorsomorphin (DM) as well as knockdown of Smad1/5 and Bmp4, all enhanced CXCL12 production. Chromatin immunoprecipitation identified the SMAD-binding element in the CXCL12 promoter to which SMAD4 binds. When deleted, increased CXCL12 promoter activity was observed, and NGN or DM no longer affected Cxcl12 expression. Interestingly, BMP7 infusion resulted in mobilization of only short-term HSCs, likely because BMP7 affected CXCL12 expression only in osteoblasts but not in other niche components. Hence, we describe SMAD-dependent BMP signaling as a novel regulator of CXCL12 production in the BM niche, influencing HSPC homing, engraftment and mobilization.
Keywords: Hematopoietic Stem Cells, niche, mobilization, homing, CXCL12, SMAD signaling, osteoblasts
Introduction
Hematopoietic stem cell (HSC) transplantation is commonly used for the treatment of various forms of cancers, bone marrow failure, hereditary metabolic disorders and severe congenital immuno-deficiencies 1. Within the bone marrow (BM), HSCs interact with their microenvironment wherein they reside, also termed as “niche”. HSCs were initially proposed to be associated with osteoblasts 2, whereas subsequently many HSCs were shown to be associated with the sinusoidal endothelium 3. Hematopoietic reconstitution following transplantation requires that HSCs home efficiently into the BM niches. Secretion of multiple cytokines and growth factors from cells in the BM niche regulate HSC maintenance and differentiation 4. HSCs also express a series of adhesion receptors for which ligands are expressed on different cell types in the BM, allowing homing and retention of transplanted HSCs 5. Alterations in adhesion receptor expression or their interaction with corresponding ligands within the niche, not only leads to HSC mobilization, but also to poor HSC maintenance 6.
The chemokine CXCL12 and its G protein-coupled receptor (GPCR), CXCR4, play major roles in hematopoietic stem/progenitor cell (HSPC) migration into the BM as well as their retention within the niche 7-11. Mx-cre based conditional deletion of CXCR4 from adult murine HSCs, demonstrated that CXCR4 is important for the maintenance of primitive HSCs 10. This interaction is also important for homing of human and murine HSCs following transplantation in irradiated hosts 10, 11. A CXCL12 chemokine gradient, produced by BM niche cells, attracts transplanted HSCs, which first attach to and roll over the endothelium, followed by their penetration of the tissue via trans-endothelial migration 12. Depending upon the cell type wherein it is expressed, CXCL12 affects maintenance of HSCs and lineage-committed progenitors differently 13, 14.
Although an extensive body of work exists that has evaluated the turnover and inactivation of secreted CXCL12, less is known regarding the regulation of Cxcl12 gene expression. CXCL12 expression is elevated by hypoxic conditions, as a result of HIF-1α binding to its promoter 15. Inflammatory stimuli like IL-1 and IL-6 induce CXCL12 expression in a CCAAT/enhancer binding protein β (c/EBPβ)-dependent manner 16. In addition, the promoter region of Cxcl12 contains binding sites for Sp1, AP1, NFκB, PARP1, among others 17.
Bone Morphogenetic Proteins (BMPs) are major regulators of mesoderm specification and play important roles in the development of the hematopoietic system 18, 19. In addition, they play important roles in the formation and homeostasis of bone tissue, which constitute a crucial BM niche 20. Although it is well known that BMPs can modulate bone homeostasis in postnatal life 21, 22, and that the modulation of bone mass affects adult hematopoiesis 2, 23-25, it is not known if BMP-mediated changes in osteoblast biology directly affect HSPC function. Earlier, TGF-β was shown to affect Cxcl12 expression in stromal cell lines 26. Here, we demonstrate that the regulation of CXCL12 expression within the BM niche by SMAD-dependent BMP signaling affects homing and engraftment of HSCs, as well as mobilization of hematopoietic progenitors.
Materials and methods
Animals
Six to eight week old C57BL/6J-CD45.2 (Centre d’Elevage R. Janvier, Le Genest-St Isle, France), B6.SJL-PTPRCA-CD45.1 (Charles River Laboratories, Raleigh, NC) and Col1a1-GFP mice (driven by the 2.3kb rat procollagen type 1, alpha 1 (Col1a1) promoter, generated by David Rowe 27) were bred and maintained in the animal facility at KU Leuven. During the experiments, mice were maintained in isolator cages, fed with autoclaved acidified water and irradiated food ad libitum. All experiments were approved by the Institutional Ethics Committee.
Cell culture
Primary BM stroma and ST2 cells were used to determine the effect of SMAD signaling on CXCL12 expression. The cells were maintained in RPMI 1640 medium (Gibco laboratories, Grand Island, NY) containing 10% fetal bovine serum (FBS; Gibco laboratories) and antibiotics (1% penicillin/streptomycin; Gibco laboratories). 25,000 cells were cultured in each well of 24 well plates for 48h and the effect of BMP7/Dorsomorphin (DM; Sigma-Aldrich)/noggin (NGN; R&D systems Inc., Minneapolis) was analyzed by performing quantitative RT-PCR (qRT-PCR). Alternatively, the cells were first transfected with the pGL3-basic vector containing a control or mutated CXCL12 promoter, and the effect of BMP7 (R&D Systems)/DM/Noggin was analyzed by performing dual luciferase assay.
FACS isolation of cellular components of the HSC niche
The cellular components of the BM niche were sorted using a system reported earlier 28. The detailed procedure can be found in the Supplementary methods.
qRT-PCR analysis
qRT-PCR was performed using standard protocols. The details of the procedures, reagents and the list of primers used are provided in the Supplementary methods.
esiRNA mediated knockdown of gene expression
The endoribonuclease-prepared siRNA targeting Smad1, Smad5 and Bmp4 were generated in the lab using published protocols 29. Briefly, the target region for esiRNA production was identified using the web server DEQOR 30, which assists in identifying regions in a gene that show high silencing capacity based on the base pair composition whilst assessing possible off target effects. As esiRNAs are a mixture of siRNAs, the off target effects are reduced, compared with the use of single siRNAs. The template for in vitro transcription was generated using cDNA from ST2 cells and specifically designed primers with a 5′ T7 promoter sequence were used to amplify the cDNA fragment (primer sequences in Supplementary table). The tagged cDNA fragment was then used as a template for in vitro transcription using T7 enzyme and the resulting dsRNA was digested using GST-RNase III. The digested dsRNA was purified using Q-Sepharose beads and was directly used to transfect ST2 cells for gene knockdown.
Luciferase Assay
A 3kb DNA sequence upstream of the Cxcl12 transcription start site was cloned upstream of the Luciferase gene in the pGL3-basic-vector (Promega, Madison, WI). ST2 cells were transfected with 5 μg of the plasmid containing the CXCL12 promoter as well as 0.5 μg of the control vector containing Renilla Luciferase (pRL-TK; Promega) and cultured with DM or Noggin. Firefly and Renilla Luciferase activities were assayed with the dual luciferase assay system (Promega) and Firefly Luciferase activity was normalized to Renilla Luciferase activity, as suggested by the manufacturer. All experiments were carried out in triplicate and repeated 3 times with consistent results.
Chromatin immunoprecipitation (ChIP) assay
ST2 cells were used to identify the binding site of SMAD4 to the CXCL12 promoter. ST2 cells were cultured and processed for qChIP following a protocol published earlier 31 with some modifications. Details of the procedures are provided in the Supplementary methods.
Site directed mutagenesis
The Smad Binding Element (SBE) identified to be important for SMAD4 binding to the CXCL12 promoter was deleted using the Phusion site-directed mutagenesis kit (Thermo Scientific, Hudson, NH) according to the manufacturer’s instructions. For PCR we used the 5_-phosphorylated primers listed in the Supplementary table. The PCR product was circularized with T4 DNA ligase and used for transforming E-coli competent cells. The resultant plasmids were sequenced to confirm the correct deletion of the targeted SBE.
Immuno-blotting
Immuno-blotting was performed using standard protocols and reagents. Details of the procedures and antibodies used are provided in the Supplementary methods. Band intensities were quantified using ImageJ 1.32 software (National Institutes of Health, Bethesda, MD) after densitometric scanning of the films, and normalized to β-actin or Histone-H3.
ELISA
ELISA was performed to quantify levels of CXCL12 in plasma from peripheral blood and BM of the mice infused with PBS/BMP7 (0.5mg/kg)/NGN (2mg/kg). To obtain BM plasma, hind limbs of mice were flushed with 200μl PBS. Cells were pelleted down and the plasma was used to quantify CXCL12 levels using the mouse CXCL12 ELISA immunoassay (R&D System, Minneapolis, MN) following the manufacturer’s instructions.
In vitro migration assays
In vitro trans-well migration assays were performed as described before with slight modifications 32. Details of the procedure are provided in the Supplementary methods section.
Homing assays
The homing potential of Lin− BM cells in PBS/BMP7/NGN/DM (200 μg/mouse) infused mice was analyzed using protocols used by us before 33. Details of the protocols are provided in Supplementary methods.
Serial transplantation
50,000 freshly isolated total BM cells from CD45.1 mice were transplanted into sub-lethally irradiated (5Gy) PBS or NGN (2mg/kg) infused (24h prior to transplantation) C57BL/6J-CD45.2 mice. For engraftment studies with mobilized HSPCs, peripheral blood (PB) cells were collected from CD45.1 mice injected with either PBS or BMP7 (0.5mg/kg) 24h prior. Mononuclear cells (MNCs) isolated from 1ml of PB were injected along with 1×105 competitor CD45.2 BM cells into lethally irradiated (10Gy) C57BL/6J-CD45.2 mice. PB chimerism analysis was performed every 4 weeks. After 12 weeks, primary recipients were sacrificed, BM harvested, and 1×106 cells were grafted in two secondary lethally irradiated CD45.2 mice. After 3 months, chimerism in secondary recipients was evaluated. Mice with more than 1% multi-lineage chimerism were considered engrafted.
Flowcytometry
PB and BM chimerism and multi-lineage analysis was performed by flowcytometry. Details of the procedures as well the reagents used for staining are provided in the Supplementary methods.
Statistical analysis
Data are shown as mean ± SEM. Statistical analysis was performed using a 2-tailed Student’s t-test. P-values less than 0.05 were considered statistically significant. Significance of the data and differences between groups were also assessed using one-way ANOVA (with post hoc comparisons using Dunnett’s test) using a statistical software package (GraphPad Prism 5; San Diego, CA).
Results
Systemic inhibition of SMAD-dependent BMP signaling leads to enhanced homing and engraftment of HSPCs
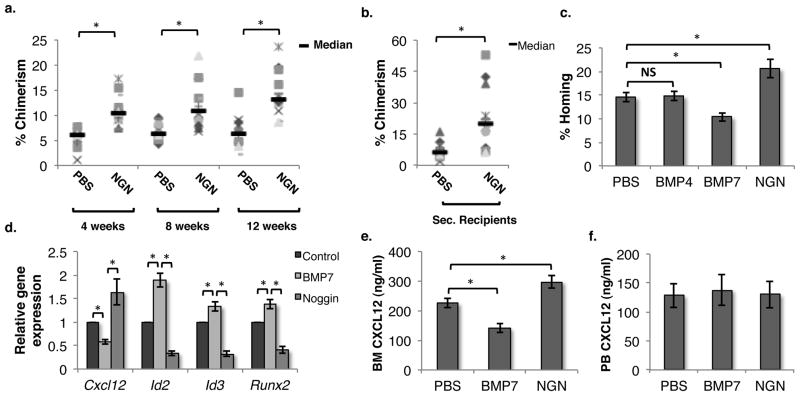
To assess the role of SMAD-dependent BMP signaling in adult hematopoiesis, we intravenously infused sub-lethally irradiated CD45.2 mice with PBS/NGN. NGN binds to BMPs and inhibits the binding to their receptors 34. 50,000 BM cells from CD45.1 mice were transplanted in the treated CD45.2 mice to assess any change in engraftment in response to alteration of BMP signaling (Fig. 1a,b). We observed a consistent increase in donor-derived chimerism at 4, 8 and 12 weeks post-transplantation in primary recipients that were pre-infused with NGN. In addition, analyses of the secondary recipients showed a 3-fold increase in long-term donor-derived chimerism with BM derived from NGN injected mice. These studies demonstrate that systemic inhibition of SMAD signaling leads to a significantly enhanced long-term repopulation ability of HSCs.
Figure 1. Inhibition of SMAD signaling in BM leads to enhanced engraftment of transplanted HSCs.
(A) 50,000 BM cells derived from CD45.1 mice were transplanted into sub-lethally irradiated CD45.2 animals pre-infused (24h prior to transplantation) with either PBS or NGN (Noggin). Donor derived chimerism was analyzed every 4 weeks. (B) After 12 weeks, primary recipients were sacrificed and 1 million BM cells were transplanted in lethally irradiated CD45.2 secondary recipients. PB chimerism was analyzed after 12 weeks. n=12, * p<0.05 (C) 100,000 BM cells derived from CD45.1 mice were injected in lethally irradiated CD45.2 hosts pre-infused with PBS, BMP4, BMP7 or NGN. The fraction of transplanted CFU-Cs that homed into the BM after 16h of transplantation was assessed. n=8, * p<0.05 (D) The effect of BMP7/NGN on Cxcl12 expression in non-hematopoietic BM cells was assessed by qRT-PCR. Mice were injected with PBS/BMP7/NGN, and after 24h, the lin−CD45− fraction of total BM cells was isolated using MACS. Expression of SMAD target genes (Id2, Id3 and Runx2) as well as Cxcl12 was analyzed by qRT-PCR. n=6, * p<0.05 (E,F) Twenty four hours after BMP7/NGN infusion, PB and BM plasma was isolated and levels of CXCL12 in the plasma was quantified by ELISA and compared with PBS injected controls. n=6–8, * p<0.04
To determine if the increased engraftment of HSPCs in NGN infused mice was due to enhanced homing, total BM cells or lin− cells were injected in lethally irradiated animals and the total number of colony forming cells (CFCs) homed within the BM after 16h of transplantation was enumerated and compared with the number of CFCs injected (Fig. 1c). The fraction of transplanted CFCs that homed into the BM was significantly higher in animals pre-treated with NGN. On the other hand, infusion of BMP7 but not BMP4 led to decreased homing of the transplanted HSPCs. We next examined the expression of key hematopoiesis regulators expressed within the niche in response to NGN infusion (Suppl. Fig. 1a). Twenty-four hours following infusion of PBS/NGN in mice, BM cells were isolated from the hind limb bones of mice following crushing and treatment with collagenase I. MACS-separated lin−CD45− BM niche cells were used to quantify gene expression by qRT-PCR. Amongst all the genes analyzed (Cd44, Cxcl12, Icam1, Vcam1, Mmp7, Mmp9, Upa, Ang1, Opn, Scf, Ccl2), we found that the expression of Cxcl12 was enhanced up to 2-fold following NGN infusion (Suppl. Fig. 1a). We repeated these experiments to analyze the status of SMAD signaling by assessing the expression of SMAD target genes in addition to Cxcl12 expression following the infusion of PBS/BMP7/NGN (Fig. 1d). Expression of the SMAD target genes Id2, Id3 and Runx2 increased following BMP7 injection, while their expression decreased upon injection of NGN compared with PBS injected controls, confirming modulation of SMAD-mediated BMP signaling in niche cells in response to the proteins injected (Fig. 1d). In addition, infusion of BMP7 increased the expression of SMAD target genes while decreasing the expression of Cxcl12 in lin−CD45− BM cells, contrary to the results from NGN infusion. We then tested if infusion of dorsomorphin (DM), a specific inhibitor of SMAD phosphorylation 35, mimicked the effect of NGN. We observed that infusion of DM also resulted in increased Cxcl12 expression in lin−CD45− BM cells (Suppl. Fig. 1b). This was also associated with an enhanced proportion of transplanted CFCs that homed to the BM within 16h after transplantation (Suppl. Fig. 1c). We also assessed the secretion of CXCL12 protein by BM niche cells by performing ELISA on BM plasma (Fig. 1e). Consistent with gene expression, NGN and BMP7 also affected the levels of CXCL12 protein in BM plasma. We did not find any significant change in the CXCL12 levels in peripheral blood (PB; Fig. 1f).
Inhibition of SMAD-mediated BMP signaling enhances CXCL12 expression in BM stromal cells
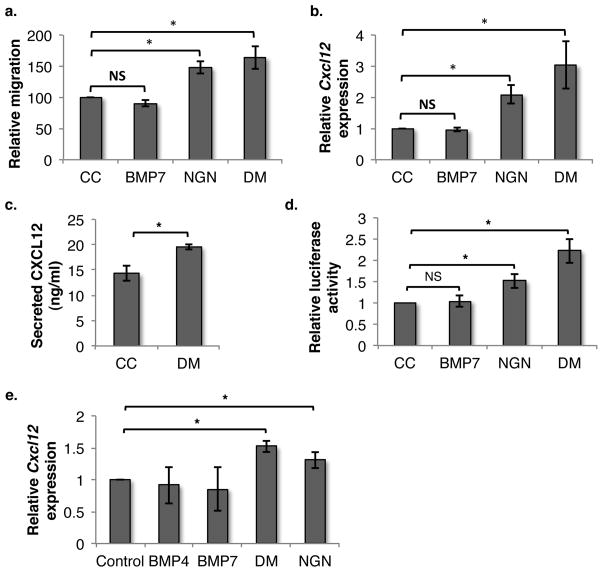
As CXCL12 is the most important chemokine known to recruit HSCs to their niche, we addressed whether its altered expression could have led to increased migration of HSPCs towards NGN-treated niche cells. We used the murine BM stroma-derived cell line ST2 as an in vitro model and performed migration assays using a trans-well system (Fig. 2a). We assessed the chemotactic migration of lineage-depleted BM cells, labeled with PKH-26, towards ST2 cells either or not treated with NGN or DM. DM is a very potent small molecule inhibitor of BMP signaling and acts by selectively inhibiting BMP type I receptors. Inhibition of BMP signaling using DM led to increased migration of HSPCs towards ST2 cells (Fig. 2a). Addition of NGN also induced increased chemo-attraction of HSPCs to ST2 cells, suggesting autocrine regulation of the pathways involved. As intravenous (iv) infusion of BMP7 inhibited homing of transplanted HSPCs, we also analyzed the migration potential of HSPCs towards BMP7 treated ST2 cells. However, unlike the in vivo effects of BMP7 on homing of HSPCs, we did not find any effect of BMP7 on migration in vitro.
Figure 2. Inhibition of SMAD-mediated BMP signaling enhances CXCL12 expression in BM stromal cells.
(A) ST2 cells cultured with BMP4/BMP7/NGN/DM were used to assess migration of HSPCs using a trans-well system n=5, * p=0.02 (NGN= noggin; DM = dorsomorhin). (B) The effect of SMAD signaling on Cxcl12 expression was analyzed by qRT-PCR on ST2 cells cultured with/without BMP4/BMP7 or inhibitors DM/NGN. n=9, * p=0.003 (C) ST2 cell were cultured with/without DM for 48h and culture supernatant was used to quantify secreted CXCL12 by ELISA. n=4, *p=0.04 (D) The CXCL12 promoter was cloned upstream of the luciferase gene in the pGL3 plasmid and ST2 cells were transfected with the vector. Effect of BMP7 or NGN/DM on Cxcl12 expression in transfected cells was assessed by Luciferase assay. n=4, * p<0,05 (E) Primary BM stromal cells were expanded and cultured in the presence of BMP4/BMP7/DM/NGN and the effect on Cxcl12 expression was compared with control by performing qRT-PCR. n=4, * p<0,05
As infusion of BMP7/NGN led to altered SMAD signaling and concomitantly changed the expression of Cxcl12 in the BM niche, we examined whether Cxcl12 expression was similarly altered in treated ST2 cells. Cxcl12 expression in ST2 cells, cultured for 2 days in the presence of the BMP inhibitors NGN or DM, was compared with that in carrier control by qRT-PCR (Fig. 2b). A significant increase in Cxcl12 expression in ST2 cells was observed upon inhibition of SMAD signaling by DM and NGN. We extended these findings by measuring the levels of CXCL12 protein in supernatant of ST2 cells treated with or without DM (Fig. 2c). Consistent with the gene expression data, we found increased levels of CXCL12 secretion by the ST2 cells cultured with DM compared with control. These results were further confirmed by performing luciferase promoter assays wherein the CXCL12 promoter was cloned upstream of the luciferase gene of the pGL3 vector. ST2 cells were transfected with the vector containing the CXCL12 promoter, and luciferase activity following treatment with carrier control or the inhibitors DM/NGN was assessed using a dual luciferase assay (Fig. 2d). Both NGN and DM enhanced CXCL12 promoter-mediated luciferase expression even if the effect of NGN was less pronounced than DM. In addition to ST2 cells, we used primary BM stromal cells to assess if Cxcl12 expression was affected by altered SMAD signaling, by performing qRT-PCR (Fig. 2e). As for ST2 cells, inhibition of SMAD-mediated BMP signaling by DM or NGN also induced increased expression of Cxcl12 in primary BM stromal cells. These results demonstrate that inhibition of BMP signaling enhances the expression of Cxcl12 in cells that belong to the BM niche. Moreover, the effect of NGN on Cxcl12 expression suggested that the SMAD-mediated BMP signaling was already active in stromal cells, either due to the presence of BMPs in the fetal bovine serum (FBS) or due to production of BMPs by these cells.
Expression of Cxcl12 in ST2 cells is controlled in autocrine fashion
While we demonstrated that inhibition of SMAD-dependent BMP signaling in ST2 cells enhanced the chemo-attraction of HSPCs and increased CXCL12 production, stimulation via BMP7 addition did not have these effects in vitro. Yet, in our in vivo studies, systemic induction of SMAD signaling by BMP7 infusion resulted in increased expression and secretion of CXCL12. We hypothesized that this discrepancy could be attributed to the differences in the expression pattern of BMP receptors in BM niche cells and ST2 cells.
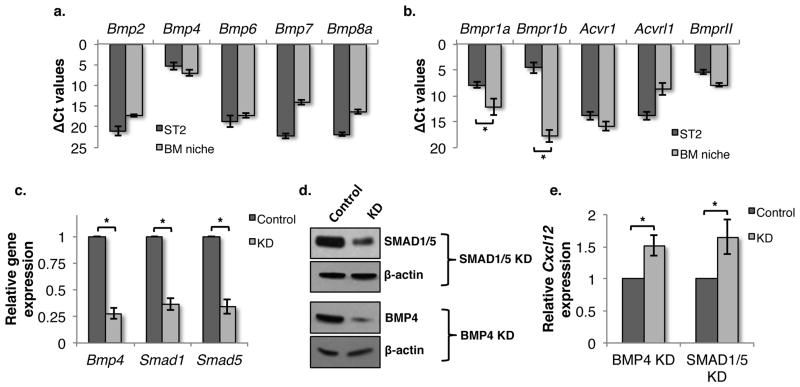
To address this, we first evaluated the expression of Bmp2, Bmp4, Bmp6, Bmp7 and Bmp8a in ST2 and primay BM derived lin−CD45− cells (representative of the BM niche) by qRT-PCR (Fig. 3a). We found high levels of Bmp4 expression in both ST2 cells and in BM niche cells. We also analyzed the expression of various BMPR genes (Bmpr1a, Bmpr1b, Acvr1, Acvrl1 and BmprII) 36 in ST2 and niche cells (Fig. 3b). We found that Bmpr1a and Bmpr1b, known to be responsible for BMP4 binding, were highly expressed in ST2 cells but not in Lin−CD45− BM niche cells. This may therefore explain why we found an effect of BMP4 in the ST2 cell-based in vitro assay system, but not following systemic infusion. Of note, we did not find high levels of transcripts for the ACVR1 gene, the major type-I receptor for BMP7 37 in Lin−CD45− niche cells, even though BMP7 inhibited homing and CXCL12 levels.
Figure 3. SMAD-mediated autocrine BMP signaling is responsible for regulation of Cxcl12 expression.
(A) Transcript levels of various Bmp’s and (B) Bmpr’s in ST2 cells and BM derived lin−CD45− cells were analyzed by qRT-PCR. (C,D) Expression of Smad1, Smad5 and Bmp4 in ST2 cells was inhibited using esiRNA-mediated knockdown. Efficiency of knockdown was quantified using qRT-PCR (C) and (D) immuno-blotting. n=4, * p<0.02 (E) Effect of Bmp4 and Smad1/5 knockdown on Cxcl12 expression in ST2 cells was assessed using a luciferase promoter assay. n=4, * p<0.04
The observation that blocking SMAD signaling via DM and NGN induced CXCL12 expression in ST2 cells, suggested that CXCL12 expression was regulated in an autocrine fashion, and likely via the production of BMP4 by ST2 cells that binds to the BMPR1A and BMPR1B receptors, all co-expressed in ST2 cells. To further substantiate this, we used anti-BMP4 antibodies to neutralize BMP4 present in the FBS or secreted by ST2 cells and assessed its effect on Cxcl12 expression (Suppl. Fig. 2). We found that addition of neutralizing antibodies against BMP4 significantly increased Cxcl12 expression. We confirmed these results by performing knockdown studies for Bmp4 as well as Smad1 and Smad5 using an endonuclease prepared siRNA (esi-RNA) approach, which has been reported to reduce off-target effects compared with siRNAs 38. We confirmed the knockdown of Bmp4, Smad1 and Smad5 by qRT-PCR (Fig. 3c) and western blotting (Fig. 3d, quantitation in Suppl. Fig. 3). Knockdown of Bmp4, Smad1 or Smad5, resulted in decreased expression of Cxcl12 (Fig. 3e), confirming the hypothesis that autocrine BMP4 signaling is responsible for regulating CXCL12 expression in ST2 cells.
Identification of SMAD binding elements responsible for regulation of Cxcl12 expression
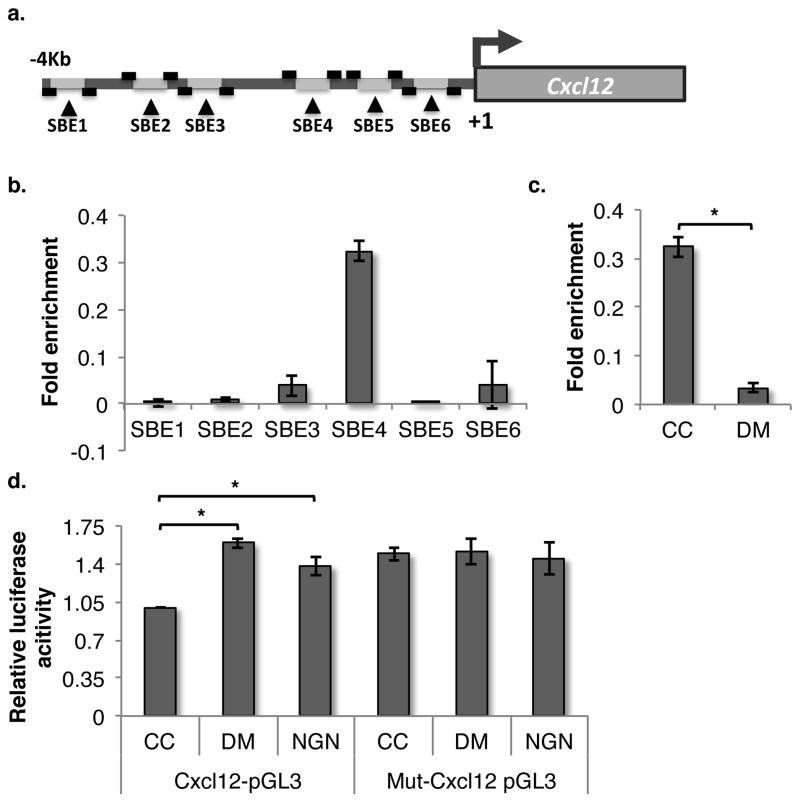
We identified 6 SMAD binding elements (SBEs; CAGACA or TGTCTG) in a region 4kb upstream of the transcription start site of the CXCL12 gene (Fig. 4a). We performed ChIP assays to determine to which of these SBEs in the CXCL12 promoter SMAD4 binds in response to BMP signaling in ST2 cells. ChIP for SMAD4 using the sheared chromatin was performed and the immuno-precipitated DNA was used as template to identify the binding sites for SMAD4 using primers flanking all 6 SBEs (Suppl. table). For each ChiP experiment, signals from isotype control antibody were subtracted from IP signals. Recovery of the amplified sequences was quantified as percent of input chromatin. Out of the 6 SBEs, we found one site, 2769bp upstream of the start site, significantly enriched in the immuno-precipitated DNA (Fig. 4b). Importantly, addition of DM that inhibits phosphorylation of R-SMADs also inhibited binding of SMAD4 to this SBE site in the CXCL12 promoter (Fig. 4c). To confirm the importance of this SBE site for SMAD4 binding, the sequence was deleted from the promoter construct using site-directed mutagenesis. The mutated promoter was cloned upstream of the luciferase gene in the pGL3 vector, and transfected in ST2 cells. Luciferase assays demonstrated that deletion of the SBE at 2769bp upstream of the transcriptional start site in the CXCL12 promoter prevented promoter activation (Fig. 4d). Moreover, addition of DM or NGN no longer affected the function of the CXCL12 promoter in ST2 cells. These experiments thus confirmed regulation of CXCL12 expression by SMAD signaling as deletion of the SBE responsible for SMAD4 binding led to increase in basal expression levels of Cxcl12.
Figure 4. Identification of SMAD binding element responsible for regulation of BMP-mediated CXCL12 expression.
(A) Graphical representation of CXCL12 promoter region that contains 6 putative SBEs. Primers were designed to detect each of the individual SBEs following ChIP analysis. (B) Levels of the 6 SBEs from CXCL12 promoter immunoprecipitated using SMAD4 antibodies measured following PCR amplification of DNA fragments. Signals from isotype controls antibodies used for each ChiP were subtracted from the SMAD4 IP signals and the fold enrichment ratios of ChIP enriched versus total input DNA were represented. (C) Effect of DM treatment of ST2 cells on SMAD4 binding to SBE4 as quantified by qChIP analysis. n=4, * p=0.01 (D) The SBE4 was deleted from the promoter of CXCL12 by PCR based mutagenesis, and the mutated (Mut-Cxcl12 pGL3) or wild-type (Cxcl12-pGL3) promoter was transfected in ST2 cells to assess the effect of DM and NGN on Cxcl12 expression in ST2 cells vector by luciferase assays. n=4, * p<0.04
Stimulation of SMAD signaling in BM leads to decrease in Cxcl12 expression in the BM niche and mobilization of ST-HSCs
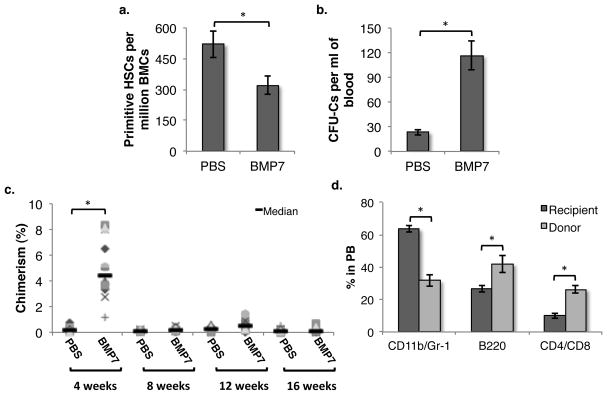
As we observed decreased levels of CXCL12 in BM plasma in response to BMP7 infusion, we examined whether mobilization of HSPCs was also affected. First, we enumerated the number of primitive HSCs, defined as CD150+CD48− KLS cells (SLAM KLS), in the BM of mice 24h after infusion of PBS or BMP7 (Fig 5a). BMP7 injection resulted in a decrease in the frequency of primitive HSCs in the BM. To determine if there was any change in circulating HSPCs in the PB, we performed methylcellulose colony assays and enumerated the frequency of CFCs in PB following PBS or BMP7 injection (Fig. 5b). We observed a 4.96±1.53 fold (p=0.003) increase in the frequency of circulating CFCs in PB of mice infused with BMP7.
Figure 5. Stimulation of SMAD signaling in the bone marrow leads to decreased Cxcl12 expression in the BM niche and mobilization of ST-HSCs.
Mice were treated with PBS/BMP7 by iv injection in the tail vein. (A) After 24h, the frequency of primitive HSCs in the BM was assessed by performing flowcytometric detection of CD150+CD48− KLS cells. (B) Following PBS/BMP7 infusion, the frequency of circulating HSPCs was quantified by methylcellulose based colony formation assays. The number of CFCs per ml of PB was plotted. n=8, *p=0.007. (C) PB derived MNCs from PBS/BMP7 injected CD45.1 mice were transplanted along with 100,000 total BM cells from CD45.2 mice in lethally irradiated CD45.2 mice. Donor derived chimerism was analyzed every 4 weeks. n=12, * p=0.0015 (D) Multi-lineage potential of the donor derived cells was examined by using antibodies against CD11b/Gr-1 (macrophage/granulocyte), B-220 (B-lymphocytes), CD4/CD8 (T-lymphocytes) in addition to CD45.1/CD45.2. Proportion of differentiated cells in each lineage within CD45.1 cell fraction of PB after 4 weeks of transplantation is presented. n=8, * p<0.05.
As BMP7 increased, even if relatively modestly, the number of circulating CFCs, we assessed the ability of HSPCs mobilized by BMP7 to reconstitute hematopoiesis in lethally irradiated hosts. MNCs derived from 1ml of PB of CD45.1 mice that were injected intravenously with either PBS/BMP7 24 h earlier, along with 100,000 total BM cells from CD45.2 mice were transplanted in lethally irradiated CD45.2 animals. Chimerism was analyzed every 4 weeks (Fig. 5c). After 4 weeks, we observed engraftment of the PB derived cells from BMP7 treated but not control mice in all mice transplanted. Surprisingly, PB derived cells from BMP7 treated mice did not contribute to hematopoiesis from 8 weeks onwards, indicating that BMP7 mobilizes short-term (ST) HSPCs but not long-term (LT) HSCs. After 16 weeks we sacrificed the primary recipients to analyze BM repopulation by PB cells from BMP7 or control treated mice. We did not find any contribution of cells from BMP7 or control treated mice to the hematopoietic compartment in the BM (Suppl. Fig. 4a) confirming the hypothesis that the transplanted cells were indeed ST-HSPCs. Interestingly, multi-lineage analysis from the donor derived cells revealed a bias towards lymphoid engraftment. The proportion of both B- and T-cells derived from donor-derived cells was significantly higher than the recipient’s derived cells (Fig. 5d, Suppl. Fig. 4b). On the other hand, myeloid differentiation potential of the donor-derived cells was lower than the recipient.
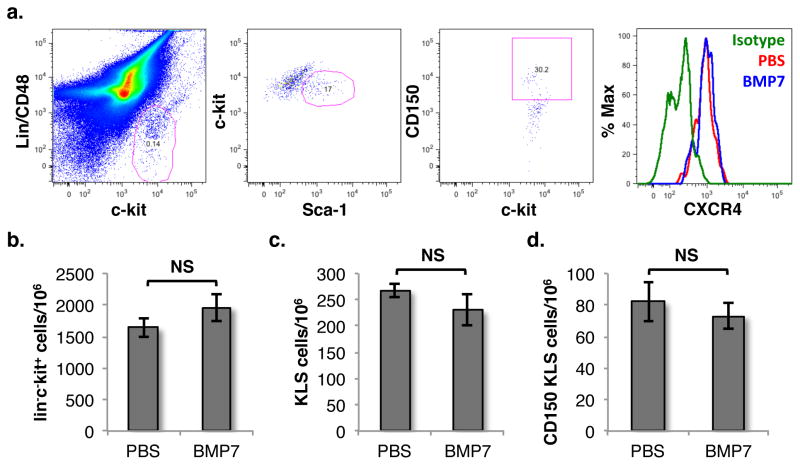
We next examined whether there was any effect of BMP7 infusion on the frequency of HSCs within the BM or CXCR4 expression on these cells, which could also lead to an increased proportion of circulating HSPCs in the PB (Fig. 6). We harvested BM cells from PBS/BMP7 infused mice and assessed the expression of CXCR4 on SLAM KLS cells (Fig. 6a). We did not observe any change in the expression of CXCR4 on these cells. We also assessed the expression of CXCR4 on HSCs that migrated in chemotactic response to ST2 cells cultured with or without NGN (Suppl. Fig. 5). We did not find any change in CXCR4 expression on HSCs that migrated towards NGN treated ST2 cells as compared with control cells. Finally, we also assessed if there was any effect NGN infusion on the frequency of hematopoietic progenitors as well as HSCs within the BM (Fig. 6b–d). We could not detect any change in the frequency of HSCPs (lin−c-kit+), KLS or SLAM KLS cells following BMP7 infusion. This rules out the possibility that mobilization of hematopoietic progenitors was caused by an increased number of HSC within the BM or by changes in the expression of CXCR4 expression on these cells.
Figure 6. BMP7 infusion does not change expression of CXCR4 expression or frequency of HSCs.
(A) Mice with infused with PBS/BMP7 and BM cells were harvested after 24h. CXCR4 expression on lin−c-kit+Sca-1+CD48−CD150+ (SLAM KLS) cells was analyzed by flowcytometry using specific antibodies. (B–D) Number of HSPCs, KLS cells and SLAM KLS cells per million of the total BM cells was quantified by flowcytometry. n=6, ns p>0.05
BMP7 affects Cxcl12 expression specifically in osteoblasts within the BM niche
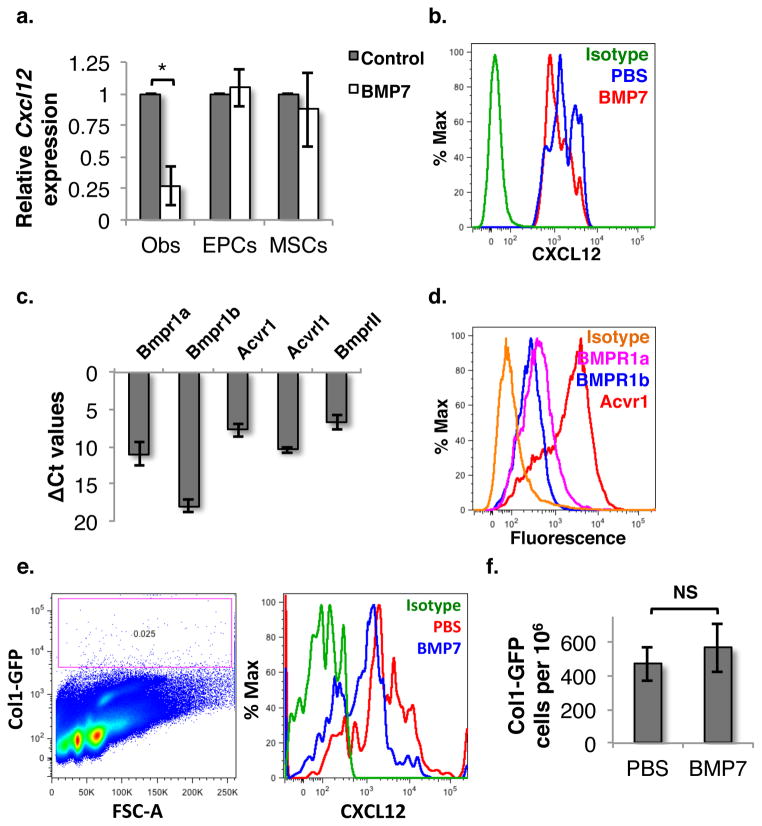
As it has been demonstrated that deletion of Cxcl12 expression in different cell types of the BM niche affects different subsets of HSPCs, we assessed in which cell type within the BM niche Cxcl12 expression changed in response to infusion of BMP7. Control or BMP7 treated animals were sacrificed and BM cells were isolated following crushing the hind limb bones and treating with collagenase I. Cellular components of the HSC niche were sorted based on a compliment of cell surface antigens known to be expressed on these subpopulations (Suppl. fig. 6, upper panel): Mesenchymal stromal/stem cells (lin−CD45−CD31−CD51+Sca-1+; MSCs), osteoblasts (lin−CD45−CD31−CD51+Sca-1−; OBs) and endothelial progenitor cells (lin−CD45−CD31+; EPCs), as published earlier 28. The sorted cells were used for RNA isolation followed by amplification and quantitation of expression of Cxcl12 using qPCR (Fig. 7a). These studies demonstrated that expression of Cxcl12 in EPCs and MSCs was unchanged in response to BMP7 infusion, whereas BMP7 treatment significantly decreased Cxcl12 expression in osteoblasts. Using FACS we confirmed that CXCL12 protein levels were lower in osteoblasts (lin−CD45−CD31−CD51+Sca-1−) from BMP7 injected mice compared with control mice (Fig. 7b). These results were also confirmed by using collagen type I (Col1) as the marker for osteoblasts. Col1-specific antibodies were used to identify osteoblast population in lin−CD45−CD31−Sca-1− cells (Suppl. Fig. 6 lower panel).
Figure 7. BMP7 infusion results in decreased Cxcl12 expression specifically in osteoblasts.
(A) BM cells were harvested from mice treated with or without BMP7 for 24h. Cellular components of HSC niche; endothelial progenitor cells (EPCs), mesenchymal stroma/stem cells (MSCs) and osteoblasts (Obs) were isolated by FACS. Total RNA was isolated from EPCs, MSCs and Obs, and subjected to qRT-PCR for Cxcl12 and levels compared between cells derived from BMP7 treated or control mice (n=3, p=0.04). (B) CXCL12 expression in Obs in BM cells of mice infused with PBS/BMP7 was analyzed by flowcytometry (n=4). (C) Gene expression levels of BMP receptors assessed by qRT-PCR on amplified RNA obtained from osteoblasts sorted from BM. (D) Flowcytometric analysis confirming expression of the BMP7 receptor Acvr1 in comparison to BMPR1a and BMPR1b in osteoblasts. (E,F) Col1a1-GFP mice infused with/without BMP7, were sacrificed and BM cells were harvested by enzymatic digestion. Expression of CXCL12 in Col1a1-GFP+ osteoblasts (E) as well their proportion in BM cells (F) was examined by flowcytometry using specific antibodies.
We also assessed the expression of different BMPR genes on osteoblastic cells by qRT-PCR (Fig. 7c) and flowcytometry (Fig. 7d). Sorted osteoblastic cells from control mice were subjected to qRT-PCR to examine the expression of various BMPR genes. We found that amongst all type I BMPRs analyzed, Acvr1 showed the highest expression. We confirmed that ACVR1 was expressed on the cell surface of osteoblasts by flowcytometry (Fig. 7d).
To confirm the effect of BMP signaling on CXCL12 expression, specifically in osteoblasts, we used Col1a1-GFP reporter mice. First, we used this system to confirm the phenotypic markers of osteoblasts used in experiments described above. We found that 73.42±4.52% of Lin−CD45−CD31−CD51+Sca-1− BM cells were also Col1a1-GFP+ (Suppl. Fig. 7a) confirming that these phenotypic markers successfully identified osteoblasts. Next, mice were infused with either PBS or BMP7, and after 24h all BM cells were harvested by enzymatic digestion of crushed bones. The expression of CXCL12 in Col1a1-GFP+ osteoblasts was detected by flowcytometry (Fig. 7e). As in earlier experiments, we observed a decrease in CXCL12 expression in Col1a1-GFP+ cells following BMP7 infusion. We also assessed whether BMP7 treatment led to any change in the number of osteoblasts (Fig. 7f). We could not detect a significant change in the number of Col1a1-GFP+ osteoblasts following BMP7 infusion as compared with the PBS infused animals within the timespan of the experiment. We also used BrdU incorporation assays to assess the proliferation of osteoblasts (Suppl. Fig. 7b). Consistent with the fact that the osteoblast number was unchanged, we did not see any change in their proliferation status following BMP7/DM infusion. Therefore, the effect of altered BMP signaling on CXCL12 expression within the BM niche could not be linked to changes in the absolute number of osteoblasts. Thus, we conclude that osteoblasts exhibit significantly decreased CXCL12 expression following BMP7 infusion in vivo.
Discussion
Although it is well established that BMP signaling is of great importance during fetal hematopoiesis, the role of BMP signaling in adult hematopoiesis is less well understood. Others and we have demonstrated that SMAD-dependent BMP signaling is dispensable for postnatal HSPC functions 33, as elimination of SMAD1 and SMAD5 expression in HSPCs did not affect their ability to sustain hematopoiesis upon transplantation 39. However, BMPs are present in the BM microenvironment and it has been shown that conditional inactivation of the BMP receptor type IA (BMPRIA) causes expansion of osteoblasts as well as HSPCs, indicating an indirect effect of BMPs on adult hematopoiesis 40. We here demonstrated that infusion of BMP7 systemically led to decreased homing and engraftment of transplanted HSCs while infusion of NGN showed the opposite effect. Screening for important hematopoiesis regulators within niche cells identified altered expression of Cxcl12 in lin−CD45− BM cells (containing all HSC niche cells). The chemokine CXCL12 mediates chemotactic migration of transplanted HSPCs towards the BM niche supporting their homing 9. Deletion or inhibition of CD26, an enzyme that cleaves CXCL12, in vivo, leads to increased homing and engraftment of transplanted HSCs 28, 41.
Activation of SMAD signaling via infusion of BMP7 (but not BMP4) led to decreased Cxcl12 expression and secretion in the BM niche. Correspondingly, we detected increased numbers of mobilized HSPCs in the peripheral blood, as demonstrated by increased frequency of CFCs in the blood following BMP7 infusion. HSPCs mobilized by G-SCF are widely used clinically as a source for transplantation 42. In healthy allogeneic stem cell donors, G-CSF results in variable mobilizing efficiency 43 and mobilization of HSPCs to the blood in patients for autologous transplantation is especially poor 44, 45. In fact, patients who received previous chemo- and/or radio-therapy have a 20–25% risk of failing to mobilize HSPCs efficiently 46. Multiple mobilization attempts in patients have been shown to adversely affect their quality of life, not to mention the cost of treatment 47. Small molecules such as Plerixafor (AMD3100) that interfere with the interaction of HSPCs with their microenvironment have been reported to aid in G-CSF mediated mobilization 48, 49. Ramirez et al. showed that BIO5192, a small molecule inhibitor of VLA-4, leads to up to 30-fold increase in PB HSPCs 50. When used along with Plerixafor, it increased the efficacy of HSPC mobilization 17-fold more, demonstrating the importance of targeting multiple pathways for mobilization. The finding that BMP7 also enhances CFC mobilization may therefore be of significant clinical importance.
We used mouse BM stroma-derived ST2 cells (in addition to OP9 and ac11 cells; data not presented in this manuscript) as in vitro model of BM niche cells to understand the underlying processes that lead to alteration of Cxcl12 expression by SMAD-mediated BMP signaling. ST2 cells have been shown to support both myelopoiesis and lymphopoiesis 51. We found that BMP4 is highly expressed by ST2 cells, and by using neutralizing antibodies we found that BMP4 regulates Cxcl12 expression in an autocrine SMAD-dependent manner. This was confirmed by knockdown of Bmp4 or Smad1/5 expression in ST2 cells, which resulted in enhanced Cxcl12 expression. Interestingly, while BMP4 secreted by ST2 cells affected the expression of Cxcl12, infusion of BMP4 did not affect Cxcl12 expression in vivo; in contrast, BMP7 infusion did affect Cxcl12 expression in vivo. This could be due to differences in the expression of the BMPRs between ST2 cells and BM niche cells. While ST2 cells expressed Bmpr1a and Bmpr1b as type I BMP receptors, osteoblasts within the BM niche expressed significantly higher levels of Acvr1, which is known to act as the main receptor for BMP7 37. These findings likely explain why infusion of BMP7 only affects CXCL12 expression in osteoblasts, while addition of BMP7 to ST2 cells that do not express ACVR1 does not affect CXCL12 expression.
Upon ligand binding, the R-SMADs (SMAD1/5) are phosphorylated by BMP type I receptors and bind to the common SMAD (SMAD4) 52. This complex is then translocated to the nucleus and controls transcription of various genes, either by binding to the promoter regions themselves or by associating with other DNA binding transcription factors 53. The crystal structure of the complex formed between the SMAD DNA binding domain, MH1, and SMAD binding elements (SBEs) identified the minimal SBE sequence to be GTCT (or its complementary CAGA) 54. In many instances SBEs have been reported to have 1–2 extra bases 55. Hence, we used CAGACA to determine if SBEs are present in the CXCL12 promoter region. We identified 6 SBEs in the promoter region of Cxcl12. The SBE at 2769bp upstream of the transcriptional start site in the CXCL12 promoter is responsible for SMAD-mediated repression of Cxcl12 expression in ST2 cells.
The CXCL12-CXCR4 signaling axis plays not only a major role in retention of HSCs in the BM but also in maintaining the HSC pool 10, which explains why disruption of the CXCR4/CXCL12 axis leads to HSPC mobilization and HSC exhaustion 56. Our experiments showed mobilization of only ST-HSPCs in response to infusion of BMP7. Two recent reports suggest the importance of the cell type in which CXCL12 is expressed on HSC maintenance 13, 14. These studies demonstrate that various cells in the BM including mesenchymal stromal cells, perivascular stromal cells, endothelial cells, osteoblasts as well as some hematopoietic cells express Cxcl12. Conditional elimination of Cxcl12 from Tie2+ endothelial cells in the BM affects HSC maintenance. When Cxcl12 expression was eliminated from leptin receptor (Lepr)-positive perivascular stromal cells, no effect on HSC maintenance was seen even if there was a significant increase in circulating HSPCs, along with higher numbers of HSPCs in spleen. Cxcl12 deletion from Prx1-positive mesenchymal stromal cells significantly decreased HSC maintenance in the BM. However, elimination of Cxcl12 expression in Osx-positive osteoprogenitors or Col1a1[2.3kb]-positive osteoblasts did not affect overall HSC maintenance but had a specific effect on lymphoid progenitor numbers 13, 14. Our data supported these results as the hematopoietic progenitors mobilized following BMP7 infusion demonstrated lymphoid bias when transplanted in irradiated hosts.
Altogether, our data presents a novel mechanism by which CXCL12 expression is regulated in the BM niche. We presented evidence that by increasing CXCL12 production by inhibition of SMAD signaling within the BM niche, it was possible to increase homing and engraftment potential of transplanted HSPCs. Contrary to this, decrease in CXCL12 expression led to mobilization of at least the ST-HSPCs.
Supplementary Material
Supplementary figure 1. (A) The effect of NGN on the expression of various known hematopoiesis related genes in non-hematopoietic BM cells was assessed by qRT-PCR. Mice were injected with PBS/NGN, and after 24h, the lin−CD45− fraction of total BM cells was isolated using MACS. Expression of genes such as Cd44, Cxcl12, Icam1, Vcam1, Mmp7, Mmp9, Upa, Ang1, Opn, Scf, Ccl2 as well as Cxcl12 was analyzed by qRT-PCR. n=4, * p=0.034 (B) Primary BM stromal cells cultured with BMP7/DM were analyzed for expression of Cxcl12 by performing qRT-PCR. n=4, * p,0.05 (C) Proportion of transplanted CFCs homed within 16h of transplantation was compared between DM infused and control animals. n=8, * p=0.041
Supplementary figure 2. The effect of SMAD signaling on Cxcl12 expression was analyzed by qRT-PCR on ST2 cells cultured with/without neutralizing antibodies against BMP4. n=4, * p=0.02
Supplementary figure 3. Expression of Smad1, Smad5 and Bmp4 in ST2 cells was inhibited using esiRNA-mediated knockdown. Efficiency of knockdown was quantified using immuno-blotting. Change in protein expression was quantified by measuring band intensity using ImageJ software. n=4, * p<0.04
Supplementary figure 4. PB derived MNCs from PBS/BMP7 injected CD45.1 mice were transplanted along with 100,000 total BM cells from CD45.2 mice in lethally irradiated CD45.2 mice. After 16 weeks, mice were sacrificed and BM derived cells were analyzed for presence of donor derived cells by flowcytometry (A) as well as multi-lineage engraftment in PB was analyzed using (B) using antibodies against CD45.1/CD45.2 in addition to CD11b/Gr-1 (macrophage/granulocyte), B-220 (B-lymphocytes), CD4/CD8 (T-lymphocytes) (4 weeks data shown here suppl fig. 4B) (n=8).
Supplementary figure 5. Expression of CXCR4 on the HSCs, identified as lin−c-kit+Sca-1+CD48−CD150+, migrated towards NGN treated versus control ST2 cells was analyzed by flowcytometry.
Supplementary figure 6. PBS or BMP7 treated animals were sacrificed and BM cells were isolated following crushing the hind limb bones and treating with Collagenase I. (Upper panel) Cellular components of the HSC niche were sorted on the basis of their phenotype; Mesenchymal stem cells (MSCs; lin−CD45−CD31−CD51+Sca-1+), osteoblasts (OBs; lin−CD45−CD31−CD51+Sca-1−) and endothelial progenitor cells (EPCs; lin−CD45−CD31+). (Lower panel) Collagen I was used as specific marker to identify osteoblasts within lin−CD45−CD31−Sca-1− population.
Supplementary figure 7. (A) BM cells from Col1a1-GFp mice were harvested by flushing followed by enzymatic treatment of the crushed bone pieces. Flowcytometry analysis was performed to examine the GFP expression in osteoblasts identified by phenotypic markers (lin−CD45−CD31−CD51+Sca-1−). (B) Col1a1-GFP mice were infused with PBS/BMP7/DM in addition to BrdU. BM cells were harvested after 24h and BrdU incorporation in Col1-GFP+ fraction of lin−CD45−CD31− cells from the three groups was analyzed by flowcytometry.
Acknowledgments
The work was supported by an FWO grant (1.2.665.11.N.00) to S Khurana; FWO funding (G085111N), NIH-PO1-CA-65493-06, Odysseus funding, CoE and GOA/11/012 funding from KU Leuven, and the Vanwayenberghe fund to CM Verfaillie. The authors wish to thank Prof. An Zwijsen for critical review of the accurateness of the manuscript. David Rowe (University of Connecticut Health Center) and Cristina LoCelso (Imperial College London) are acknowledged for providing Col1a1-GFP mice.
Footnotes
Author Contribution: S.K. Designed and performed the experiments, analyzed the data, wrote the manuscript; A.M. performed the experiments, analyzed the data; R.Y. performed the experiments, analyzed the data; S.S. Performed FACS analyses and some of the murine homing and engraftment studies; T.N. designed esiRNA based experiments, produced esiRNAs; M.P. conceived, supervised and analyzed ChIP experiments; C.M. provided materials and reviewed the manuscript; C.V. supervised all the research and edited the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
References
- 1.Gratwohl A, Baldomero H, Aljurf M, et al. Hematopoietic stem cell transplantation: a global perspective. Jama. 2010;303:1617–1624. doi: 10.1001/jama.2010.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 3.Kiel MJ, Yilmaz OH, Iwashita T, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 4.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 5.Taichman RS. Blood and bone: two tissues whose fates are intertwined to create the hematopoietic stem-cell niche. Blood. 2005;105:2631–2639. doi: 10.1182/blood-2004-06-2480. [DOI] [PubMed] [Google Scholar]
- 6.Lemoli RM, D’Addio A. Hematopoietic stem cell mobilization. Haematologica. 2008;93:321–324. doi: 10.3324/haematol.12616. [DOI] [PubMed] [Google Scholar]
- 7.Dar A, Kollet O, Lapidot T. Mutual, reciprocal SDF-1/CXCR4 interactions between hematopoietic and bone marrow stromal cells regulate human stem cell migration and development in NOD/SCID chimeric mice. Exp Hematol. 2006;34:967–975. doi: 10.1016/j.exphem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Broxmeyer HE, Orschell CM, Clapp DW, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aiuti A, Webb IJ, Bleul C, et al. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama T, Kohara H, Noda M, et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Lapidot T, Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16:1992–2003. doi: 10.1038/sj.leu.2402684. [DOI] [PubMed] [Google Scholar]
- 12.Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006;116:1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceradini DJ, Kulkarni AR, Callaghan MJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 16.Calonge E, Alonso-Lobo JM, Escandon C, et al. c/EBPbeta is a major regulatory element driving transcriptional activation of the CXCL12 promoter. J Mol Biol. 2010;396:463–472. doi: 10.1016/j.jmb.2009.11.064. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Moruja C, Alonso-Lobo JM, Rueda P, et al. Functional characterization of SDF-1 proximal promoter. J Mol Biol. 2005;348:43–62. doi: 10.1016/j.jmb.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Winnier G, Blessing M, Labosky PA, et al. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 19.Marshall CJ, Kinnon C, Thrasher AJ. Polarized expression of bone morphogenetic protein-4 in the human aorta-gonad-mesonephros region. Blood. 2000;96:1591–1593. [PubMed] [Google Scholar]
- 20.Cao X, Chen D. The BMP signaling and in vivo bone formation. Gene. 2005;357:1–8. doi: 10.1016/j.gene.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu XB, Li Y, Schneider A, et al. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J Clin Invest. 2003;112:924–934. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazzerro E, Pereira RC, Jorgetti V, et al. Skeletal overexpression of gremlin impairs bone formation and causes osteopenia. Endocrinology. 2005;146:655–665. doi: 10.1210/en.2004-0766. [DOI] [PubMed] [Google Scholar]
- 23.Kollet O, Dar A, Shivtiel S, et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 24.Lymperi S, Ersek A, Ferraro F, et al. Inhibition of osteoclast function reduces hematopoietic stem cell numbers in vivo. Blood. 2011;117:1540–1549. doi: 10.1182/blood-2010-05-282855. [DOI] [PubMed] [Google Scholar]
- 25.Visnjic D, Kalajzic Z, Rowe DW, et al. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- 26.Wright N, de Lera TL, Garcia-Moruja C, et al. Transforming growth factor-beta1 down-regulates expression of chemokine stromal cell-derived factor-1: functional consequences in cell migration and adhesion. Blood. 2003;102:1978–1984. doi: 10.1182/blood-2002-10-3190. [DOI] [PubMed] [Google Scholar]
- 27.Kalajzic I, Kalajzic Z, Kaliterna M, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 28.Khurana S, Margamuljana L, Joseph C, et al. Glypican-3-mediated inhibition of CD26 by TFPI: a novel mechanism in hematopoietic stem cell homing and maintenance. Blood. 2013;121:2587–2595. doi: 10.1182/blood-2012-09-456715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kittler R, Heninger AK, Franke K, et al. Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods. 2005;2:779–784. doi: 10.1038/nmeth1005-779. [DOI] [PubMed] [Google Scholar]
- 30.Henschel A, Buchholz F, Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pistoni M, Verrecchia A, Doni M, et al. Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a. Embo J. 2010;29:1215–1224. doi: 10.1038/emboj.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zigmond SH, Hirsch JG. Leukocyte locomotion and chemotaxis. New methods for evaluation, and demonstration of a cell-derived chemotactic factor. J Exp Med. 1973;137:387–410. doi: 10.1084/jem.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khurana S, Buckley S, Schouteden S, et al. A novel role of BMP4 in adult hematopoietic stem and progenitor cell homing via Smad independent regulation of integrin-alpha4 expression. Blood. 2013;121:781–790. doi: 10.1182/blood-2012-07-446443. [DOI] [PubMed] [Google Scholar]
- 34.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 35.Yu PB, Hong CC, Sachidanandan C, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller TD, Nickel J. Promiscuity and specificity in BMP receptor activation. FEBS Lett. 2012;586:1846–1859. doi: 10.1016/j.febslet.2012.02.043. [DOI] [PubMed] [Google Scholar]
- 37.Macias-Silva M, Hoodless PA, Tang SJ, et al. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J Biol Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 38.Kittler R, Surendranath V, Heninger AK, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- 39.Singbrant S, Karlsson G, Ehinger M, et al. Canonical BMP signaling is dispensable for hematopoietic stem cell function in both adult and fetal liver hematopoiesis, but essential to preserve colon architecture. Blood. 2010;115:4689–4698. doi: 10.1182/blood-2009-05-220988. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 41.Christopherson KW, 2nd, Hangoc G, Mantel CR, et al. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305:1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 42.Petit I, Szyper-Kravitz M, Nagler A, et al. G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat Immunol. 2002;3:687–694. doi: 10.1038/ni813. [DOI] [PubMed] [Google Scholar]
- 43.Stroncek DF, Clay ME, Smith J, et al. Changes in blood counts after the administration of granulocyte-colony-stimulating factor and the collection of peripheral blood stem cells from healthy donors. Transfusion. 1996;36:596–600. doi: 10.1046/j.1537-2995.1996.36796323058.x. [DOI] [PubMed] [Google Scholar]
- 44.Kobbe G, Sohngen D, Bauser U, et al. Factors influencing G-CSF-mediated mobilization of hematopoietic progenitor cells during steady-state hematopoiesis in patients with malignant lymphoma and multiple myeloma. Ann Hematol. 1999;78:456–462. doi: 10.1007/s002770050598. [DOI] [PubMed] [Google Scholar]
- 45.Nademanee A, Sniecinski I, Schmidt GM, et al. High-dose therapy followed by autologous peripheral-blood stem-cell transplantation for patients with Hodgkin’s disease and non-Hodgkin’s lymphoma using unprimed and granulocyte colony-stimulating factor-mobilized peripheral-blood stem cells. J Clin Oncol. 1994;12:2176–2186. doi: 10.1200/JCO.1994.12.10.2176. [DOI] [PubMed] [Google Scholar]
- 46.Stiff PJ. Management strategies for the hard-to-mobilize patient. Bone Marrow Transplant. 1999;23 (Suppl 2):S29–33. doi: 10.1038/sj.bmt.1701671. [DOI] [PubMed] [Google Scholar]
- 47.Boeve S, Strupeck J, Creech S, et al. Analysis of remobilization success in patients undergoing autologous stem cell transplants who fail an initial mobilization: risk factors, cytokine use and cost. Bone Marrow Transplant. 2004;33:997–1003. doi: 10.1038/sj.bmt.1704486. [DOI] [PubMed] [Google Scholar]
- 48.DiPersio JF, Stadtmauer EA, Nademanee A, et al. Plerixafor and G-CSF versus placebo and G-CSF to mobilize hematopoietic stem cells for autologous stem cell transplantation in patients with multiple myeloma. Blood. 2009;113:5720–5726. doi: 10.1182/blood-2008-08-174946. [DOI] [PubMed] [Google Scholar]
- 49.Flomenberg N, Devine SM, Dipersio JF, et al. The use of AMD3100 plus G-CSF for autologous hematopoietic progenitor cell mobilization is superior to G-CSF alone. Blood. 2005;106:1867–1874. doi: 10.1182/blood-2005-02-0468. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez P, Rettig MP, Uy GL, et al. BIO5192, a small molecule inhibitor of VLA-4, mobilizes hematopoietic stem and progenitor cells. Blood. 2009;114:1340–1343. doi: 10.1182/blood-2008-10-184721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa M, Nishikawa S, Ikuta K, et al. B cell ontogeny in murine embryo studied by a culture system with the monolayer of a stromal cell clone, ST2: B cell progenitor develops first in the embryonal body rather than in the yolk sac. Embo J. 1988;7:1337–1343. doi: 10.1002/j.1460-2075.1988.tb02949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 53.Whitman M. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y, Wang YF, Jayaraman L, et al. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 55.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 56.Levesque JP, Hendy J, Takamatsu Y, et al. Disruption of the CXCR4/CXCL12 chemotactic interaction during hematopoietic stem cell mobilization induced by GCSF or cyclophosphamide. J Clin Invest. 2003;111:187–196. doi: 10.1172/JCI15994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. (A) The effect of NGN on the expression of various known hematopoiesis related genes in non-hematopoietic BM cells was assessed by qRT-PCR. Mice were injected with PBS/NGN, and after 24h, the lin−CD45− fraction of total BM cells was isolated using MACS. Expression of genes such as Cd44, Cxcl12, Icam1, Vcam1, Mmp7, Mmp9, Upa, Ang1, Opn, Scf, Ccl2 as well as Cxcl12 was analyzed by qRT-PCR. n=4, * p=0.034 (B) Primary BM stromal cells cultured with BMP7/DM were analyzed for expression of Cxcl12 by performing qRT-PCR. n=4, * p,0.05 (C) Proportion of transplanted CFCs homed within 16h of transplantation was compared between DM infused and control animals. n=8, * p=0.041
Supplementary figure 2. The effect of SMAD signaling on Cxcl12 expression was analyzed by qRT-PCR on ST2 cells cultured with/without neutralizing antibodies against BMP4. n=4, * p=0.02
Supplementary figure 3. Expression of Smad1, Smad5 and Bmp4 in ST2 cells was inhibited using esiRNA-mediated knockdown. Efficiency of knockdown was quantified using immuno-blotting. Change in protein expression was quantified by measuring band intensity using ImageJ software. n=4, * p<0.04
Supplementary figure 4. PB derived MNCs from PBS/BMP7 injected CD45.1 mice were transplanted along with 100,000 total BM cells from CD45.2 mice in lethally irradiated CD45.2 mice. After 16 weeks, mice were sacrificed and BM derived cells were analyzed for presence of donor derived cells by flowcytometry (A) as well as multi-lineage engraftment in PB was analyzed using (B) using antibodies against CD45.1/CD45.2 in addition to CD11b/Gr-1 (macrophage/granulocyte), B-220 (B-lymphocytes), CD4/CD8 (T-lymphocytes) (4 weeks data shown here suppl fig. 4B) (n=8).
Supplementary figure 5. Expression of CXCR4 on the HSCs, identified as lin−c-kit+Sca-1+CD48−CD150+, migrated towards NGN treated versus control ST2 cells was analyzed by flowcytometry.
Supplementary figure 6. PBS or BMP7 treated animals were sacrificed and BM cells were isolated following crushing the hind limb bones and treating with Collagenase I. (Upper panel) Cellular components of the HSC niche were sorted on the basis of their phenotype; Mesenchymal stem cells (MSCs; lin−CD45−CD31−CD51+Sca-1+), osteoblasts (OBs; lin−CD45−CD31−CD51+Sca-1−) and endothelial progenitor cells (EPCs; lin−CD45−CD31+). (Lower panel) Collagen I was used as specific marker to identify osteoblasts within lin−CD45−CD31−Sca-1− population.
Supplementary figure 7. (A) BM cells from Col1a1-GFp mice were harvested by flushing followed by enzymatic treatment of the crushed bone pieces. Flowcytometry analysis was performed to examine the GFP expression in osteoblasts identified by phenotypic markers (lin−CD45−CD31−CD51+Sca-1−). (B) Col1a1-GFP mice were infused with PBS/BMP7/DM in addition to BrdU. BM cells were harvested after 24h and BrdU incorporation in Col1-GFP+ fraction of lin−CD45−CD31− cells from the three groups was analyzed by flowcytometry.