Abstract
It is now evident that many nuclear hormone receptors can modulate target gene expression. REV-ERBα, one of the nuclear hormone receptors with the capacity to alter clock function, is critically involved in lipid metabolism, adipogenesis, and the inflammatory response. Recent studies suggest that REV-ERBα plays a key role in the mediation between clockwork and inflammation. The purpose of the current study was to investigate the role of REV-ERBα in the regulation of interleukin-6 (il6) gene expression in murine macrophages. REV-ERBα agonists, or overexpression of rev-erb α in the murine macrophage cell line RAW264 cells, suppressed the induction of il6 mRNA following a lipopolysaccharide (LPS) endotoxin challenge. Also, rev-erb α overexpression decreased LPS-stimulated nuclear factor κB (NFκB) activation in RAW264 cells. We showed that REV-ERBα represses il6 expression not only indirectly through an NFκB binding motif but also directly through a REV-ERBα binding motif in the murine il6 promoter region. Furthermore, peritoneal macrophages from mice lacking rev-erb α increased il6 mRNA expression. These data suggest that REV-ERBα regulates the inflammatory response of macrophages through the suppression of il6 expression. REV-ERBα may therefore be identified as a potent anti-inflammatory receptor and be a therapeutic target receptor of inflammatory diseases.
1. Introduction
The human genome contains 48 nuclear hormone receptor genes, comprising a large family of ligand-dependent transcription factors. In contrast with most classic receptors, nuclear hormone receptors modulate transcription by binding directly to DNA, and ligand interactions occur primarily within the cell cytosol or nucleus. Nuclear hormone receptors are now recognized as key intermediaries between the molecular clock machinery and a wide array of physiological processes [1]. In particular, REV-ERBα, one of the nuclear hormone receptors encoded by nr1d1, is a crucial regulator of lipid, lipoprotein metabolism, and inflammation [1].
REV-ERBα, one of the key clock genes, is a part of the clock machinery and plays an important role in maintaining the proper rhythm of circadian timing [2]. REV-ERBα binds as a monomer to the retinoic acid receptor-related orphan receptor (ROR) response elements (ROREs) composed of a 6 bp A/T-rich sequence immediately preceding a site with the core motif of (A/G)GGTCA [3]. It also binds as a homodimer to the RevDR2 response element, which is composed of a 6 bp A/T-rich sequence immediately preceding a site with a tandem repeat of two (A/G)GGTCA motifs spaced by two nucleotides [4].
Our recent work has demonstrated that REV-ERBα suppresses chemokine (C-C motif) ligand 2 (ccl2) gene expression directly through a RORE in the ccl2 promoter region [5]. These results implicate REV-ERBα as a critical intermediary between the core clockwork and inflammatory pathways. Gibbs et al. [6] have demonstrated that the administration of a REV-ERBα ligand or a genetic knockdown of rev-erb α expression is effective at modulating the production and release of the proinflammatory cytokine interleukin-6 (IL6). Furthermore, Journiac and coworkers [7] have shown that 2 and 3 putative ROREs have also been found in the il6 promoter region of mice and rats, respectively [7]. However, it is unclear whether the putative ROREs in the murine il6 promoter are sensitive to REV-ERBα regulation. In some cases, there are several similarities and differences in the inflammatory response to endotoxin in mice and humans [8]. Therefore, it is important to demonstrate the impact of REV-ERBα on il6 gene in murine immune cells as well as humans.
Results from the current study showed that REV-ERBα directly and indirectly suppresses il6 gene expression in macrophages through a RORE and a nuclear factor κB response element (NFκBRE), respectively, in the murine il6 promoter region. Furthermore, we observed increases in il6 gene expression in peritoneal macrophages from mice lacking rev-erb α. REV-ERBα may therefore be a key link between the clockwork and inflammation.
2. Materials and Methods
2.1. Animals
C57BL/6J mice and B6.Cg-Nr1d1<tm1Ven>/LazJ mice were obtained from Sankyo Labo Service (Tokyo, Japan) and Jackson Laboratories (Bar Harbor, ME), respectively. The mice were housed in plastic cages and reared at 23°C with a 12 h light/dark cycle. Food and water were available ad libitum. All animals were cared for in accordance with the Guiding Principles for the Care and Use of Animals approved by the Council of the Physiological Society of Japan, based upon the Declarations of Helsinki, 1964.
2.2. Preparation and Culture of Peritoneal Macrophages
Peritoneal macrophages were collected from 2-month-old C57BL/6J mice and rev-erb α −/− mice and cultured as described previously [5, 9–11]. The cells from C57BL/6J mice were treated with or without REV-ERBα agonist, 2 or 20 μM GSK4112 (Sigma Aldrich), for 16 h in the absence or presence of 1 μg/mL lipopolysaccharide (LPS) from Escherichia coli 055 (Sigma Aldrich, St. Louis, MO). GSK4112 was dissolved with DMSO, and the control cells were treated using the same volume of DMSO. The cells from rev-erb α −/− mice were stimulated with or without LPS for 24 h.
2.3. Cell Line Culture
The murine macrophage cell line RAW264 (RCB0535) was purchased from RIKEN Cell Bank (Ibaraki, Japan) and cultured as described previously [5, 9, 10, 12]. To study the effects of REV-ERBα agonists on il6 gene expression, the cells were treated with or without 20 μM GSK4112 or 1 μg/mL LPS for 16 h.
2.4. Real-Time Quantitative PCR (qPCR)
Total cellular RNA was prepared from peritoneal macrophages using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and from RAW264 cells using RNAiso reagent (Takara bio, Siga, Japan). Extracted RNA was reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) with random primers. The reaction mixture was amplified in a Power SYBR Green Master Mix (Applied Biosystems) using a 7500 Real-Time PCR System (Applied Biosystems) with 200 nM oligonucleotide primers (forward and reverse). The oligonucleotide sequences used for qPCR were as follows: il6: 5′-GAT GGA TGC TAC CAA ACT GGA-3′ (forward), 5′-CCA GGT AGC TAT GGT ACT CCA GAA-3′ (reverse); β actin (actb, internal control), 5′-AAG GCC AAC CGT GAA AAG AT-3′ (forward), and 5′-GTG GTA CGA CCA GAG GCA TAC-3′ (reverse). The expression of the target gene was normalized to the housekeeping gene actb.
2.5. rev-erbα or rorα Plasmid Constructs and Stable Transfection
A stable rev-erb α transfectant (RAWrev) and the control cell line (RAWvecB) and a stable ror α transfectant (RAWror) and the control cell line (RAWvecA) were established as described previously [5].
2.6. Western Blot Analysis
Nucleic and cytosolic protein was extracted as described previously [5, 10, 13]. Extracted proteins were separated by SDS-PAGE and then transferred to a polyvinylidene difluoride membrane (Millipore, Milford, MA). Membranes were blocked with 5% nonfat dried milk in TBST and then immunoblotted with rabbit polyclonal Abs against NFκB p65 (sc-372, Santa Cruz Biothechnology, Santa Cruz, CA),α-Tublin (α-Tub, ab7291, Abcam, Cambridge, UK), or TATA binding protein (TBP, ab51841, Abcam). Thereafter, HRP-conjugated donkey anti-rabbit or anti-mouse IgG secondary Abs (GE Healthcare Japan, Tokyo, Japan) was applied. The immunoreactivity was visualized with an ECL reagent (Bio-Rad, Hercules, CA).
2.7. EMSA
Nuclear extracts were prepared as described [5, 10, 13]. The murine NFκB consensus oligonucleotide probe (5′-AGT TGA GGG GAC TTT CCC AGG C-3′) was labeled with biotin. The nuclear protein (2.5 μg) and labeled oligonucleotide probe (20 fmol) were incubated in 10 mM HEPES-KOH, pH 7.8, 50 mM KCl, 0.2 mM EDTA, 10% glycerol, 1 μg poly(dI-dc), 0.05% NP-40, and 5 mM DTT at room temperature for 20 min, electrophoresed in 4.5% polyacrylamide gels, transferred onto a nylon membrane (Biodyne, Pall Corporation, Pensacola, FL), and UV cross-linked. To detect the signals, a Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, Rockford, IL) was used according to the manufacturer's protocol.
2.8. Luciferase Reporter Assay
For the analysis of the promoter activity of the NFκB-responsive promoter reporter luciferase construct, the cells were transfected with pNFκB-Luc (Clontech, Palo Alto, CA) using a LipofectAMINE Reagent (Invitrogen, Carlsbad, CA), and luciferase activity was determined using a Luciferase Assay System Kit (Promega, Madison, WI).
For the analysis of il6 promoter activity, the murine il6 promoter (distal fragment, −1029 to +31; proximal fragment, −649 to +31) was amplified from mouse genomic DNA (Promega) using an LA Taq polymerase (Takara bio) and was subcloned into pCR-XL-TOPO vector (Invitrogen). The subcloned fragments were digested at KpnI/XhoI sites and cloned into pGL3 vector (Promega) at the corresponding sites. The cells were transiently transfected by using a LipofectAMINE Reagent with distal or proximal constructs containing the luciferase reporter gene, and luciferase activity was determined with a Dual Luciferase Assay System Kit (Promega). Activity was normalized relative to an internal cotransfected constitutive control (Renilla luciferase expression vector, pRL-TK vector, Promega), as described [5, 10, 12].
2.9. Mutagenesis
The il6 promoter mutant construct was made by using a QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) as described [5, 12]. The proximal RORE (−529 to −518) was mutated from AAA CTC AGG TCA to AAA CTC AGG CCT by using the mutant primers 5′-CTG AAA AAA CTC AGG CCT GAA CAT CTG TAG-3′ (forward) and 5′-CTA CAG ATG TTC AGG CCT GAG TTT TTT CAG-3′ (reverse) for the distal and proximal il6 promoter constructs mutagenesis (underline, mutant sequences). The NFκBRE (−91 to −82) was mutated from GGG ATT TTC C to GGG CCC TTC C by using the mutant primers 5′-GAT TTT TAT CAA ATG TGG GCC CTT CCC ATG AGT CTC-3′ (forward) and 5′-GAG ACT CAT GGG AAG GGC CCA CAT TTG ATA AAA ATC-3′ (reverse) for the proximal il6 promoter constructs mutagenesis.
2.10. Statistical Analysis
The results were expressed as the means ± S.E. When two means were compared, a Student's t-test for unpaired samples was conducted. For more than two groups, the statistical significance of the data was assessed by ANOVA. When significant differences were found, individual comparisons were made between groups by using the t-statistic and adjusting the critical value according to the Tukey-Kramer method. Differences were considered significant at P < 0.05.
3. Results
3.1. REV-ERBα Agonists Suppress il6 Induction following LPS Stimulation
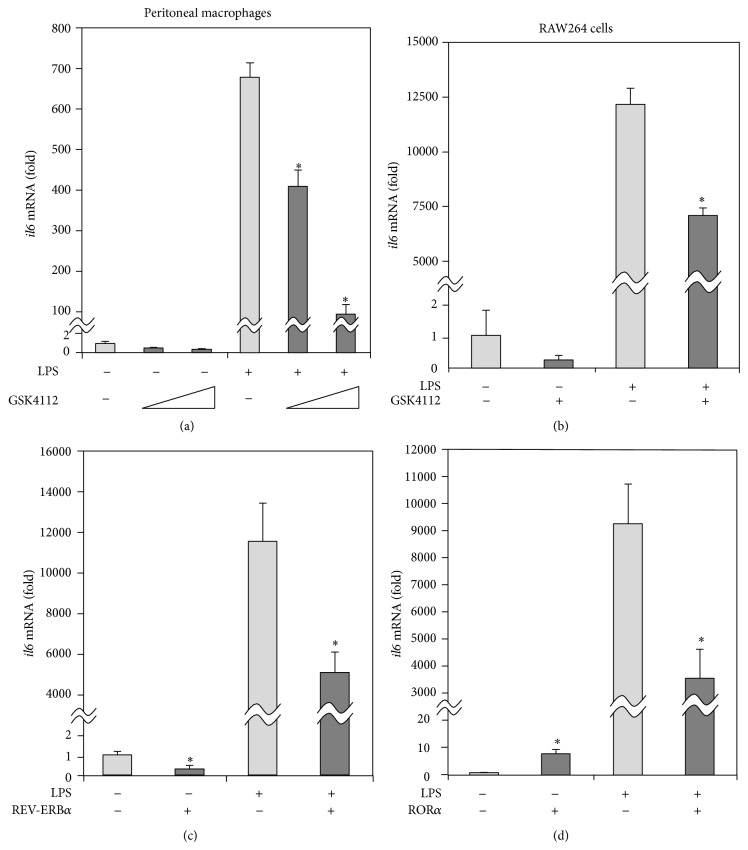
To determine the role of REV-ERBα in inflammatory responses, we analyzed the effects of the REV-ERBα agonist, GSK4112, on the gene expression of il6 as a crucial inflammatory molecular element in macrophages. The induction of il6 mRNA after LPS stimulation was dose-dependently repressed by the addition of GSK4112 in peritoneal macrophages (Figure 1(a)). Furthermore, as shown in Figure 1(b), qPCR analysis confirmed that GSK4112 treatment also decreased the induction of il6 mRNA after LPS stimulation in murine macrophage cell line RAW264 cells as well as peritoneal macrophages. These data suggest that activation of REV-ERBα led to the suppression of il6 gene induction in macrophages.
Figure 1.
REV-ERBα represses il6 gene induction following a LPS challenge in macrophages. (a) Peritoneal macrophages were harvested as adherent cells from 2-month-old C57BL/6J mice and were either untreated or treated with 1 μg/mL LPS or 2 or 20 μM GSK4112 for 16 h. (b) Murine macrophage cell line RAW264 cells were either untreated or treated with 1 μg/mL LPS or 20 μM GSK4112 for 16 h. (c) RAW264 cells transfected with or without rev-erb α were either untreated or treated with 1 μg/mL LPS for 24 h. (d) RAW264 cells transfected with or without ror α were either untreated or treated with 1 μg/mL LPS for 24 h. The gene expression of il6 was analyzed by qPCR. For normalization, actb mRNA was used. The data are presented as the means ± S.E. (n = 3-4). * P < 0.05 versus cells treated with LPS and without GSK4112 or versus vector control.
3.2. rev-erbα Overexpression Represses il6 Expression
To investigate the potential role of REV-ERBα in il6 expression in macrophages, a stable rev-erb α transfectant (RAWrev) and the control cell line (RAWvecB) were established using RAW264 cells [5]. As seen in Figure 1(c), overexpression of rev-erb α repressed the gene expression of il6 in both the absence and presence of LPS, suggesting that REV-ERBα was involved in the suppression of il6 gene expression in macrophages.
3.3. rorα Overexpression Enhances il6 Expression
REV-ERBα is known to cross-talk with RORα (orphan nuclear receptor encoded by nr1f1), another of the clock genes that has similar DNA binding specificity to REV-ERBα and competes for the binding of REV-ERBα [14–16]. Whereas REV-ERBα represses transcription from these sites of the target genes, RORα acts as a transcriptional activator [3, 4, 17]. From these findings, we hypothesized that RORα might positively regulate il6 expression and established a stable ror α transfectant (RAWror) and the control cell line (RAWvecA) using RAW264 cells [5]. Interestingly, overexpression of ror α enhanced the gene expression of il6 in the absence of LPS, whereas it repressed the gene expression of il6 in the presence of LPS (Figure 1(d)), indicating that regulation of il6 gene expression by RORα is different between nonactivated and activated states in macrophages.
3.4. REV-ERBα Suppressed NFκB Activity
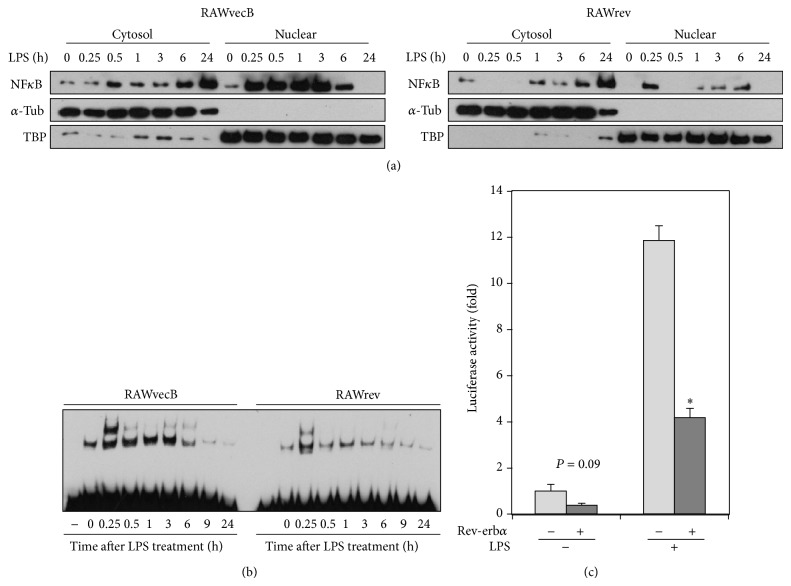
The il6 gene contains a functional κB element in its promoter region [18]. Thus, activation of NFκB leads to the transcription of this proinflammatory gene. Therefore, we next investigated whether REV-ERBα regulates NFκB activity in macrophages. As shown in Figure 2(a), cytosolic expression and LPS-induced nuclear translocation of NFκB subunit p65 were attenuated in RAWrev cells, compared with RAWvecB cells. No marked change in inhibitory κB (IκB) expression was observed between RAWrev and RAWvecB cells (data not shown). Furthermore, LPS-induced NFκB activation in RAWrev cells was markedly lower than that in RAWvecB cells (Figure 2(b)). In addition, REV-ERBα attenuated the promoter activity of the NFκB-responsive promoter reporter luciferase construct in both the absence and presence of LPS in macrophages (Figure 2(c)). These results strongly suggest that REV-ERBα suppresses LPS-enhanced NFκB activity in macrophages.
Figure 2.
REV-ERBα represses NFκB activation in macrophages. (a) Nuclear and cytoplasmic fractions of each cell untreated or treated with LPS for various durations were analyzed by Western blot for p65 NFκB,α-Tub, and TBP. Detection ofα-Tub and TBP is used as marker of nuclear and cytoplasmic fractions, respectively. (b) Each cell was stimulated with LPS for varying durations and NFκB activation was analyzed by EMSA. Data shown are representative of three separate experiments. (c) Each cell was transiently transfected with NFκB-responsive promoter reporter luciferase construct and luciferase activities in each cell stimulated either with or without LPS for 24 h were analyzed. The data are presented as the means ± S.E. from sextuplicate cultures. * P < 0.01 versus vector control.
3.5. REV-ERBα Represses the Activity of the Murine il6 Promoter
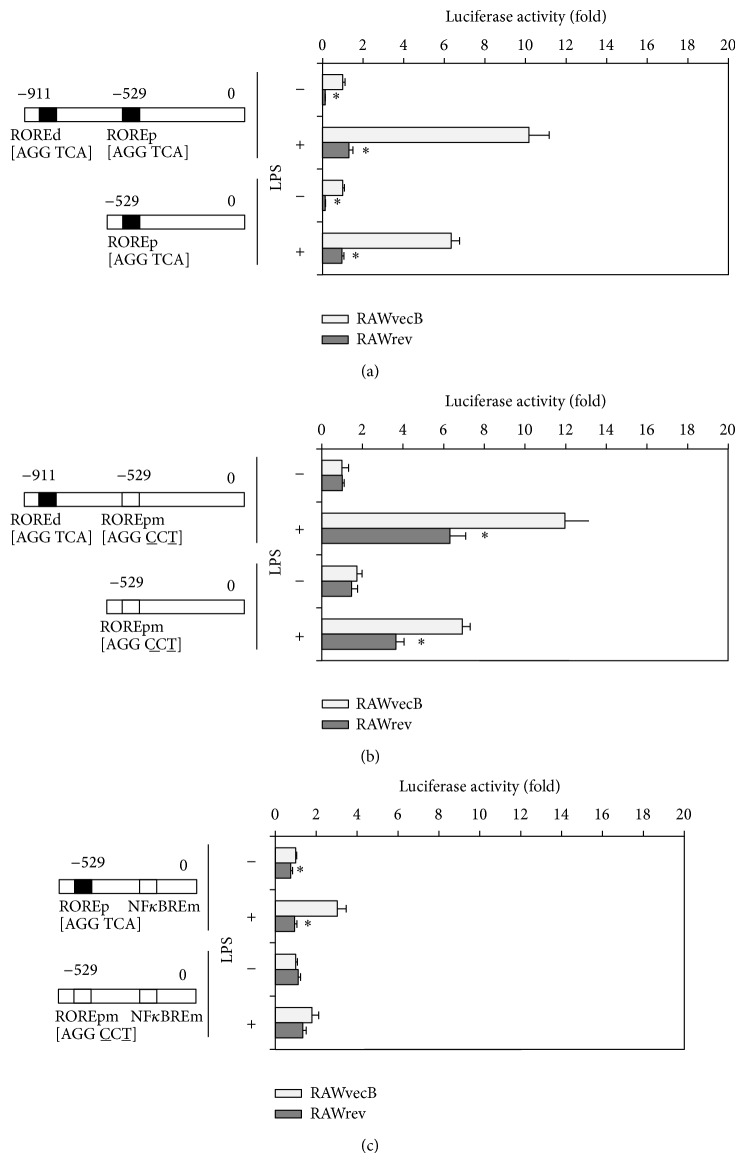
Two putative ROREs have been found in the mouse il6 promoter sequence [7]. Therefore, to determine whether the putative ROREs in the il6 promoter are sensitive to REV-ERBα regulation, we cloned il6 promoters with different lengths—a distal promoter that included one RORE located in the distal region and one RORE located in the proximal region, and a proximal promoter that included one RORE located in the proximal region—into a luciferase reporter vector. Then, these two constructs were transiently transfected into cell lines, RAWrev and RAWvecB cell lines. The activities of each longitudinal promoter in RAWrev cells were considerably lower than those in RAWvecB cells in both the absence and presence of LPS (Figure 3(a)). We next investigated whether two ROREs in the il6 promoter are necessary for REV-ERBα-mediated repression. As shown in Figure 3(b), the mutation of the proximal RORE abolished the repression of the promoter activities in RAWrev transfected with the distal construct as well as the proximal construct in the absence of LPS. These results suggest a critical role for the proximal RORE in REV-ERBα-mediated repression of il6 expression. However, the mutation of the proximal RORE still repressed the promoter activities in RAWrev transfected with the distal construct as well as the proximal construct in the presence of LPS (Figure 3(b)), indicating that REV-ERBα repressed il6 promoter activity through other transcriptional regulators such as NFκB, which was independent of the direct binding of REV-ERBα to the RORE in the promoter.
Figure 3.
REV-ERBα represses il6 promoter activity, independent of the inhibition of NFκB signaling. (a) RAWrev and RAWvecB cells were transiently transfected with luciferase reporter construct containing either a distal or a proximal construct of il6 promoter. After treatment with or without 1 μg/mL LPS for 24 h, luciferase activities were determined. ROREd, distal RORE; ROREp, proximal RORE. (b) The AGGTCA half-site in the proximal RORE was changed to AGGCCT by site-directed mutagenesis of nucleotides −518 (A to T) and −520 (T to C), and luciferase activities of each cell either untreated or treated with LPS for 24 h were determined. ROREpm, proximal RORE mutant. (c) The GGGATTTTCC half-site in the NFκBRE was changed to GGGCCCTTCC by site-directed mutagenesis of nucleotides −86 (T to C), −87 (T to C), and −88 (A to C), and luciferase activities of each cell either untreated or treated with LPS for 24 h were determined. NFκBREm, NFκBRE mutant. Luciferase values were normalized using Renilla luciferase. The data are presented as the means ± S.E. from sextuplicate cultures. * P < 0.05 versus vector control.
3.6. REV-ERBα Represses il6 Promoter Activity, Independent of NFκB
To dissect the effect of NFκB on il6 promoter activity, we used point-mutated variants in the response element of NFκB. The activity of an il6 promoter containing an NFκBRE mutated construct in RAWrev cells was lower than that in RAWvecB cells in both the absence and presence of LPS (Figure 3(c)). These results show that REV-ERBα repressed il6 promoter activity, independent of NFκB. A double mutation of RORE and NFκBRE completely abrogated the suppression of the promoter activity in RAWrev cells in both the absence and presence of LPS (Figure 3(c)). These results suggest both a direct and an indirect repression of the il6 promoter activity by REV-ERBα.
3.7. RORα Enhances the Activity of Murine il6 Promoter
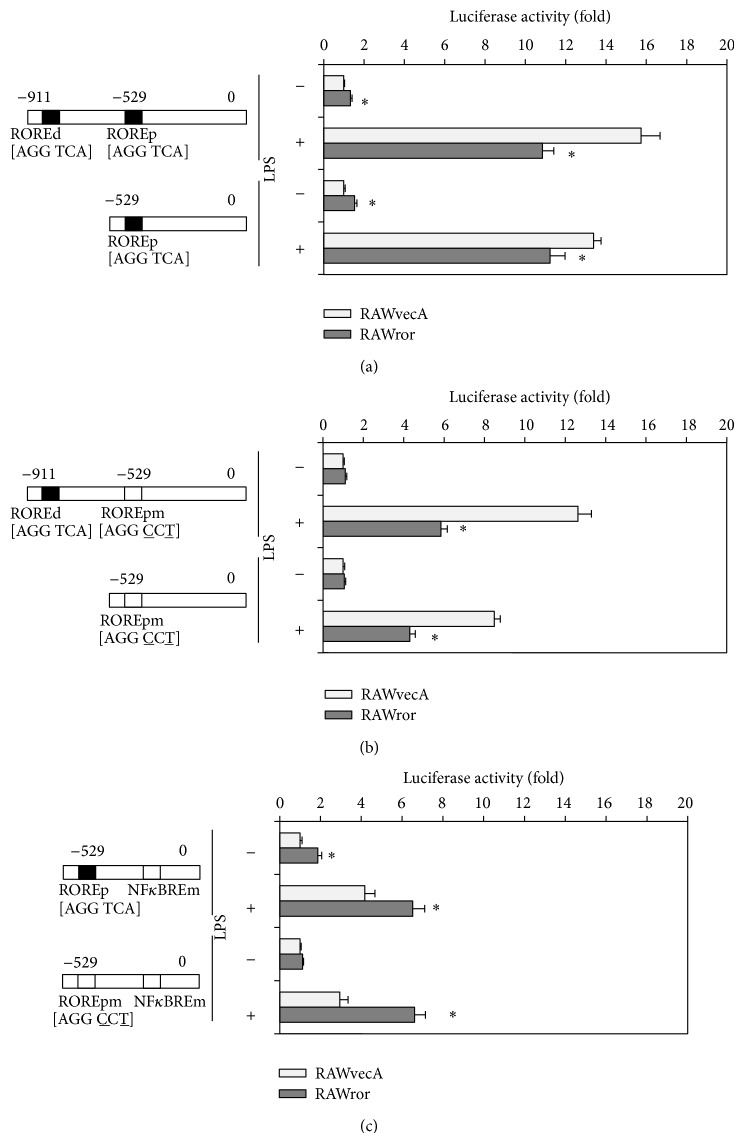
Because RORα activates target genes via ROREs in their promoters, we reasoned that RORα might positively regulate il6 promoter activity. Therefore, we transiently transfected the distal and the proximal il6 promoter constructs into RAWror and RAWvecA cells. The activity of each of the liner promoter in the RAWror cells was considerably higher than that in RAWvecA cells in the absence of LPS, whereas that in RAWror cells was lower than that in RAWvecA cells in the presence of LPS (Figure 4(a)). We also investigated whether two ROREs in the il6 promoter were essential for RORα-mediated enhancement of il6 expression. The mutation of a proximal RORE abrogated the enhancement of the promoter activities in RAWror cells transfected with either distal or proximal construct in the absence of LPS (Figure 4(b)), suggesting that the positive regulatory effects of RORα on the il6 expression are mainly dependent on the proximal RORE in the il6 promoter. However, the mutation of the proximal RORE additionally repressed the promoter activities in RAWror transfected with the distal construct as well as the proximal construct in the presence of LPS (Figure 4(b)). Therefore, we hypothesized that RORα also suppressed il6 promoter activity via the inhibition of NFκB-induced transactivation after LPS stimulation as is the case with REV-ERBα. In fact, the activity of an il6 promoter containing an NFκBRE mutated construct in RAWror cells was higher than that in RAWvecA cells in both the absence and presence of LPS (Figure 4(c)). These results show that RORα activated il6 promoter activity through proximal RORE in nonactivated cells, whereas it indirectly repressed the activity through negative regulation of NFκB signaling in activated cells. A double mutation of RORE and NFκBRE showed no changes in the promoter activity between RAWror and RAWvecA cells in the absence of LPS (Figure 4(c)). However, in the presence of LPS, il6 promoter activity of RAWror cells is lower than that of RAWvecA cells, suggesting that RORα also repressed il6 promoter activity through other transcriptional regulators than NFκB.
Figure 4.
Effect of RORα on il6 promoter activity. (a) RAWror and RAWvecA cells were transiently transfected with luciferase reporter construct containing either a distal or a proximal construct of il6 promoter. After treatment with or without 1 μg/mL LPS for 24 h, luciferase activities were determined. ROREd, distal RORE; ROREp, proximal RORE. (b) The AGGTCA half-site in the proximal RORE was changed to AGGCCT by site-directed mutagenesis of nucleotides −518 (A to T) and −520 (T to C), and luciferase activities of each cell either untreated or treated with LPS for 24 h were determined. ROREpm, proximal RORE mutant. (c) The GGGATTTTCC half-site in the NFκBRE was changed to GGGCCCTTCC by site-directed mutagenesis of nucleotides −86 (T to C), −87 (T to C), and −88 (A to C), and luciferase activities of each cell either untreated or treated with LPS for 24 h were determined. NFκBREm, NFκBRE mutant. Luciferase values were normalized using Renilla luciferase. The data are presented as the means ± S.E. from sextuplicate cultures. * P < 0.05 versus vector control.
3.8. Peritoneal Macrophages from rev-erbα Knockout Mice Display Increases in the il6 Gene Expression
To test whether results observed in the in vitro study are physiologically relevant, we investigated the effects of a rev-erb α deficiency on il6 expression in peritoneal macrophages using rev-erb α −/− mice. As shown in Figure 5, il6 gene expression in the absence of LPS in the peritoneal macrophages of rev-erb α −/− mice was significantly higher than that in wild-type mice. The induction of the il6 gene following a LPS challenge in the peritoneal macrophage of mice lacking rev-erb α also was relatively higher (P = 0.08) than that found in wild-type mice. These results show that il6 expression is negatively regulated by REV-ERBα in vivo as well as in vitro.
Figure 5.
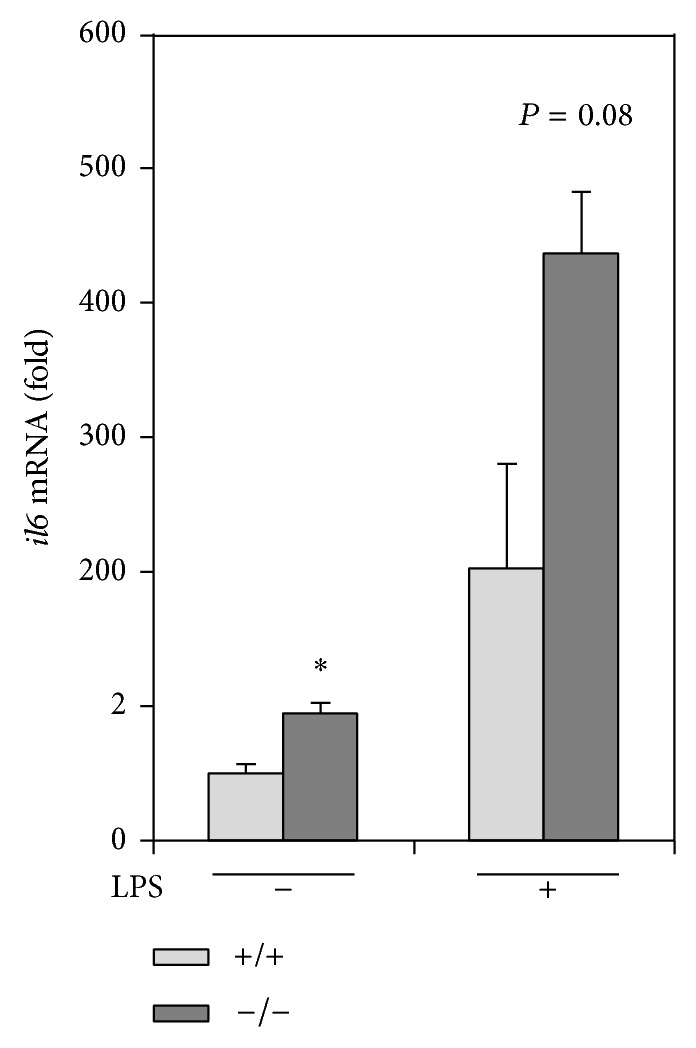
Peritoneal macrophages from rev-erb α −/− mice increase il6 gene expression. Peritoneal macrophages were harvested as adherent cells from 2-month-old rev-erb α −/− mice and their wild-type (+/+) mice. The cells were treated with or without 1 μg/mL LPS for 24 h. The gene expression of il6 was analyzed by qPCR. For normalization, actb mRNA was used. The data are presented as the means ± S.E. (n = 3). * P < 0.05 versus rev-erb α +/+ mice.
4. Discussion
Until recently, REV-ERBα was considered to be a constitutively active nuclear orphan receptor, although heme has now been shown to bind reversibly to the receptor and to drive ligand-dependent activity [19]. This implies that REV-ERBα is responsive to the cellular redox state and perhaps to gaseous signaling molecules such as NO and CO through interactions with heme [20]. In addition, REV-ERBα acts as a transrepressor for a number of genes, including bmal1 [14], apolipoprotein AI (apoAI) [21], apoCIII [22], fibrinogen-β [23], plasminogen activator inhibitor type 1 (pai1) [24], il6 [7], and ccl2 [5], which indicates that the nuclear hormone receptor plays an important role in the regulation of metabolism, the cardiovascular system, and inflammation.
Recently, we demonstrated that REV-ERBα negatively regulates the inflammatory function of macrophages through the direct repression of ccl2 expression [5]. Furthermore, we showed that REV-ERBα suppresses not only intracellular signals such as extracellular signal-regulated protein kinase (ERK) and p38 mitogen-activated protein kinase (p38 MAPK), which is known as CCL2 and the receptor chemokine (C-C motif) receptor 2- (CCR2-) activated signaling pathways, but also the inflammatory functions of macrophages such as adherent and migratory activities, the activation of which is known to be dependent on CCL2-mediated ERK and p38, respectively [5, 25]. These observations identified the nuclear hormone receptor REV-ERBα as an anti-inflammatory receptor and a therapeutic target in inflammatory disease.
As in the previous report, for the current study, we analyzed the role of REV-ERBα in the gene expression of inflammatory cytokine il6 in murine macrophages. We confirmed that REV-ERBα agonist GSK4112 inhibits the induction of the il6 gene in murine peritoneal macrophages and in murine macrophage cell line RAW264 cells following LPS stimulation. Our results are consistent with the recently published results by Gibbs et al. [6] who demonstrated that GSK4112 abolishes the induction of inflammation-related genes, including il6, following a LPS challenge, using primary human monocyte-derived macrophages. In the current study, the overexpression of rev-erb α also revealed that REV-ERBα contributes to the negative regulation of il6 expression in macrophages. By contrast, peritoneal macrophages from mice lacking rev-erb α increase il6 gene expression. Reporter assay and site-directed mutagenesis identified a critical role for the proximal RORE in the murine il6 promoter in REV-ERBα-mediated repression of il6 expression. We also showed that REV-ERBα represses il6 expression not only directly through a RORE but also indirectly through an NFκBRE in the murine il6 promoter. These results strongly suggest that REV-ERBα functions as a repressor of inflammatory response in macrophages via the inhibition of the target genes, including ccl2 and il6.
REV-ERBα has been known to cross-talk with RORα, an orphan nuclear receptor encoded by nr1f1, that has similar DNA binding specificity to REV-ERBα, acts as a constitutive transcriptional activator, and thus competes with the binding of REV-ERBα [3, 4, 14–17, 26]. Furthermore, Journiac and coworkers [7] have shown that REV-ERBα and/or RORα directly bind to a RORE in the human il6 promoter and acts as a transrepressor and a transactivator of il6 gene expression, respectively. We also confirmed that RORα overexpression in murine macrophage cell line enhances il6 gene expression and the promoter activity through RORE in its promoter region without any exogenous LPS stimulation, whereas it suppressed il6 gene induction and the promoter activity, at least partly, via the inhibition of NFκB-induced transactivation after LPS stimulation. These results suggest that RORα transactivates il6 expression by interacting with a RORE in the promoter of murine macrophages, whereas RORα negatively regulates il6 expression through the NFκB signaling in murine macrophages. From these observations, in resting cultured macrophages, a dual regulation also pertains to the il6 promoter activity; REV-ERBα potently represses il6 promoter activity, whereas RORα potently enhances il6 promoter activity through murine il6 promoter as well as human [7]. The dual regulation seems to have an advantage in the modulation of inflammatory responses of macrophages, although evidence for in vivo relevance is clearly lacking.
Patients with rheumatoid arthritis (RA) report daily variations in their symptoms, experiencing greater joint pain, stiffness, and functional disability in the mornings, which is accompanied by fluctuations in circulating IL6 concentration [27, 28]. Some asthma patients experience nighttime exacerbations that can be attributed to not only daily variations in lung physiology but also increased bronchial responsiveness at night [29]. Macrophages exhibit a rhythmic expression of rev-erb α , are capable of cell-autonomous gene oscillation in culture, and display a robust circadian gating in their responses to exogenous inflammatory stimulation [6, 30–32]. Therefore, it seems likely that REV-REBα expressed in macrophages plays an important role in the regulation of the diurnal fluctuation of several inflammatory diseases as well as in the production and secretion of these inflammatory and/or anti-inflammatory factors. Further studies are needed to clarify the in vivo relationship between REV-ERBα and inflammatory diseases. Taken together, the results of the current study indicate that REV-ERBα is a potent anti-inflammatory receptor and a therapeutic target for inflammatory diseases.
5. Conclusion
We demonstrated that a circadian clock gene, REV-ERBα, represses il6 expression not only indirectly through an NFκB binding motif but also directly through a REV-ERBα binding motif in the murine il6 promoter region. Overexpression of rev-erb α in murine macrophage cell line suppressed il6 induction and NFκB activity following a LPS endotoxin challenge. The present study also showed that peritoneal macrophages from mice lacking rev-erb α display increases in il6 expression. These data suggest that REV-ERBα regulates the inflammatory response of macrophages through the suppression of il6 expression. REV-ERBα may therefore be a key link between clockwork and inflammation.
Acknowledgments
This work was supported, in whole or in part, by the Japanese Ministry of Education, Culture Sports, Science and Technology (to Takako Kizaki and Hideki Ohno) and the Nakatomi foundation (to Shogo Sato).
Abbreviations
- NFκBRE:
Nuclear factor κB response element
- ROR:
Retinoic acid receptor-related orphan receptor
- RORE:
ROR response element.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Bechtold D. A., Gibbs J. E., Loudon A. S. I. Circadian dysfunction in disease. Trends in Pharmacological Sciences. 2010;31(5):191–198. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 3.Harding H. P., Lazar M. A. The orphan receptor Rev-ErbAα activates transcription via a novel response element. Molecular and Cellular Biology. 1993;13(5):3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding H. P., Lazar M. A. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Molecular and Cellular Biology. 1995;15(9):4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato S., Sakurai T., Ogasawara J., Takahashi M., Izawa T., Imaizumi K., Taniguchi N., Ohno H., Kizaki T. A circadian clock gene, Rev-erb, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. Journal of Immunology. 2014;192(1):407–417. doi: 10.4049/jimmunol.1301982. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs J. E., Blaikley J., Beesley S., Matthews L., Simpson K. D., Boyce S. H., Farrow S. N., Else K. J., Singh D., Ray D. W., Loudon A. S. I. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Journiac N., Jolly S., Jarvis C., Gautheron V., Rogard M., Trembleau A., Blondeau J., Mariani J., Vernet-der Garabedian B. The nuclear receptor RORα exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21365–21370. doi: 10.1073/pnas.0911782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland S., Warren H. S., Lowry S. F., Calvano S. E., Remick D. Acute inflammatory response to endotoxin in mice and humans. Clinical and Diagnostic Laboratory Immunology. 2005;12(1):60–67. doi: 10.1128/CDLI.12.1.60-67.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kizaki T., Maegawa T., Sakurai T., Ogasawara J., Ookawara T., Oh-ishi S., Izawa T., Haga S., Ohno H. Voluntary exercise attenuates obesity-associated inflammation through ghrelin expressed in macrophages. Biochemical and Biophysical Research Communications. 2011;413(3):454–459. doi: 10.1016/j.bbrc.2011.08.117. [DOI] [PubMed] [Google Scholar]
- 10.Shirato K., Kizaki T., Sakurai T., Ogasawara J., Ishibashi Y., Iijima T., Okada C., Noguchi I., Imaizumi K., Taniguchi N., Ohno H. Hypoxia-inducible factor-1α suppresses the expression of macrophage scavenger receptor 1. Pflugers Archiv European Journal of Physiology. 2009;459(1):93–103. doi: 10.1007/s00424-009-0702-y. [DOI] [PubMed] [Google Scholar]
- 11.Kizaki T., Oh-Ishi S., Ookawara T., Yamamoto M., Izawa T., Ohno H. Glucocorticoid-mediated generation of suppressor macrophages with high density FcγRII during acute cold stress. Endocrinology. 1996;137(10):4260–4267. doi: 10.1210/en.137.10.4260. [DOI] [PubMed] [Google Scholar]
- 12.Kizaki T., Suzuki K., Hitomi Y., Taniguchi N., Saitoh D., Watanabe K., Onoé K., Day N. K., Good R. A., Ohno H. Uncoupling protein 2 plays an important role in nitric oxide production of lipopolysaccharide-stimulated macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(14):9392–9397. doi: 10.1073/pnas.142206299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kizaki T., Ookawara T., Iwabuchi K., Onoe K., Day N. K., Good R. A., Maruyama N., Haga S., Matsuura N., Ohira Y., Ohno H. Age-associated increase of basal corticosterone levels decreases ED2high, NF-κBhigh activated macrophages. Journal of Leukocyte Biology. 2000;68(1):21–30. [PubMed] [Google Scholar]
- 14.Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Lino M., Shigeyoshi Y., Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 15.Guillaumond F., Dardente H., Giguère V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. Journal of Biological Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 16.Harding H. P., Atkins G. B., Jaffe A. B., Seo W. J., Lazar M. A. Transcriptional activation and repression by RORα, an orphan nuclear receptor required for cerebellar development. Molecular Endocrinology. 1997;11(11):1737–1746. doi: 10.1210/me.11.11.1737. [DOI] [PubMed] [Google Scholar]
- 17.Forman B. M., Chen J., Blumberg B., Kliewer S. A., Henshaw R., Ong E. S., Evans R. M. Cross-talk among RORα1 and the Rev-erb family of orphan nuclear receptors. Molecular Endocrinology. 1994;8(9):1253–1261. doi: 10.1210/me.8.9.1253. [DOI] [PubMed] [Google Scholar]
- 18.May M. J., Ghosh S. Signal transduction through NF-κB. Immunology Today. 1998;19(2):80–88. doi: 10.1016/S0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 19.Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ . Nature Structural and Molecular Biology. 2007;14(12):1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teboul M., Gréchez-Cassiau A., Guillaumond F., Delaunay F. How nuclear receptors tell time. Journal of Applied Physiology. 2009;107(6):1965–1971. doi: 10.1152/japplphysiol.00515.2009. [DOI] [PubMed] [Google Scholar]
- 21.Vu-Dac N., Chopin-Delannoy S., Gervois P., Bonnelye E., Martin G., Fruchart J., Laudet V., Staels B. The nuclear receptors peroxisome proliferator-activated receptor α and rev-erbα mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. Journal of Biological Chemistry. 1998;273(40):25713–25720. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- 22.Raspé E., Duez H., Mansén A., Fontaine C., Fiévet C., Fruchart J., Vennström B., Staels B. Identification of Rev-erbα as a physiological repressor of apoC-III gene transcription. Journal of Lipid Research. 2002;43(12):2172–2179. doi: 10.1194/jlr.M200386-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Chauvet C., Bois-Joyeux B., Fontaine C., Gervois P., Bernard M., Staels B., Danan J. The gene encoding fibrinogen-β is a target for retinoic acid receptor-related orphan receptor α . Molecular Endocrinology. 2005;19(10):2517–2526. doi: 10.1210/me.2005-0153. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Yin L., Lazar M. A. The orphan nuclear receptor Rev-erbα regulates circadian expression of plasminogen activator inhibitor type 1. Journal of Biological Chemistry. 2006;281(45):33842–33848. doi: 10.1074/jbc.M607873200. [DOI] [PubMed] [Google Scholar]
- 25.Ashida N., Arai H., Yamasaki M., Kita T. Distinct signaling pathways for MCP-1-dependent integrin activation and chemotaxis. The Journal of Biological Chemistry. 2001;276(19):16555–16560. doi: 10.1074/jbc.M009068200. [DOI] [PubMed] [Google Scholar]
- 26.Migita H., Morser J., Kawai K. Rev-erbα upregulates NF-ρB-responsive genes in vascular smooth muscle cells. The FEBS Letters. 2004;561(1–3):69–74. doi: 10.1016/S0014-5793(04)00118-8. [DOI] [PubMed] [Google Scholar]
- 27.Straub R. H., Cutolo M. Circadian rhythms in rheumatoid arthritis: implications for pathophysiology and therapeutic management. Arthritis & Rheumatism. 2007;56(2):399–408. doi: 10.1002/art.22368. [DOI] [PubMed] [Google Scholar]
- 28.Perry M. G., Kirwan J. R., Jessop D. S., Hunt L. P. Overnight variations in cortisol, interleukin 6, tumor necrosis factor α and other cytokines in people with rheumatoid arthritis. Annals of the Rheumatic Diseases. 2009;68(1):63–68. doi: 10.1136/ard.2007.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferraz E., Borges M. C., Terra-Filho J., Martinez J. A. B., Vianna E. O. Comparison of 4 AM and 4 PM bronchial responsiveness to hypertonic saline in asthma. Lung. 2006;184(6):341–346. doi: 10.1007/s00408-006-0017-0. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi M., Shimba S., Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biological & Pharmaceutical Bulletin. 2007;30(4):621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 31.Keller M., Mazuch J., Abraham U., Eom G. D., Herzog E. D., Volk H., Kramer A., Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silver A. C., Arjona A., Walker W. E., Fikrig E. he circadian clock controls toll-like recept or 9-mediated innate and adaptive immunity. Immunity. 2012;36(2):251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]