Abstract
In plants, sugar transporters are involved not only in long-distance transport, but also in sugar accumulations in sink cells. To identify members of sugar transporter gene families and to analyze their function in fruit sugar accumulation, we conducted a phylogenetic analysis of the Malus domestica genome. Expression profiling was performed with shoot tips, mature leaves, and developed fruit of “Gala” apple. Genes for sugar alcohol [including 17 sorbitol transporters (SOTs)], sucrose, and monosaccharide transporters, plus SWEET genes, were selected as candidates in 31, 9, 50, and 27 loci, respectively, of the genome. The monosaccharide transporter family appears to include five subfamilies (30 MdHTs, 8 MdEDR6s, 5 MdTMTs, 3 MdvGTs, and 4 MdpGLTs). Phylogenetic analysis of the protein sequences indicated that orthologs exist among Malus, Vitis, and Arabidopsis. Investigations of transcripts revealed that 68 candidate transporters are expressed in apple, albeit to different extents. Here, we discuss their possible roles based on the relationship between their levels of expression and sugar concentrations. The high accumulation of fructose in apple fruit is possibly linked to the coordination and cooperation between MdTMT1/2 and MdEDR6. By contrast, these fruits show low MdSWEET4.1 expression and a high flux of fructose produced from sorbitol. Our study provides an exhaustive survey of sugar transporter genes and demonstrates that sugar transporter gene families in M. domestica are comparable to those in other species. Expression profiling of these transporters will likely contribute to improving our understanding of their physiological functions in fruit formation and the development of sweetness properties.
Keywords: Malus, fruit sugar, EDR6, SWEET, genome, sugar transporter
Introduction
In plants, soluble sugars [i.e., sucrose (Suc), monosaccharides, and polyols] are essential molecules that not only provide energy and building blocks for growth and development, but also constitute osmotic, nutrient, and signal molecules (Ruan, 2014). In fruit crops, soluble sugars are also central to quality; their accumulation during the maturation process largely determines sweetness at harvest.
In multicellular organisms, the movement of sugar is an essential part of long-distance transport for assimilates from source to sink organs, storage of carbon, regulation of osmotic potential and turgor, and the cellular exchange of carbon and energy (Slewinski, 2011). Biochemical and molecular researchers have argued that hexoses or sucrose are transported into the chloroplast (Weber et al., 2000), the vacuoles (Martinoia et al., 2000), and the Golgi apparatus (Wang et al., 2006). As relatively large and polar solutes, soluble sugars require proteins to facilitate efficient diffusion across membranes. Not only the loading and the unloading of the conducting complex, but also the allocation of sugars into source and sink cells, are controlled by sugar transporters that mediate the movement of Suc (sucrose transporter: SUT) (Ruan, 2014), reducing monosaccharides (monosaccharide transporter or hexose transporter: MST or HT) (Büttner, 2010; Slewinski, 2011), or sugar alcohols [sorbitol (Sor), mannitol, xylitol, et al.] (Noiraud et al., 2001; Gao et al., 2005; Fan et al., 2009). This model is depicted in Figure 1.
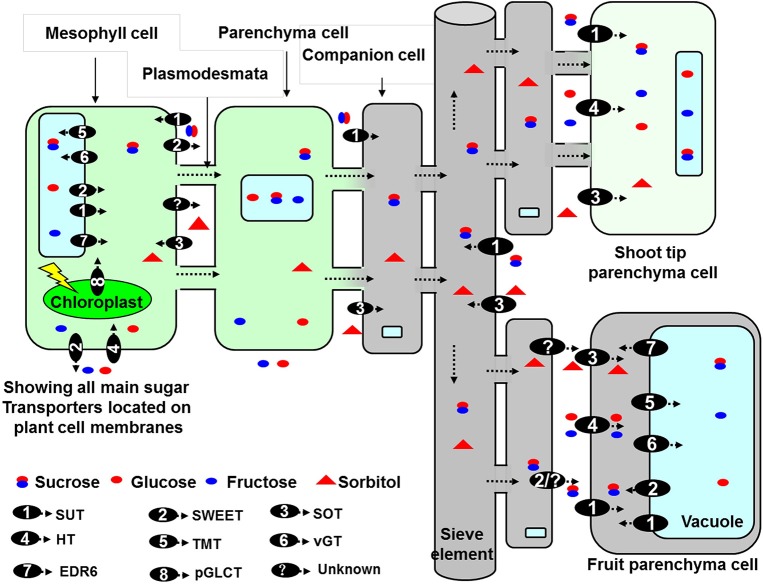
Figure 1.
Model for sugar transport in apple (Zhang et al., 2004; Doidy et al., 2012; Li et al., 2012; Chardon et al., 2013; Ruan, 2014). Sorbitol (Sor) and sucrose (Suc), both of which are synthesized in mesophyll cell of source leaves, are loaded into phloem via symplast pathway. After translocating to SE-CC of fruit, both are unloaded into fruit parenchyma cells via apoplast pathway. In apple cells, four families of transporters are implicated in the distribution of Suc, Sor, fructose (Fru), and glucose (Glc): sucrose transporter (SUT), SWEET, sorbitol transporter (SOT), and monosaccharide transporter (MST) [including hexose transporter (HT), tonoplast membrane transporter (TMT), vacuolar glucose transporter (vGT), ERD six-like transporters (EDR6), and plastid glucose transporters (pGLCT)]. At the plasma membrane, all of SUTs, SOTs, and HTs have been characterized as H+/sugar importers, and transport influx of extracellular Suc, Sor, and hexose. By contrast, SWEETs function as energy-independent uniporters that mainly mediate sugar efflux. At the vacuolar membrane, vGTs, and TMTs function as sugar/H+ antiporters transporting sugars into the vacuole, while EDR6 and SWEET IV subfamilies are involved in energy-independent sugar efflux from the vacuole. Especially, in fruit cells these special tonoplast transporters may be involved in accumulating Fru, Glc, and Suc in vacuole. Additionally, pGLCTs may also play a role in efflux of sugars from plastids.
Efficient movement of sugars across membranes requires the operation of multiple transporters, which will have different energetic and kinetic properties suited for efficient unloading into cell wall spaces, uptake of Suc or other sugars leaked or locally transported via the apoplasm, loading from the cytosol into storage vacuoles, and the fine-tuning of sugar fluxes for homoeostasis and interactions with other proteins for sugar sensing and signaling (Slewinski, 2011; Ruan, 2014). To achieve this, multiple families for genes encode these transporters.
Since the cloning of the first MST (Sauer and Tanner, 1989), SUT (Riesmeier et al., 1992), and polyol transporter in plants (Noiraud et al., 2001), many genes in those families have been isolated from various species. For example, the complete Arabidopsis genome contains nine SUT-like sequences (Ruan, 2014) plus a monosaccharide transporter (-like) gene family that has 53 members in seven subfamilies (Büttner, 2010). The Vitis vinifera (grapevine) genome has four SUTs and 59 MSTs (Afoufa-Bastien et al., 2010). Evolutionary analysis of plant MSTs has revealed seven ancient subfamilies in land plants (Slewinski, 2011). Recently identified SWEET proteins in a distinct transporter family account for 17 members in Arabidopsis and 21 in rice. These members can transport Suc or glucose (Glc) (Chen et al., 2010, 2012) or fructose (Fru) (Chardon et al., 2013; Klemens et al., 2013), and are involved in loading (Chen et al., 2012), sugar storage (Chardon et al., 2013), nectar production (Lin et al., 2014), and interactions between plants and fungi (Chen et al., 2010).
Knowledge is gradually increasing about the intracellular distribution of sugar transporters and their roles in regulating this transport, signaling, and homeostasis in model herbaceous plants, e.g., Arabidopsis. Three families of transporters—SUTs, MSTs, and SWEETs—are mainly implicated in the distribution of sugars within most plant cells (reviewed by Doidy et al., 2012). At the plasma membrane, most transporters have been characterized as H+/sugar importers. However, ZmSUT1 (Carpaneto et al., 2005) and AtSUC4 (Schneider et al., 2011) also mediate the active efflux of Suc. By contrast, SWEETs function as energy-independent uniporters that mediate sugar influx and/or efflux (Chen et al., 2010). Both AtSWEET11and 12 have been localized to the plasma membrane (Chen et al., 2012) whereas AtSWEET17 occurs in the tonoplast membrane, where it transports Fru (Chardon et al., 2013). At the vacuolar membrane, the MST subfamilies, vacuolar glucose transporter (vGT), and tonoplast membrane transporter (TMT) function as sugar/H+ antiporters that load sugars into the vacuole (Wormit et al., 2006; Aluri and Büttner, 2007; Schulz et al., 2011). Proteins of the MST subfamily of ERD six-like transporters (ERD6 or ESL1) are likely involved in energy-independent sugar efflux from the vacuole (Poschet et al., 2011; Klemens et al., 2014).
Research data have also suggested that expression of sugar transporters might be regulated at the transcriptional level by distinct but usually converging signaling pathways that depend upon either developmental and environmental cues or metabolic and hormonal signals. Despite the progress made in identifying genes that encode sugar transporters, little is known about the roles and transcriptional regulation of these genes, especially in crop plants. It is unknown how different transporter orthologs modulate sugar distribution and homeostasis in plant cells, and how they control sugar accumulations in storage tissues and cells. Therefore, analysis of these orthologs in different species might help improve our understanding of their biological functions.
Apple (Malus domestica Borkh.), a member of the Rosaceae family, is among the most important commercial fruit crops grown worldwide. Apple and other Rosaceae tree fruits synthesize sorbitol (Sor) and Suc in source leaves. Both are then translocated to and utilized in fruit, with Sor accounting for approximately 60–70% of the photosynthates produced in the leaves. They are loaded via the symplasmic pathway for transport in the phloem (Reidel et al., 2009). After being unloaded from SE-CC (sieve elements and companying cells) complexes into the cell wall space of apple fruit (Zhang et al., 2004), Sor is taken up into the cytosol of parenchyma cells by a sorbitol transporter (SOT) located on the plasma membrane. Meanwhile, Suc is directly transported into parenchyma cells by SUT on the plasma membrane, or else first converted to Fru and Glc by cell wall invertase and then transported into parenchyma cells by hexose transporters (HT) (Figure 1; Zhang et al., 2004; Fan et al., 2009; Li et al., 2012). In the cytoplasm of the mesocarp cells, sucrose and hexoses must be transported into the vacuole via tonoplastic transporters (Figure 1). Compared with sink organs in model plants that import and metabolize only sucrose (e.g., Arabidopsis, Solanum tuberosum, and Populus), apple is unique in its metabolism and accumulation of sugars. More than 80% of the total carbon flux goes through fructose (because almost all of the Sor and half of the Suc are converted to Fru) (Li et al., 2012). Consequently, the characteristics of sugar transporters can differ between apple and other plants. Roles in unloading and changes in expression during fruit development have been preliminarily reported for MdSOT (Gao et al., 2005) and MdSUT1 (Fan et al., 2009), and Li et al. (2012) identified the some members of gene families encoding transporters (including MdSUT, MdTMT, and MdvGT) and analyzed the relationship of their transcripts with sugar accumulation during fruit development of “Greensleeves” apple, but there was no exhaustive knowledge on apple sugar transporter based on genome, especially for MST and SWEET families. Identification and characterization of these transporter genes in Malus are important steps in understanding the roles of these proteins in growth and development as well as the process of sugar accumulation in the fruit.
Here, we identified SUT, MST, and SWEET genes in the M. domestica genome (Velasco et al., 2010) through phylogenetic analysis and compared them primarily with Arabidopsis transporters. Real-time PCR was used to determine expression patterns in different tissue types, and to examine the relationship between relative transcript abundances and sugar accumulation over time. Our objective was to devise a useful approach for investigating the function of sugar transporters and the development of sweetness traits in apple fruit.
Materials and methods
Plant materials
Nine-year-old “Gala” apple trees (Malus domestica) grafted onto rootstock M. sieversii were trained as a central leader system and grown at the spacing of 3 (row) × 4 (interval) m in North-South rows in an experimental orchard at the Horticultural Experimental Station of Northwest A & F University, Yangling, China. Fungicides and pesticides were sprayed at regular intervals throughout the growing season to protect the plants from diseases and insects. At 16, 34, 55, 75, 98, and 122 days after bloom (DAB), fruits were sampled from the south side of the tree canopy between 3:00 and 4:00 p.m., under full sun exposure. On each collection date, six apples were from 3 trees and pooled for one replicate, with five replications in all from in total 15 trees. The fruits were immediately weighed, cut into small pieces after removing the core, and frozen on-site in liquid nitrogen (2-min interval between harvest and freezing). To compare the expression patterns of related genes in source and sink tissues, we also collected mature leaves and shoot tips at 34 DAB. All frozen samples were stored at −80°C.
Identification of candidate genes
Candidate genes were identified by performing a BlastP analysis against the apple gene set (amino acids) in the Malus Genome Database (http://www.rosaceae.org) (Velasco et al., 2010) from the “Fondazione Edmund Mach Istituto Agrario San Michele All'Adige,” Italy, or IASMA (http://www.rosaceae.org/tools/ncbi_blast). As query, we used sequences for Arabidopsis thaliana polyol/monosaccharide transporter (PMT) and sugar transporters [including STP (sugar transport protein) or HT, EDR6 (early-responsive to dehydration), TMT, vGT, pGLCT (plastid glucose transporter), and SWEET family members] that were obtained from The Arabidopsis Information Resource (http://arabidopsis.org/). Additionally, reported MdSOTs (Li et al., 2011) and MdSUT (Fan et al., 2009) were also employed as query. An E-value of 1.00E-10 was set as the threshold. Putative candidate gene sequences were retrieved from the Malus Genome Database (http://www.rosaceae.org/species/malus/malus_x_domestica/genome_v1.0). To confirm their expression in apple and their reliability as putative candidates, we used their corresponding sequences in a Blast search against the Malus EST database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). A predicted gene was considered expressed when ESTs in the Malus transcriptome showed high similarity, i.e., length >300 bp and identity score >98%, were identified. After comparing between each predicted gene and its EST-constructed contig or EST, we re-conducted the Blast analysis with divergent genes against all predictions in apple (nucleic acids) (http://www.rosaceae.org/tools/ncbi_blast). Here, EST-constructed contigs or ESTs were used so that a sequence concordant with the EST would be found in all predictions.
Sequence similarities and phylogeny analyses
Multiple alignments of the transporter proteins were obtained using DNAMAN software. To construct the phylogenetic tree, we downloaded the full-length amino acid sequences of PMT, sugar transporters, and SWEET family sequences from Arabidopsis and grapevine in the NCBI protein database (http://www.ncbi.nlm.nih.gov/guide/). Those sequences were then aligned with the integrated MUSCLE alignment program in MEGA5 (Molecular Evolutionary Genetics Analysis) with default parameters. Phylogenetic analysis was performed via the Neighbor-Joining method, using MEGA5 software and bootstrap tests replicated 1000 times. In addition, the subcellular localizations of candidate genes were predicted with TargetP software (http://www.cbs.dtu.dk/services/TargetP) and the WoLF PSORT version of PSORT II (http://wolfpsort.org/).
Information about chromosome lengths and gene locations have previously been reported for the apple genome (Velasco et al., 2010). Locations and transcriptional directions were assigned to the chromosomes by MapDraw software (Liu and Meng, 2003).
Analysis of mRNA expression
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to analyze the expression of all selected genes (Supplementary Data Excel Files1–4). Total RNA was extracted from samples by the modified CTAB method (Gasic et al., 2004), and DNase was used to remove DNA before reverse-transcription. After sequence similarities were examined, gene-specific primers (Supplementary Data Excel Files 5) were designed from the coding sequences of apple genes, using Primer5 software. Primer specificity was determined by RT-PCR and Melt Curve analysis. Afterward, qRT-PCR was performed with an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer's protocol. Amplified PCR products were quantified by an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) and an iQ SYBR Green Supermix kit (Bio-Rad). Actin (CN938023) transcripts were used to standardize the different cDNA samples throughout the test (Li et al., 2012). For all samples, total RNA was extracted into five tubes from five replicates and then mixed in a tube used for reverse-transcription. The qRT-PCR experiments were done with three technical replicates. Data were analyzed by the ddCT method in iQ5 2.0 standard optical system analysis software.
Measurements of soluble sugars
Soluble sugars and hexose phosphates were obtained and derivatized as described by Wang et al. (2010). Briefly, samples (0.09–0.11 g) were extracted in 1.4 mL of 75% methanol, with ribitol added as the internal standard. After the non-polar metabolites were fractionated into chloroform, 2 μl of the polar phase was transferred into 2.0-mL Eppendorf vials for measurements of the metabolites (Sor, Fru, Glc, Suc, galactose: Gal and myo-inositol) of each sample. They were dried under vacuum without heating and then derivatized sequentially with methoxyamine hydrochloride and N-methyl-N-trimethylsilyl-trifluoroacetamide (Lisec et al., 2006). Afterward, the metabolites were analyzed with a Shimadzu GCMS-2010SE (Shimadzu Corporation, Tyoto, Japan). These metabolites were identified by comparing their fragmentation patterns with those from a mass spectral library generated on our GC/MS system, and from an annotated quadrupole GC–MS spectral library downloaded from the Golm Metabolome Database (http://csbdb.mpimp-golm. mpg.de/csbdb/gmd/msri/gmd_msri.html. Quantifications were based on standard curves generated for each metabolite and internal standard.
Results
Sorbitol transporters in the malus domestica genome
Using AtPMT and reported MdSOT sequences as queries, we conducted a BlastP search to identify candidate genes in Malus. In all, 40 consensus gene sets containing major facilitator superfamily (MFS) domains were selected from the “Gold Delicious” apple genome database, with an EXP cutoff <1.00E-10. Based on pair-wise comparisons of these predicted genes against Vitis vinifera and peach genome databases, as well as the TrEMBL database, all genes were highly similar to orthologs in Vitis and peach. In addition, 30 selected genes showed very high homology with reported MdSOT (Supplementary Data Excel File 1—Sheet 1). To confirm their expression in apple and to examine the reliability of putative candidate gene sequences, we used the corresponding sequences of candidate genes for a BLAST search against the Malus EST database. Sixteen predicted genes (Supplementary Data Excel File 1—Sheet 2) had high similarities in EST sequences (score > 300 bp, identity > 98%). However, the sequences of six selected genes diverged from their corresponding EST-constructed contigs. A concordant sequence with an EST was found in all predictions from either the Malus genome database or GenBank. For MdSOT1, −4, and −5, we were unable to find concordant sequences in all predicted genes. Based on reports of functional PMTs and SOTs, we set thresholds for the number of transmembrane domains(≥8) and the length of coding amino acids (>300 a.a.). After re-screening, we selected 27 genes as possible members of the MdSOT or MdPMT family. All candidates from the AtPMT families were aligned to elucidate their evolutionary relationships, and were named according to our phylogenetic tree (Figure 2; Supplementary Data Excel File 1).
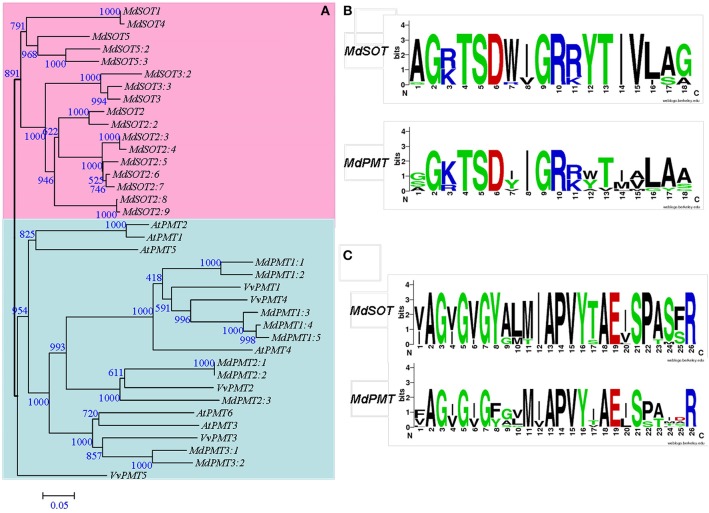
Figure 2.
Phylogenetic analysis (A) of Malus domestica polyol transporter families with Vitis vinifera and Arabidopsis thaliana, and conserved properties of transporter Motif 1 (B) and 2 (C) in MdPMT and MdSOT subfamily. Tree was constructed via Neighbor-Joining method with 1000 bootstrap replications. Accession numbers for all Malus genes are listed in Supplementary Data Excel File 1. Accession numbers for Vitis genes were reported by Afoufa-Bastien et al. (2010). Accession numbers for A. thaliana: AT2G16120.1/ATPMT1, AT2G16130.1/ATPMT2, AT2G20780.1/ATPMT4, AT2G18480.1/AtPMT3, AT3G18830.1/ATPMT5, and AT4G36670.1/ATPMT6.
This phylogenetic tree revealed two distinct groups, with the first containing all reported MdSOTs, but no PMTs from Arabidopsis or Vitis. In this group, MdSOT1 had the highest similarity with MdSOT4 and shared the same clade with MdSOT5. The apple genome showed two newly identified members (MdSOT5.2 and −5.3) with high similarity to MdSOT5. MdSOT3 was very similar to MdSOT3.2 and −3.3 within a small, discrete clade. Eight genes shared high similarity with the amino acid sequence of MdSOT2, clustering in the same clade with MdSOT3. These included MdSOT2.8 and−2.9, both of which had 99.4% similarity.
In the second group, 10 candidate genes were clustered with the PMTs of Arabidopsis and Vitis. TheMdPMT1 subfamily contained five members that had high homology and were in the same clade as AtPMT4, VvPMT1, and VvPMT4. In addition, theMdPMT2 subfamily had three members that were orthologs of VvPMT2. Both MdPMT3.1 and −3.2 had high similarity with AtPMT3, AtPMT6, and VvPMT3, and shared the same clade (Figure 2).
The approximate gene positions and transcriptional directions of candidate genes were marked on the physical map of chromosomes. In all, 17MdSOTs were located on four apple chromosomes (chr1, 3, 12, and 17). They included 11 on chr12 (including six in the MdSOT2 subfamily) (Supplementary Figure 1) plus three on chr17. On chr12, 10 MdSOTs were in tandem at approximately 449 kb. From the MdPMT subfamilies, two pairs of MdPMTs showed tandem duplication, including MdPMT2.1-MdPMT2.2 on chr2 and MdPMT1.4-MdPMT1.5 on chr9. MdPMT2.3 was located on chr15 while single MdPMT1 members were found on chr5, 10, and 17, and two MdPMT3 members occurred on chr2 and chr15.
Sucrose transporters in the malus domestica genome
Using AtSUT or AtSUC protein sequences as query, we found 15 candidate gene sets in the Malus genome, including five predicted genes with high concordant EST sequences. Two sets were divergent with their corresponding ESTs; concordant sequences with ESTs were found in all predictions from the Malus database (Supplementary Data Excel File 2). Based on the reported functionality of SUT or SUC genes, we set the thresholds at ≥6 for the number of transmembrane domains and >200 a.a. for the length of coding amino acids. Nine genes were selected as candidates for the MdSUT family, including five MdSUTs we had described previously (Li et al., 2012).
Phylogenetic analysis divided the nine candidate MdSUTs into three groups that incorporated MdSUT1, MdSUT2, and MdSUT4 clades (Supplementary Figure 2; Supplementary Data Excel File 2). Three members of the MdSUT1 group shared the same clade with VvSUC27 but did not cluster in the same clade with those in the AtSUT1 group (including AtSUC1, AtSUC2, and AtSUC9) that show high-affinity uptake in dicot plants (Ayre, 2011). MdSUT2.1 and −2.2 were 93.6% similar and were clustered in the SUT2 clade with AtSUT2/SUC3 and VvSUT2. The SUT4 clade contained four MdSUT4 members that were similar toAtSUC4 and VvSUC1.
The physical map presented nine MdSUTs on seven apple chromosomes. These included three MdSUT4 members with tandem duplications at the end of chr8, plus MdSUT4.1 on chr15 (Figure S1). Single members of the MdSUT1 subfamily were located on chr10, chr9, and chr5, while MdSUT2.1 and −2.2 were found on chr16 and chr13, respectively (Figure S1).
Monosaccharide transportersin the malus domestica genome
We found 114 MST candidate gene sets in the Malus genome when AtHT, AtSTP, AtEDR6, AtTMT, or other sugar transporter protein sequences were used as query (Supplementary Data Excel File 3). Of these, 27 predicted genes had high-concordance EST sequences in the Malus EST database. Although five sets were divergent with the corresponding ESTs, concordant sequences with ESTs were found in all predictions of the Malus genome database. Simultaneously, 12 sets matched more reasonable splicing sequences in all predictions of that database. Based on the reported functionality of AtSTP, AtHT, and other genes, we set the threshold at >6 for the number of transmembrane domains and at >800 bp for the length of coding amino acids. Ultimately, 30 genes were selected as candidates for the MdHT family, including eight MdEDR6s, five MdTMTs, three MdvGTs, and three MdpGLTs. All candidates that were homologous to family members in the A. thaliana or V. vinifera genomes were aligned to elucidate evolutionary relationships. These were then named according to our phylogenetic tree (Supplementary Data Excel File 3).
Thirty candidate MdHTs were divided into four groups. The first, named the MdHT1 subfamily, contained 11 genes (Figure 3). MdHT1.1 through MdHT1.3 clustered with AtSTP13 and VvHT5. By contrast, MdHT1.4through MdHT1.11 did not cluster with any Arabidopsis or Vitis genes. This MdHT1 subfamily included MdHT1.1, which shared 95.3% similarity to the amino acid sequence ofMdHT1.2, as well as MdHT1.10 and −1.11, with 95.9% similarity. In the second group, five members of the MdHT2 subfamily clustered with AtSTP14, AtSTP7, VvHT13, and VvHT3, while both MdHT2.1 and −2.2 had higher homology with AtSTP14 and VvHT13. The protein sequences of MdHT2.3 and −2.4 differed by only two amino acids. In the third group, MdHT3 subfamily members MdHT3.1through MdHT3.3 were highly homologous with VvHT4 and AtSTP3, while three other MdHT3s were highly conserved and clustered with VvHT2 and AtSTP5. The eight MdHT4 subfamilies that comprised the fourth group were separated into two subgroups. The first had four relatively conserved members clustered with four AtSTPs, based on their amino acid sequences. The second subgroup contained MdHT4.5 and −4.6 with 89.8% similarity in a separated branch, while MdHT4.7 and −4.8 shared 93.8% similarity with AtSTP1 (Figure 3; Supplementary Data Excel File 3).
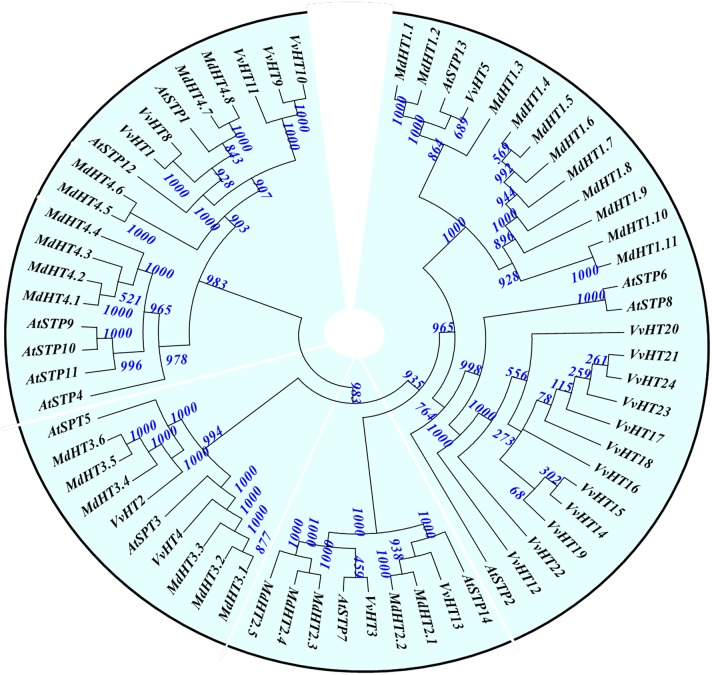
Figure 3.
Phylogenetic analysis of Malus domestica MdHT families with Vitis vinifera and Arabidopsis thaliana. Tree was constructed via Neighbor-Joining method with 1000 bootstrap replications. Accession numbers for all Malus genes are listed in Supplementary Data Excel File 3. Accession numbers for Vitis genes were reported by Afoufa-Bastien et al. (2010). Accession numbers for A. thaliana: AT1G11260.1/AtSTP1, AT1G07340.1/AtSTP2, AT5G61520.1/AtSTP3, AT3G19930.1/AtSTP4, AT1G34580.1/AtSTP5, AT3G05960.1/AtSTP6, AT4G02050.1/AtSTP7, AT5G26250.1/AtSTP8, AT1G50310.1/AtSTP9, AT3G19940.1/AtSTP10, AT5G23270.1/AtSTP11, AT4G21480.1/AtSTP12, AT5G26340.1/AtSTP13, and AT1G77210.1/AtSTP14.
Within the Malus genome, 29 MdHT families were located on 10 chromosomes while MdHT4.6occurred on an unanchored contig. Five MdHT1 members—MdHT1.1, −1.4, −1.5, −1.6, and −1.10—were in tandem on chr9 at approximately 66 kb while the other five MdHT1s were in tandem on chr13 at approximately 95 kb. MdHT1.3 was located on chr7 along with MdHT3.3. For the other MdHT subfamilies, three pairs of MdHTs showed tandem duplications, including MdHT2.3-MdHT2.4 on chr2, MdHT4.1-MdHT4.2 on chr15, and MdHT3.5-MdHT3.6 on chr13 (Supplementary Figure 1). Based on the chromosomal evolution described by Velasco et al. (2010), two pairs of very similar MdHT were located on homologous chromosomes, including MdHT2.1 (chr2)-MdHT2.2 (chr15) and MdHT4.7 (chr5)-MdHT4.8 (chr10) (Supplementary Figure 1).
For other MST families, MdTMT1 on chr6 had 13 ESTs in the Malus EST database, and showed high homology with VvTMT1 (Figure 4). Both MdTMT2 and −3 were 90.4% similar in their amino acid sequences, and clustered with VvTMT2 and AtTMT2. The MdTMT4 and MdTMT5 tandems on chr9 were highly homologous to AtTMT3. MdvGT1 (on chr7) was 93.2% similar to MdvGT2 (chr1), and was orthologous to VvvGT1 and AtvGT1. MdvGT3 (chr11) was orthologous to VvvGT2 and AtvGT3. Chromosome 3 had four MdpGLT genes, i.e., MdpGLT1 and MdpGLT2 with high homology to VvpGLT1 and AtpGLT (At16150), as well as MdpGLT3 (homologous to VvpGLT2 and At05030, a putative AtpGLT) and MdpGLT4 (homologous to VvpGLT4 and At67300) (Figure 4; Supplementary Data Excel File 3).
Figure 4.
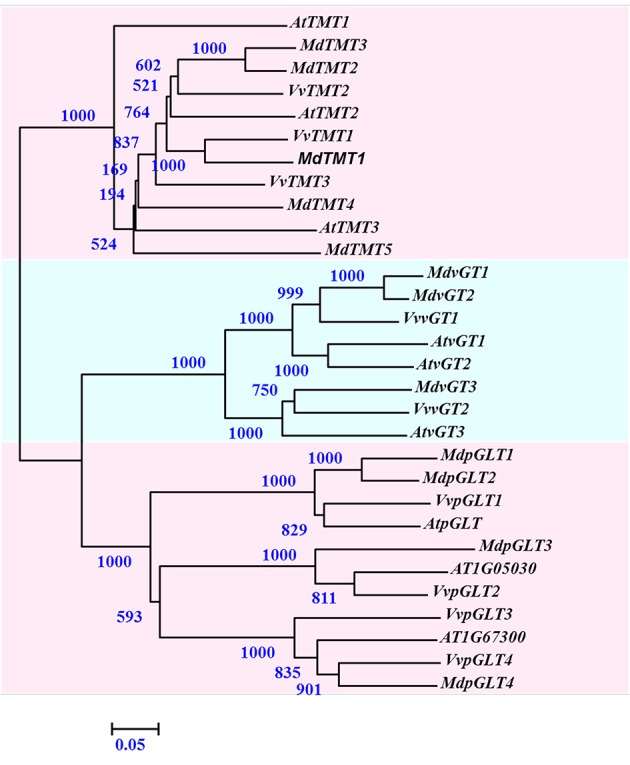
Phylogenetic analysis of Malus domestica MdTMT, MdvGT, and MdpGLT families with Vitis vinifera and Arabidopsis thaliana. Tree was constructed via Neighbor-Joining method with 1000 bootstrap replications. Accession numbers for all Malus genes are listed in Supplementary Data Excel File 3. Accession numbers for Vitis genes were reported by Afoufa-Bastien et al. (2010). Accession numbers for A. thaliana: AT1G20840.1/AtTMT1, AT4G35300.1/AtTMT2, AT3G51490.2/AtTMT3, AT5G16150.1/AtpGLT, AT3G03090.1/AtvGT1, AT5G17010.1/AtvGT2, and AT5G59250.1/AtvGT3.
When AtEDR6 protein sequences were used as query, we found eight MdEDR6 families in the Malus genome. Five predicted genes had highly concordant EST sequences, especially MdEDR6.5. Results of phylogenetic analysis with Arabidopsis AtEDR6-like families indicated that eight candidate MdEDR6s could be divided into three groups (Figure 5; Supplementary Data Excel File 3). For example, five members—MdEDR6.1 through MdEDR6.5—showed high similarity and shared the same clade with AtEDR6L-7, whereas MdEDR6.6 had high homology with AtEDR6L-1. Both MdEDR6.7 and −6.8 were clustered with AtEDR6L-5 and AtEDR6L-6. However, 14 AtEDR6L families plus AtEDR6 (At1g08930) were clustered in clades that had no Malus genes (Figure 5). Four members—MdEDR6.2 through MdEDR6.5—were in tandem at approximately 19 kb at the tail end of chr11. MdEDR6.1 occurred on chr3, MdEDR6.6 and −6.7 were found on chr12, and MdEDR6.8 was on chr8 (Supplementary Figure 1).
Figure 5.
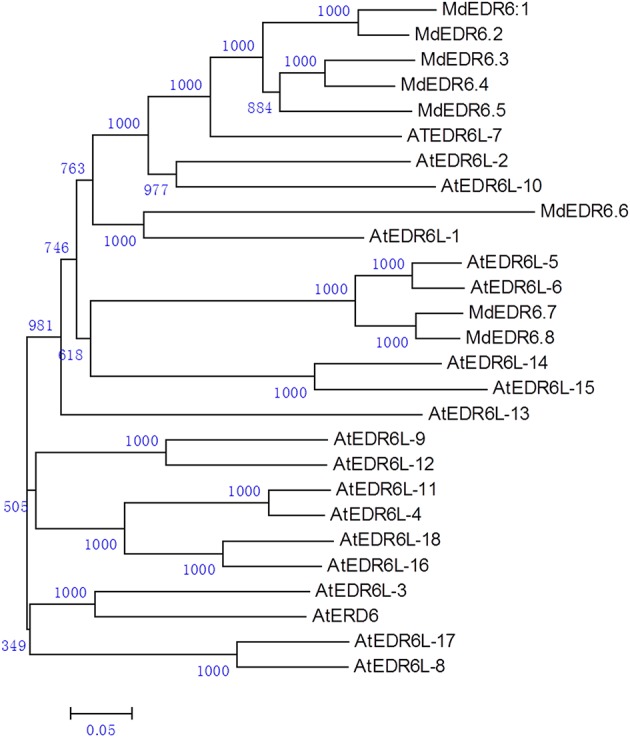
Phylogenetic analysis of Malus domestica MdEDR6 families with Arabidopsis thaliana. Tree was constructed via Neighbor-Joining method with 1000 bootstrap replications. Accession numbers for all Malus genes are listed in Supplementary Data Excel File 3. Accession numbers for A. thaliana: AT1G08930.1/AtERD6, AT1G54730.2/AtEDR6L-1, AT5G18840.1/AtEDR6L-2, AT1G08920.2/AtEDR6L-3, AT3G05160.1/AtEDR6L-4, AT1G19450.1/AtEDR6L-5, AT1G75220.1/AtEDR6L-6, AT2G48020.2/AtEDR6L-7, AT1G08900.1/AtEDR6L-8, AT3G05155.1/AtEDR6L-9, AT3G05150.1/AtEDR6L-10, AT3G05165.1/AtEDR6L-11, AT3G05400.1/AtEDR6L-12, AT3G20460.1/AtEDR6L-13, AT4G04750.1/AtEDR6L-14, AT4G04760.1/AtEDR6L-15, AT5G27350.1/AtEDR6L-16, AT1G08890.1/AtEDR6L-17, and AT5G27360.1/AtEDR6L-18.
MdSWEET genes in the malus domestica genome
The Malus genome contained 33 candidate SWEET gene sets, which were revealed when 17 AtSWEET protein sequences were used as query. Of these, 16 had high concordant EST sequences in the Malus EST database. Five sets diverged in their corresponding ESTs, but concordant sequences with ESTs were found in all predictions for the Malus genome database (Supplementary Data Excel File 4). Another three sets had reasonably well-matched splicing sequences in all predictions. The thresholds for MdSWEET were set at >4 for the number of transmembrane domains and >200 a.a. for the length of the coding amino acids. Upon re-screening, 29 genes were identified as candidates, which were then aligned to elucidate their evolutionary relationships and named according to our phylogenetic tree (Figure 6; Supplementary Data Excel File 4).
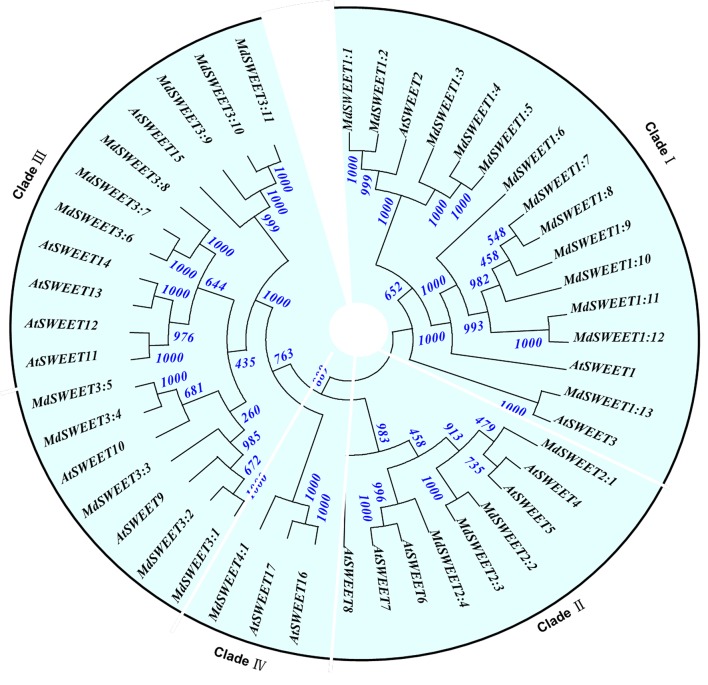
Figure 6.
Phylogenetic analysis of Malus domestica MdSWEET families with Arabidopsis thaliana. Tree was constructed via Neighbor-Joining method with 1000 bootstrap replications. Accession numbers for Malus genes are listed in Supplementary Data Excel File 4. Accession numbers for A. thaliana: AT1G21460.1/AtSWEET1, AT3G14770.1/AtSWEET2, AT5G53190.1/AtSWEET3, AT3G28007.1/AtSWEET4, AT5G6285.1/AtSWEET5, AT1G66770.1/AtSWEET6, AT4G10850.1/AtSWEET7, AT5G40260.1/AtSWEET8, AT2G39060.1/AtSWEET9, AT5G50790.1/AtSWEET10, AT3G48740.1/AtSWEET11, AT5G23660.1/AtSWEET12, AT5G50800.1/AtSWEET13, AT4G25010.1/AtSWEET14, AT5G13170.1/AtSWEET15, AT3G16690.1/AtSWEET16, and AT4G15920.1/AtSWEET17.
Phylogenetic analysis demonstrated that the 29 families could be separated into four groups: MdSWEET1, MdSWEET2, MdSWEET3, and MdSWEET4 (Figure 6), similar to what has been reported previously with Arabidopsis and rice (Chen et al., 2010). The MdSWEET1 subfamily contained 13 members. Of these, MdSWEET1.1 through MdSWEET1.5 were clustered in a clade with AtSWEET2. The amino acid sequence of MdSWEET1.2 had 43 more residues of amino acid at 5′ end compared with MdSWEET1.1. MdSWEET1.4 and −1.5 were 93.1% similar. MdSWEET6 through MdSWEET12 were homologous to AtSWEET1, and had >83.5% similarity among their amino acid sequences. As a single, AtSWEET1.13 clustered with AtSWEET3. Of the four candidates in the MdSWEET2 subfamily, MdSWEET2.1 showed high homology with AtSWEET4 and −5, but was also closely homologous to MdSWEET2.2 and −2.3. MdSWEET2.4 showed high homology with AtSWEET7 and −8. In the MdSWEET3 subfamily, MdSWEET3.1 and −3.2 (87.3% similarity) were clustered with AtSWEET9 and MdSWEET3.3. MdSWEET3.4 and −3.6 (88.1% similarity) were homologous to AtSWEET10. Both MdSWEET3.6 and −3.7 were 88.3% similar to MdSWEET3.8, and clustered with AtSWEET11 through AtSWEET14. MdSWEET3.9 clustered with AtSWEET15 and showed 85.8% similarity with MdSWEET3.10 and −3.11, both of which had the same sequence. In the fourth subfamily, MdSWEET4.1 was grouped with AtSWEET16 and −17 (Figure 6; Supplementary Data Excel File 4).
In all, these 29 MdSWEET genes were distributed on 15 chromosomes plus one anchored contig. Four pairs of MdHT arose from tandem duplications, including MdSWEET1.1-MdSWEET1.2 on chr3, MdSWEET1.9-MdSWEET1.10 on chr7, MdSWEET3.7-MdSWEET3.8 on chr14, and MdSWEET3.11-MdSWEET3.12 on chr16 (Supplementary Figure 1).
Expression profiling of sugar transporters in different tissues of apple
To determine the tissue-specific expression levels of candidate genes, we performed qRT-PCR to analyze mRNA relative abundance in mature leaves, shoot tips, young fruit (16 DAB), and completely ripened fruit (122 DAB) from “Gala” apple (Supplementary Data Excel File 6). Transcripts of 10 MdSOT and seven MdPMT subfamilies were examined. Expression of MdSOT3, MdSOT5, and MdSOT5.3 was strongest in mature leaves, whereas transcripts for MdSOT1, MdSOT3.3, and MdPMT1.1 were most abundant in ripened fruit and those of MdSOT2 and MdPMTs were at their highest levels in young fruit (Figure 7).
Figure 7.
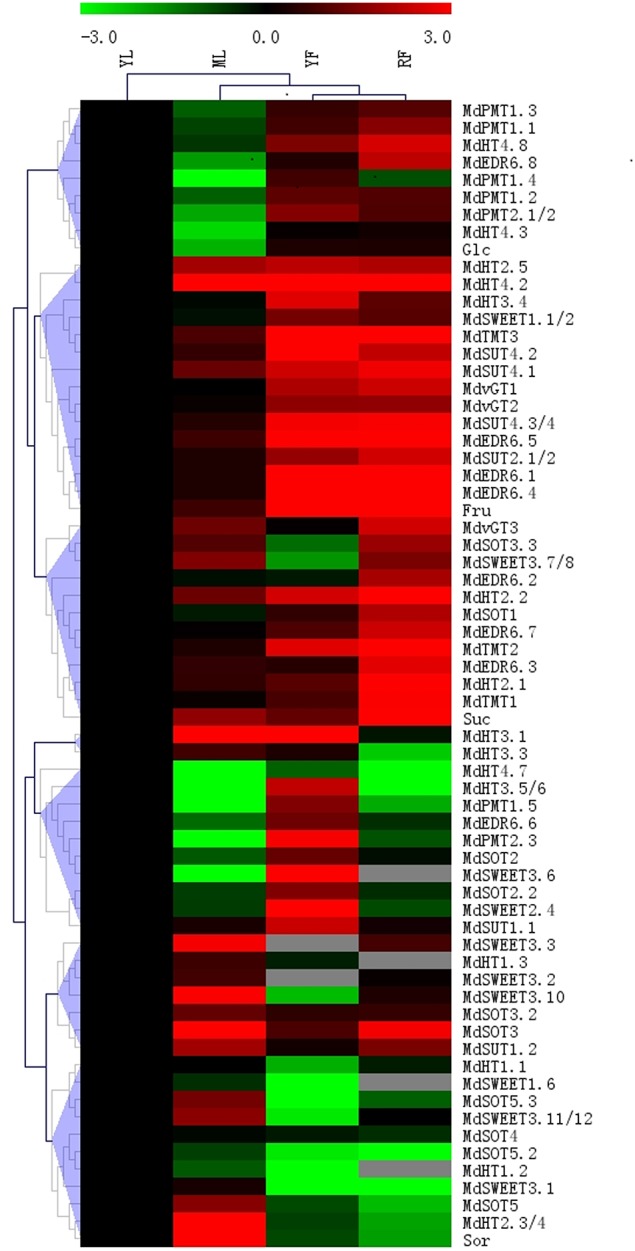
Heat map illustrating sugar concentrations and transporter expression levels in shoot tips, mature leaves, young fruit (16 DAB), and mature fruit (122 DAB) from “Gala” apple. Relative expression abundances and sugar concentration are shown in Supplementary Data Excel Files 6, 7. Fold-difference is designated as log2 value, while the data in shoot tip was set as “1” for each gene and sugar. Genes with similar profiles across arrays are grouped on top by hierarchical-clustering method.
Expression was detected for 11 members of the MdHT family. However, MdHT1.2 and −1.3 were barely detected in the fruit, especially those that were already ripened. MdHT2.1, −2.2, and −4.8 showed the highest abundance in older fruit, while MdHT3.4 and MdHT3.5/6 were most strongly detected in young fruit, MdHT2.3/4 in mature leaves, and MdHT1.2 and MdHT4.7 in young leaves (Figure 7; Supplementary Data Excel File 6).
Except for MdEDR6.6 (most abundant in young fruit), the seven MdEDR family members showed their highest levels of expression in ripened fruit. For those genes, expression was not distinctly different between young and mature leaves. Transcripts ofMdTMT1, MdTMT2, and MdvGT3 were most abundant in ripened fruit while those of MdTMT3 were highest in young fruit (Supplementary Data Excel File 6).
Using 10 pairs of primers, we detected a high level of expression by various MdSWEETs in mature leaves. Expression by MdSWEET1.6 and −3.5 was not abundant in ripened fruit or by MdSWEET3.1 and −3.2 in young fruit. Transcript levels were much higher for MdSWEET1.1/2, MdSWEET2.4, and MdSWEET3.5 in young fruit and for MdSWEET3.1 and MdSWEET3.10/11 in mature leaves (Supplementary Data Excel File 6).
Except for MdSUT1.2 in mature leaves, expression of MdSUTs was higher in fruit tissues. In particular, MdSUT1.1 and MdSUT4.2 transcripts were more abundant in the young fruit while those of MdSUT2.1/2 and MdSUT4.1 were at higher levels in ripened fruit (Supplementary Data Excel File 6).
When we investigated possible correlations between gene expression and sugars (Figure 7), we found that transcripts of MdSOT members, especially MdSOT5, paralleled that of Sor transport and accumulations. The expression of most MdEDR6 members (except MdEDR6.6 and −6.8) showed parallel trends for Suc and Fru concentrations, particularly in the fruit. Other genes, including MdTMT1, MdTMT2, MdHT2.1, and MdHT2.2, were closely associated with Fru and Suc concentrations.
The relationship between expression abundance and the concentration of main sugars during fruit development
To investigate the relationship between gene expression and sugar accumulations, we assayed fruits at different stages of development (Figure 8; Supplementary Data Excel Files 6, 7). Overall, five general patterns emerged. In the first, and largest, group, expression by 17 genes increased in parallel with higher concentrations of Fru and Suc. This was especially true for MdTMT1, MdTMT2, MdHT2.1, MdHT2.2, MdSWEET3.6/7, and five MdEDRs, for which expression was significantly and positively correlated with levels of Suc and Fru in ripened fruit (Table 1). Concentrations of Sor decreased over time in conjunction with diminished expression by 12 genes, such as MdHT3.3, MdPMT1.4, MdPMT1.5, MdSUT1.1, MdSWEET2.4, MdSOT2, and MdSOT5. In addition, Glc concentrations were significantly and positively correlated with MdvGT2 and MdSOT4 in developed fruit, while Gal concentrations were correlated with the expression of MdEDR6.6 and MdSOT5.2 (Table 1; Figure 8).
Figure 8.
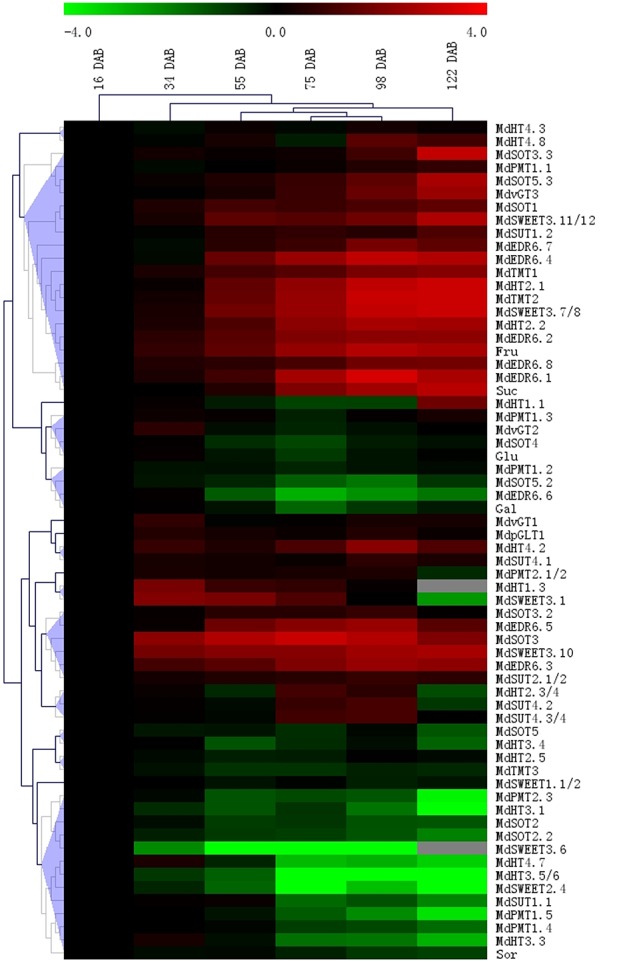
Heat map illustrating sugar concentrations and transporter expression levels in fruits of “Gala” apple at different developmental stages. Relative expression abundances, measured with qRT-PCR, are shown in Supplementary Data Excel Files 6, 7. Fold-difference is designated as log2 value, while the data in young fruit (16 DAB) s was set as “1” for each gene and sugar. Genes with similar profiles across arrays are grouped on top by hierarchical-clustering method.
Table 1.
Correlation coefficients between expression abundance and main sugars or sugar alcohols during development of apple fruit (n = 6).
| Glc | Fru | Suc | Gal | Sor | Glc | Fru | Suc | Gal | Sor | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MdHT1.1 | 0.439 | 0.241 | 0.558 | 0.234 | −0.446 | MdSWEET1.1/2 | 0.249 | −0.790 | −0.707 | 0.406 | 0.692 |
| MdHT1.2 | 0.407 | −0.842* | −0.866* | 0.737 | 0.869* | MdSWEET1.6 | 0.709 | −0.809 | −0.688 | 0.768 | 0.682 |
| MdHT1.3 | 0.177 | −0.538 | −0.691 | 0.305 | 0.528 | MdSWEET2.4 | 0.622 | −0.897* | −0.795 | 0.801 | 0.843* |
| MdHT2.1 | −0.274 | 0.958** | 0.987** | −0.587 | −0.971** | MdSWEET4.1 | −0.004 | −0.525 | −0.705 | 0.242 | 0.572 |
| MdHT2.2 | −0.452 | 0.997** | 0.948** | −0.753 | −0.968** | MdSWEET3.1 | −0.017 | 0.728 | 0.844* | −0.234 | −0.774 |
| MdHT2.3/4 | −0.651 | 0.245 | 0.012 | −0.696 | −0.133 | MdSWEET3.2 | 0.074 | 0.785 | 0.945** | −0.269 | −0.888* |
| MdHT2.5 | 0.636 | 0.003 | 0.141 | 0.385 | −0.106 | MdSWEET3.5 | 0.445 | −0.706 | −0.553 | 0.578 | 0.665 |
| MdHT4.2 | −0.262 | 0.850* | 0.689 | −0.581 | −0.790 | MdSWEET3.6/7 | −0.307 | 0.968** | 0.977** | −0.609 | −0.967** |
| MdHT4.3 | −0.134 | 0.612 | 0.515 | −0.242 | −0.485 | MdSWEET3.9 | −0.261 | 0.869* | 0.812* | −0.517 | −0.876* |
| MdHT4.7 | 0.666 | −0.907* | −0.860* | 0.844* | 0.829* | MdSWEET3.1011 | −0.058 | 0.741 | 0.888* | −0.301 | −0.820* |
| MdHT4.8 | 0.091 | 0.749 | 0.730 | −0.202 | −0.730 | MdSUT1.1 | 0.457 | −0.885* | −0.920** | 0.763 | 0.911* |
| MdHT3.1 | 0.135 | −0.808 | −0.810 | 0.371 | 0.833* | MdSUT1.2 | −0.353 | 0.761 | 0.857* | −0.524 | −0.791 |
| MdHT3.3 | 0.569 | −0.923** | −0.919** | 0.798 | 0.883* | MdSUT2.1/2 | −0.472 | 0.954** | 0.820* | −0.716 | −0.891* |
| MdHT3.4 | 0.383 | −0.445 | −0.496 | 0.341 | 0.420 | MdSUT4.1 | 0.032 | 0.723 | 0.592 | −0.263 | −0.698 |
| MdHT3.5/6 | 0.565 | −0.915* | −0.802 | 0.785 | 0.875* | MdSUT4.2 | −0.597 | 0.455 | 0.169 | −0.700 | −0.296 |
| MdEDR6.1 | −0.449 | 0.962** | 0.849* | −0.756 | −0.899* | MdSUT4.3/4 | −0.687 | 0.668 | 0.433 | −0.858* | −0.539 |
| MdEDR6.2 | −0.522 | 0.981** | 0.925** | −0.784 | −0.949** | MdSOT1 | −0.271 | 0.856* | 0.866* | −0.488 | −0.861* |
| MdEDR6.3 | −0.455 | 0.993** | 0.905* | −0.753 | −0.960** | MdSOT2 | 0.484 | −0.896* | −0.808 | 0.653 | 0.828* |
| MdEDR6.4 | −0.463 | 0.993** | 0.921** | −0.742 | −0.937** | MdSOT2.2 | 0.335 | −0.879* | −0.848* | 0.566 | 0.882* |
| MdEDR6.5 | −0.781 | 0.818* | 0.567 | −0.884* | −0.640 | MdSOT3 | −0.806 | 0.534 | 0.245 | −0.793 | −0.380 |
| MdEDR6.8 | −0.200 | 0.961** | 0.972** | −0.546 | −0.984** | MdSOT3.2 | −0.718 | 0.571 | 0.241 | −0.71 | −0.345 |
| MdEDR6.7 | −0.314 | 0.953** | 0.877* | −0.610 | −0.895* | MdSOT3.3 | 0.233 | 0.528 | 0.784 | −0.033 | −0.691 |
| MdEDR6.6 | 0.772 | −0.857* | −0.725 | 0.865* | 0.732 | MdSOT4 | 0.959** | −0.500 | −0.292 | 0.849* | 0.314 |
| MdTMT1 | −0.257 | 0.951** | 0.972** | −0.560 | −0.964** | MdSOT5 | 0.168 | −0.475 | −0.631 | 0.329 | 0.607 |
| MdTMT2 | −0.300 | 0.976** | 0.974** | −0.616 | −0.971** | MdSOT5.2 | 0.688 | −0.907* | −0.709 | 0.895* | 0.805 |
| MdTMT3 | 0.691 | −0.635 | −0.484 | 0.693 | 0.537 | MdSOT5.3 | 0.075 | 0.698 | 0.897* | −0.219 | −0.822* |
| MdvGT1 | 0.569 | 0.025 | 0.064 | 0.304 | −0.200 | MdPMT1.1 | −0.059 | 0.849* | 0.974** | −0.390 | −0.915* |
| MdvGT2 | 0.837* | −0.503 | −0.379 | 0.760 | 0.311 | MdPMT1.2 | 0.734 | −0.455 | −0.205 | 0.770 | 0.362 |
| MdvGT3 | 0.001 | 0.793 | 0.951** | −0.318 | −0.889* | MdPMT2.1/2 | −0.361 | −0.092 | −0.431 | −0.259 | 0.251 |
| MdpGLT1 | 0.165 | 0.233 | 0.025 | 0.015 | −0.213 | MdPMT2.3 | 0.427 | −0.837* | −0.837* | 0.583 | 0.810 |
| MdpGLT2 | −0.358 | 0.764 | 0.531 | −0.559 | −0.625 | MdPMT1.3 | 0.786 | 0.050 | 0.239 | 0.612 | −0.202 |
| MdpGLT3 | 0.319 | 0.678 | 0.775 | −0.067 | −0.765 | MdPMT1.4 | 0.426 | −0.947** | −0.974** | 0.729 | 0.960** |
| MdpGLT4 | −0.558 | 0.939** | 0.776 | −0.781 | −0.820* | MdPMT1.5 | 0.439 | −0.969** | −0.974** | 0.727 | 0.963** |
Significant at P ≤ 0.05;
Significant at P ≤ 0.01.
Discussion
Domesticated apple genotypes are very heterozygous, making genome sequencing difficult (Velasco et al., 2010). As a result, predicted genes in the Malus genome database contain redundant data. Using protein sequences of AtPMT and reported MdSOT (Gao et al., 2005; Fan et al., 2009) as query, we obtained 40 consensus putative SOT gene sets from the Malus database, which is more than found for apple by Velasco et al. (2010) and much more than those described for peach (Verde et al., 2013). As thresholds, we used Malus ESTs, and set sequence size and number of transmembrane domains as high as possible. Putative gene sets of insufficient size or with too few transmembrane domains did not allow one to find ESTs in the GenBank database. After screening, 27 of 40 putative SOT genes were chosen as possible members of the MdSOT and MdPMT families. The same selections were done for MSTs, SUTs, and SWEETs. Although it is not likely that these genes are the only ones that exist, we are confident that they are functional because we found all of them to be expressed in Malus, and that they represent most of the genes expressed in apple leaves, shoot tips, and fruit.
Rosaceae plants have specific sorbitol transporters
In Rosaceae, photosynthesis-derived carbohydrates are transported mainly as Sor, which is mediated by SOT, a subfamily of the PMT family (Fan et al., 2009). Compared with other plant genomes, apple has considerably more copies of key genes related to Sor metabolism, including aldose 6-P reductase (A6PR), sorbitol-dehydrogenase (SDH), and SOT (Velasco et al., 2010). Both SDH and SOT, which are highly specific to Rosaceae fruits, have evolutionarily been shown to be large families of specific paralogous genes. After clustering to elucidate the evolutionary relationship with AtPMT and VvPMT families, we were able to divide the 27 candidate genes into two groups: MdSOTs and MdPMTs, and MdSOTs group contained all reported functional MdSOTs but lacked any PMT from Arabidopsis and Vitis. Our 10 MdSOTs (including highly expressed MdSOT2, −4, and −5; Gao et al., 2005) were in tandem at approximately 449 kb on chr12. However, no MdSOTs or MdPMTs were found on chr4 and chr14, both of which share partial homology with large segments on chr12 (Velasco et al., 2010). Reported members of MdSOT families have greater substrate selectivity for Sor (Watari et al., 2004; Gao et al., 2005). All of our selected MdPMT members had lower transcript abundance in mature leaves that contained a high Sor concentration. These results implied that the segment on chr12 is related evolutionarily to Sor metabolism, and that a trend toward Sor accumulation may have been somewhat based on gene duplication that created large families of MdSOT paralogous genes. Many PMT subfamily members in Arabidopsis transport a wide range of sugar alcohols and hexoses, and may be non-specific for sorbitol (Klepek et al., 2010). Supporting this, we determined that the expression of MdPMT1.1 and -1.2 was highly correlated with Fru and Suc concentrations in developed fruit, and that expression of seven MdPMTs was relatively higher in fruits than in leaves. Further confirmation is needed for the function of MdPMT in conferring substrate selectivity for Sor or other sugar alcohols and hexoses.
Carbohydrate unloading in apple is driven by SOT, SUT, and HT families
After their synthesis in apple leaves, both Sor and Suc are passively loaded into SE-CC of the phloem via the symplasm pathway (Reidel et al., 2009), and then unloaded in the fruit to the parenchyma cells through the apoplasm pathway (Zhang et al., 2004). Therefore, both SOT and SUT may not be essential for phloem loading, but serve as proton co-transporters for phloem unloading. This theory is supported by reports of higher transcript abundances for MdSOT1 and −2 (Gao et al., 2005) and our finding of a MdSUT4 clade and four MdPMT1 members in fruit. Although AtSUT4, as an antiporter, is located on the tonoplast (Schneider et al., 2011), MdSUT4.1 is present on the plasma membrane and functions in unloading Suc to the apoplasm (Fan et al., 2009). Many MdSOTs, e.g., MdSOT3, −4, −5, and −5.3, are expressed in the vascular tissue of source leaves (Watari et al., 2004). They might not play direct roles in the movement of sugar alcohols or phloem loading. When highly expressed in the leaves, MdSOT and MdSUT may be involved in recovering leaked Suc or Sor, local transport via the apoplasm, or regulation of osmotic potential and turgor among intracellular compartments by adjusting concentrations to adapt to changes in environmental conditions, such as drought (Li et al., 2012).
Most of the HTs or STPs identified in Arabidopsis are localized to the plasma membrane and are capable of transporting hexoses, such as Glc, Fru, and Gal. In apple fruit, after Suc is unloaded, it can be hydrolyzed to Glc and Fru by cell wall invertase. MdHT is needed for moving Glc and Fru from cell wall spaces into parenchyma cells because Sor transport is inhibited by high concentrations of hexoses (Gao et al., 2003). We detected mRNA expression of MdHT2.1 and −2.2 were more abundant in the fruit and were significantly correlated with Fru and Suc concentrations. This is the same trend noted for VvHT13, a highly orthologous gene in grape (Afoufa-Bastien et al., 2010). However, their homologous gene in Arabidopsis is AtSTP14, which is galactose substrate-specific (Poschet et al., 2010). Expression of MdHT4.4 was also highly correlated with Fru concentrations in developed fruit. This gene has high homology with AtSTP9, which is Glc substrate-specific (Schneidereit et al., 2003). These results implied that the functions of these three MdHTs are different in apple when compared with their orthologs in Arabidopsis. MdHT4.8 was highly expressed in ripened fruit and showed high homology to AtSTP1, VvHT1, and VvHT8. AtSTP1 transports Glc as well as Gal, mannose, and xylose (Büttner, 2010). AtSTP1, −13, and −14 function in the recovery of hexoses generated during the process of cell wall remodeling and because of passive leakage by sugars from cells (Yamada et al., 2011). Stronger expression of MdHT2.1, −2.2, and −4.8 in ripened apple fruit may be related to the process of hexose release (including that of Gal, xylose, and Glc) due to cell wall degradation. In young fruit, transcript levels for MdHT3.5/6 and MdHT3.4 were high, and those genes showed the closest similarity to AtSTP5 (uncharacterized) in Arabidopsis and VvHT2 in Vitis. Although VvHT2 is weakly expressed in grape leaves, transcript levels are high in young berries but decline around veraison (Afoufa-Bastien et al., 2010). That pattern resembles what we observed for MdHT3.5/6 expression in apple. In tomato fruit, three HT genes are co-localized with QTLs for sugar accumulation (Prudent et al., 2011). That relationship has been further verified by RNAi knockdowns of three LeHTs (McCurdy et al., 2010) that are highly homologous to MdHT4.8, MdHT2.1/2.2, and MdHT1.1. AtSTP13, highly homologous to MdHT1.1/2, can transport Glc, Fru, and Gal in yeast (Yamada et al., 2011). Here, MdHT1.1 expression was strongly up-regulated in mature fruit whereas MdHT1.2 was barely detectable in any fruit. Future studies should focus on the roles that MdHTs have in sugar accumulation and efficient unloading of carbohydrates.
Expression of MdvGTs, MdTMts, MdEDR6, and MdSWEETs is correlated with sugar accumulations in apple fruit vacuoles
For apple, most of the Fru, Suc, and Glc are stored in the central vacuoles of parenchyma cells (Yamaki and Ino, 1992). This requires that sugars be transported from the cytosol by carrier proteins localized on the tonoplast membrane. In Arabidopsis, these vacuolar proteins are encoded by AtvGT (Aluri and Büttner, 2007), AtTMT (Wormit et al., 2006), AtEDR6 (Poschet et al., 2011), and AtSWEET17 (Chardon et al., 2013). We found three MdvGTs with high homology to three AtvGTs; their expression was much stronger in fruit than in leaves. We have previously reported (Li et al., 2012) that MdvGT1 and MdvGT2 transcript levels are highest in young fruit and are correlated with Glc concentrations in ripened fruit. MdvGT3, which presents an N-terminal extension as VvvGT2 (Afoufa-Bastien et al., 2010), is more closely related to AtvGT3 and VvvGT2. Its expression is highly correlated with Suc concentrations in fruit. By contrast, expression of MdvGT3 was stronger in mature leaves where less sugar was accumulated. Its predicted locations were primarily the chloroplasts and mitochondrial membrane. Although AtvGT1 mediates low-affinity Fru uptake when expressed in yeast vacuoles (Aluri and Büttner, 2007), we found no correlation between MdvGT1/2 expression and Fru uptake in apple vacuoles. These results suggested that MdvGT is not active in the accumulation of Fru and Suc within ripened fruit but may be involved in Glc transport.
Arabidopsis AtTMT1and AtTMT2 are equally capable of moving Glc and Suc into the vacuoles (Schulz et al., 2011). The former is associated with a rate of Fru transport that is approximately 30% of that for Glc in yeast (Wormit et al., 2006). Two of three grape TMTs—VvTMT1 and −2—are strongly expressed in post-veraison berries, a period that coincides with the major phase of hexose accumulation (Afoufa-Bastien et al., 2010). We previously reported that, although 18 predicted genes in apple genome are similar to AtTMTs, we were able to select only five MdTMTs based on ESTs, length of mRNA, and number of transmembrane domains. Transcript levels of MdTMT1 and −2, both of which share high similarity in amino acids with AtTMT1/2 and VvTMT1/2, paralleled Fru and Suc concentration, suggesting that both proteins are involved in transporting those sugars into the vacuoles. Therefore, they may be highly efficient Fru-specific vacuolar transporters in apple. This should be clarified in future examinations.
AtERD6 and AtESL1 (or AtEDR6L-3) are vacuolar exporters of Glc (Yamada et al., 2010; Poschet et al., 2011; Klemens et al., 2014). We found 16 predicted genes in the Malus genome with homology to AtEDR6. However, only eight had amino acids or trans-membrane domains that were of similar size or number. Expression by the seven selected MdEDR families was highest in ripened fruit, and was strongly and positively correlated with Fru and Suc concentrations but negatively with Glc concentrations. By contrast, MdEDR6.6 was highly expressed in young fruit. In pineapple, AcMST1 (EF460876 in GenBank) has high homology with MdEDR6.7 and −6.8 and is located on the tonoplast; its expression is highest in fruit tissues (Antony et al., 2008). If the substrate for MdEDR is specific to Glc, we expect that a lower Glc concentration in fruit vacuoles would be related to greater MdEDR6 expression in ripened fruit. Similar to AtTMT (Schulz et al., 2011), the protein encoded by MdTMT may be capable of transporting Glc and Suc into the vacuoles. Consequently, Glc moves into the vacuoles of ripened fruit along with Fru and Suc. However, because fruit sweetness has been artificially, rather than evolutionarily or naturally, selected, EDR6 may be capable of moving Glc from the vacuoles to maintain turgor and permit a greater accumulation of Fru, which is the sweetest sugar. This phenomenon has also been suggested for pineapple, where the high amount of EDR6 transcript in the leaves accommodates the efflux of Glc and Fru at night to provide substrates for dark-uptake of CO2 (Antony et al., 2008). Further characterization of MdTMT functioning would resolve this question.
The recently identified SWEET superfamily has 17 members in Arabidopsis and 21 in rice (Chen et al., 2010; Xuan et al., 2013). Here, we selected 29 genes as candidates in the Malus genome. Although the number of MdSWEET members related to special carbohydrate metabolism and higher Sor concentrations may in fact be larger in Malus, we detected the transcripts of only 10 MdSWEETs in apple leaves and/or fruit. Plant SWEET proteins are grouped into four subclades, based on amino acid homologies (Chen et al., 2010). Compared with the three AtSWEETs in Arabidopsis, 13 MdSWEET1 subfamilies are clustered in Clade I, in which SWEET proteins mainly transport monosaccharides (Chen et al., 2010). Expression of MdSWEET1.1/2 was twice as high in the fruit when compared with the leaves and shoot tips, which suggested that it may not be affected by Glc, Fru, and Suc concentrations in apple cells. Therefore, we hypothesized that MdSWEET1.1/2 encodes the only MdSWEET protein in apple for transporting Sor, serving as a uniporter with both uptake and efflux activities. Compared with MdSWEET1.1, more than 45 residues at the 5′-end possibly target MdSWEET1.2 to the tonoplasts, based on our predicted localization.
For Clade II, only MdSWEET2.4 was detected, especially in young fruit. Its Arabidopsis homolog ATSWEET8 is essential for pollen viability and is co-opted by pathogens, which likely provide an energy source and carbon at the site of infection by mediating Glc transport (Chen et al., 2010). Another gene, OsSWEET5 in rice, is located on the plasma membrane and is homologous to AtSWEET4, which encodes a Gal transporter in yeast. Those earlier observations support our finding that MdSWEET2.4 expression was correlated with Glc and Gal concentrations during fruit development.
In Clade III, SWEETs in Arabidopsis and rice transport disaccharides, mainly Suc (Chen et al., 2010, 2012). In particular, AtSWEET11 and -12 mediate the key step of Suc efflux from phloem parenchyma cells for translocation via the apoplast loading pathway (Chen et al., 2012). AtSWEET9 is essential for nectar production and can function as a Suc efflux transporter (Lin et al., 2014). However, in apple leaves, loadings of Suc and Sor occur through the symplast pathway, which does not require a SWEET gene to mediate Suc efflux from the parenchyma to extracellular spaces. In our MdSWEET3 clade, MdSWEET3.6, −3.7, and −3.8 were most similar (48%) to AtSWEET11 and 12. Expression of MdSWEET3.6/7 was higher in mature leaves and ripened fruits. Similar trends were observed for transcripts of MdSWEET3.9 and −3.10/11, which are homologous to AtSWEET15, a gene strongly induced by leaf senescence (Quirino et al., 1999). As homologs of AtSWEET9, MdSWEET3.1 and −3.2 showed the highest abundance in developed fruits whereas MdSWEET3.5 (homologous to AtSWEET10) had the highest expression in tissues undergoing rapid growth, such as shoot tips and young fruit. Different expression models have linked their functions with the maintenance of a balance in Suc efflux within various tissues and under certain conditions. In apple fruit, Suc and Sor unloading is accomplished through the apoplast pathway (Zhang et al., 2004), which requires one transporter to move Suc from SE-CC into the extracellular spaces. The MdSWEETs of Clade III, particularly MdSWEET3.5, may be the most likely candidate for this process.
Two members of Clade IV—SWEET16 and −17—are localized to the tonoplast, where they have key roles in exporting Fru from the vacuoles of Arabidopsis leaves (Chardon et al., 2013; Klemens et al., 2013). MdSWEET4.1 was most highly expressed in the leaves, where Fru accumulations were lower, while transcripts were reduced in the fruit, where Fru accumulation was high. This decrease in MdSWEET4.1 expression might explain why apple fruit would have elevated Fru concentrations, as has also been demonstrated with an Arabidopsis mutant of AtSWEET17 that accumulates Fru in its leaf vacuoles (Chardon et al., 2013).
Genes function to transport sugars in apple leaves
In mature leaves, transporters are necessary in both the plastid membranes and the tonoplasts. Plasma-membrane-localized sugar transporters (e.g., MdHTs, MdSUT, MdSWEETs) assist in exporting or importing sugars to control the osmotic balance and sugar signaling between extra- and intra-cellular compartments (Linka and Weber, 2010). In apple leaves, high expression of MdSOT3 and −5 may serve to retrieve Sor that is passively leaked into the apoplast space in source cells, or to transport Sor into the guard cells. This can help modulate stomatal apertures during periods of dehydration stress. Under drought conditions, MdSOTs are up-regulated in apple leaves (Li et al., 2011). Additionally, AtSTP1, −4, −5, and −13 are localized to the guard cells (Nørholm et al., 2006; Büttner, 2010). We detected MdHTs in our mature apple leaves, with expression being strongest forMdHT2.3/4. Future examinations will focus on their roles in mature leaves, especially under stress conditions.
Tonoplast transporters help control osmotic balance and sugar homeostasis in the cytosol by exporting or importing soluble carbohydrates in the vacuoles (Wingenter et al., 2010). We might associate this phenomenon with the high levels of expression by MdvGTs, MdTMT1, and MdSWEET4.1 in our mature apple leaves. In Arabidopsis, the study of tmt1-2::tDNA double mutants suggests that AtTMT1 and −2 have roles in both cellular carbon balance and whole-plant carbohydrate partitioning (Wingenter et al., 2010). Some members of the vGT subfamily may have functions that resemble those of the TMTs, as evidenced by their expression in similar tissues and their preference for transporting glucose and, to a lesser extent, fructose (Aluri and Büttner, 2007). Arabidopsis plants over-expressing EDR6 from pineapple accumulate fewer monosaccharides and become sensitive to low-temperature stress (Klemens et al., 2014). These reports suggest that EDR6 functions in osmotic adjustments by regulating sugar flux within cells. Similar results have been found with AtSWEET17 and AtSWEET16 (Chardon et al., 2013; Klemens et al., 2013). Therefore, we might speculate that altering the expression of MdTMT, MdvGT, MdEDR6, and MdSWEET4.1 could affect plant growth and tolerance to abiotic stresses. Future experiments could involve using these genes to regulate the sugar concentration of fruits from transgenic plants in which a special promoter has been utilized in the transformation procedure.
In conclusion, this study represents a comprehensive investigation of sugar transporter genes in a woody plant. Our gene identifications and comparative analysis with transporters from Arabidopsis and Vitis indicate their strong conservation between herbaceous and woody species, as well as the expansion of particular functional subfamilies, e.g., MdSOT, MdSWEET1, and MdTMT transporters. After investigating their expression profiles simultaneously in various tissue types, and analyzing any correlations between transcript levels and the amounts of Fru and Suc in the fruit, we propose that the high accumulation of Fru is a result of coordination and cooperation by MdTMT1/2 and MdEDR6 with reduced MdSWEET4.1 expression and greater Sor-associated Fru flux. In addition, we suggest that MdSOT1, MdSOT2, and MdSUT4 are involved in the efficient unloading of Sor and Suc in fruit. Furthermore, other transporters may be required for unloading Suc and Sor into the cell wall spaces (possibly MdSWEET1s or MdSWEET3s), loading hexose from the cell wall spaces into the cytosol for storage (MdHTs), retrieving sugars that passively leak into the apoplast space, fine-tuning sugar flux for homoeostasis, and regulating stomatal apertures. Our current findings serve as tools for elucidating the biological function of transporters in Malus domestica, particularly during fruit formation and sugar accumulation. This information will have a significant impact on our knowledge of sugar transporters in general, and the development of sweetness properties in particular. However, the possible errors in the apple genome sequence may also affect the location of pieces of DNA, so that members of a gene family may end up on the wrong scaffold and therefore on the wrong linkage group, which need further to confirm.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Programs of the National Natural Science Foundation of China (31101523) and the New Century Excellent Talents in University (NCET-12-0474). The authors are grateful to Mr. Xuanchang Fu and Mr. Zhengwei Ma for management of the apple trees.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00569/abstract
References
- Afoufa-Bastien D., Medici A., Jeauffre J., Coutos-Thevenot P., Lemoine R., Atanassova R., et al. (2010). The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 10:245. 10.1186/1471-2229-10-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluri S., Büttner M. (2007). Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. U.S.A. 104, 2537–2542. 10.1073/pnas.0610278104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony E., Taybi T., Courbot M., Mugford S. T., Smith J. A. C., Borland A. M. (2008). Cloning, localization and expression analysis of vacuolar sugar transporters in the CAM plant Ananas comosus (pineapple). J. Exp. Bot. 59, 1895–1908. 10.1093/jxb/ern077 [DOI] [PubMed] [Google Scholar]
- Ayre B. G. (2011). Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 4, 377–394. 10.1093/mp/ssr014 [DOI] [PubMed] [Google Scholar]
- Büttner M. (2010). The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. 12, 35–41. 10.1111/j.1438-8677.2010.00383.x [DOI] [PubMed] [Google Scholar]
- Carpaneto A., Geiger D., Bamberg E., Sauer N., Fromm J., Hedrich R. (2005). Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. J. Biol. Chem. 280, 21437–21443. 10.1074/jbc.M501785200 [DOI] [PubMed] [Google Scholar]
- Chardon F., Bedu M., Calenge F., Klemens P. A. W., Spinner L., Clement G., et al. (2013). Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 23, 697–702. 10.1016/j.cub.2013.03.021 [DOI] [PubMed] [Google Scholar]
- Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q., Qu X. Q., Hou B. H., Sosso D., Osorio S., Fernie A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. 10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Doidy J., Grace E., Kühn C., Simon-Plas F., Casieri L., Wipf D. (2012). Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 17, 413–422. 10.1016/j.tplants.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Fan R. C., Peng C. C., Xu Y. H., Wang X. F., Li Y., Shang Y., et al. (2009). Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 150, 1880–1901. 10.1104/pp.109.141374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Jayanty S., Beaudry R., Loescher W. (2005). Sorbitol transporter expression in apple sink tissues: implications for fruit sugar accumulation and watercore development. J. Am. Soc. Hort. Sci. 130, 261–268. [Google Scholar]
- Gao Z. F., Maurousset L., Lemoine R., Yoo S. D., Van Nocker S., Loescher W. (2003). Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 131, 1566–1575. 10.1104/pp.102.016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K., Hernandez A., Korban S. S. (2004). RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 22, 437–438 10.1007/BF02772687 [DOI] [Google Scholar]
- Klemens P. A., Patzke K., Deitmer J. W., Spinner L., Le-Hir R., Bellini C., et al. (2013). Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth and stress tolerance in Arabidopsis thaliana. Plant Physiol. 163, 1338–1352. 10.1104/pp.113.224972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemens P. A., Patzke K., Trentmann O., Poschet G., Büttner M., Schulz A., et al. (2014). Overexpression of a proton-coupled vacuolar glucose exporter impairs freezing tolerance and seed germination. New Phytol. 202, 188–197. 10.1111/nph.12642 [DOI] [PubMed] [Google Scholar]
- Klepek Y. S., Volke M., Konrad K. R., Wippel K., Hoth S., Hedrich R., et al. (2010). Arabidopsis thaliana POLYOL/MONOSACCHARIDE TRANSPORTERS 1 and 2: fructose and xylitol/H+symporters in pollen and young xylem cells. J. Exp. Bot. 61, 537–550. 10.1093/jxb/erp322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Lei H., Zhao X., Tian R., Li T. (2011). Characterization of three sorbitol transporter genes in micropropagated apple plants grown under drought stress. Plant Mol. Biol. Rep. 30, 123–130 10.1007/s11105-011-0323-4 [DOI] [Google Scholar]
- Li M., Feng F., Cheng L. (2012). Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 7:e33055. 10.1371/journal.pone.0033055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin I. W., Sosso D., Chen L. Q., Gase K., Kim S. G., Kessler D., et al. (2014). Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549. 10.1038/nature13082 [DOI] [PubMed] [Google Scholar]
- Linka N., Weber A. P. M. (2010). Intracellular metabolite transporters in plants. Mol. Plant 3, 21–53. 10.1093/mp/ssp108 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1, 387–396. 10.1038/nprot.2006.59 [DOI] [PubMed] [Google Scholar]
- Liu R. H., Meng J. L. (2003). MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. YiChuan 25, 317–321. [PubMed] [Google Scholar]
- Martinoia E., Massonneau A., Frangne N. (2000). Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol. 41, 1175–1186. 10.1093/pcp/pcd059 [DOI] [PubMed] [Google Scholar]
- McCurdy D. W., Dibley S., Cahyanegara R., Martin A., Patrick J. W. (2010). Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit. Mol. Plant 3, 1049–1063. 10.1093/mp/ssq050 [DOI] [PubMed] [Google Scholar]
- Noiraud N., Maurousset L., Lemoine R. (2001). Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13, 695–705. 10.1105/tpc.13.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm M. H. H., Nour-Eldin H. H., Brodersen P., Mundy J., Halkier B. A. (2006). Expression of the Arabidopsis high-affinity hexose transporter STP13 correlates with programmed cell death. FEBS Lett. 580, 2381–2387. 10.1016/j.febslet.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Poschet G., Hannich B., Büttner M. (2010). Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant Cell Physiol. 51, 1571–1580. 10.1093/pcp/pcq100 [DOI] [PubMed] [Google Scholar]
- Poschet G., Hannich B., Raab S., Jungkunz I., Klemens P. A., Krueger S., et al. (2011). A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol. 157, 1664–1676. 10.1104/pp.111.186825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent M., Lecomte A., Bouchet J. P., Bertin N., Causse M., Genard M. (2011). Combining ecophysiological modelling and quantitative trait locus analysis to identify key elementary processes underlying tomato fruit sugar concentration. J. Exp. Bot. 62, 907–919. 10.1093/jxb/erq318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B. F., Normanly J., Amasino R. M. (1999). Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 40, 267–278. 10.1023/A:1006199932265 [DOI] [PubMed] [Google Scholar]
- Reidel E. J., Rennie E., Amiard V., Cheng L., Turgeon R. (2009). Phloem loading strategies in three plant species that transport sugar alcohols. Plant Physiol. 149, 1601–1608. 10.1104/pp.108.134791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier J. W., Willmitzer L., Frommer W. B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. 10.1146/annurev-arplant-050213-040251 [DOI] [PubMed] [Google Scholar]
- Sauer N., Tanner W. (1989). The hexose carrier from Chlorella. cDNA cloning of aeucaryotic H+-cotransporter. FEBS Lett. 259, 43–46. 10.1016/0014-5793(89)81489-9 [DOI] [PubMed] [Google Scholar]
- Schneider S., Hulpke S., Schulz A., Yaron I., Höll J., Imlau A., et al. (2011). Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol. 14, 325–336. 10.1111/j.1438-8677.2011.00506.x [DOI] [PubMed] [Google Scholar]
- Schneidereit A., Scholz-Starke J., Büttner M. (2003). Functional characterization and expression analysis of the glucose specific AtSTP9 monosaccharide transporter in pollen ofArabidopsis. Plant Physiol. 133, 182–190. 10.1104/pp.103.026674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Beyhl D., Marten I., Wormit A., Neuhaus H. E., Poschet G., et al. (2011). Proton-driven sucrose symport and antiport is provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136. 10.1111/j.1365-313X.2011.04672.x [DOI] [PubMed] [Google Scholar]
- Slewinski T. (2011). Diverse transporters and their homologs in vascular plants: a physiological perspective. Mol. Plant 4, 641–662. 10.1093/mp/ssr051 [DOI] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42, 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Verde I., Abbott A. G., Scalabrin S., Jung S., Shu S., Marroni S., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. 10.1038/ng.2586 [DOI] [PubMed] [Google Scholar]
- Wang H., Ma F., Cheng L. (2010). Metabolism of organic acids, nitrogen and amino acids in chlorotic leaves of “Honeycrisp” apple (Malus domestica Borkh) with excessive accumulation of carbohydrates. Planta 232, 511–522. 10.1007/s00425-010-1194-x [DOI] [PubMed] [Google Scholar]
- Wang H. X., Weerasinghe R. R., Perdue T. D., Cakmakci N. G., Taylor J. P., Marzluff W. F., et al. (2006). A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol. Biol. Cell 17, 4257–4269. 10.1091/mbc.E06-01-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari J., Kobae Y., Yamaki S., Yamada K., Toyofuku K., Tabuchi T., et al. (2004). Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant Celll Physiol. 45, 1032–1041. 10.1093/pcp/pch121 [DOI] [PubMed] [Google Scholar]
- Weber A., Servaites J. C., Geiger D. R., Kofler H., Hille D., Groner F., et al. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–802. 10.1105/tpc.12.5.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingenter K., Schulz A., Wormit A., Wic S., Trentmann O., Hoermiller I. I., et al. (2010). Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol. 154, 665–677. 10.1104/pp.110.162040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A., Trentmann O., Feifer I., Lohr C., Tjaden J., Meyer S., et al. (2006). Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18, 3476–3490. 10.1105/tpc.106.047290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Y. H., Hu Y. B., Chen L. Q., Sossoa D., Ducat D. C., Hou B. H., et al. (2013). Functional role of oligomerization for bacterial and plant sugar transporters of the SWEET family. Proc. Natl. Acad. Sci. U.S.A. 110, E3685–E3694. 10.1073/pnas.1311244110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Kanai M., Osakabe Y., Ohiraki H., Shinozaki K., Yamaguchi-Shinozaki K. (2011). Monosaccharide absorption activity of Arabidopsis roots depends on expression profiles of transporter genes under high salinity conditions. J. Biol. Chem. 286, 43577–43586. 10.1074/jbc.M111.269712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Osakabe Y., Mizoi J., Nakashima K., Fujita Y., Shinozaki K., et al. (2010). Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J. Biol. Chem. 285, 1138–1146. 10.1074/jbc.M109.054288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S., Ino M. (1992). Alteration of cellular compartmentation and membrane permeability to sugars in immature and mature apple fruit. J. Am. Soc. Hort. Sci. 117, 951–954. [Google Scholar]
- Zhang L. Y., Peng Y. B., Pelleschi S. T., Fan Y., Lu Y. F., Lu Y. M., et al. (2004). Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol. 135, 574–586. 10.1104/pp.103.036632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.